Abstract
Drosophila melanogaster exhibits two expression-regulating systems that target whole, specific chromosomes: the dosage compensation system whereby the male-specific lethal complex doubles transcription of genes on the male X-chromosome and the chromosome 4-specific protein Painting of fourth, POF. POF is the first example of an autosome-specific protein and its presence raises the question of the universality of chromosome-specific regulation. Here we show that POF and heterochromatin protein 1 (HP1) are involved in the global regulation of the 4th chromosome. Contrary to previous conclusions, Pof is not essential for survival of diplo-4th karyotype flies. However, Pof is essential for survival of haplo-4th individuals and expression of chromosome 4 genes in diplo-4th individuals is decreased in the absence of Pof. Mapping of POF using chromatin immunoprecipitation suggested that it binds within genes. Furthermore, we show that POF binding is dependent on heterochromatin and that POF and HP1 bind interdependently to the 4th chromosome. We propose a balancing mechanism involving POF and HP1 that provides a feedback system for fine-tuning expression status of genes on the 4th chromosome.
Keywords: dosage compensation, gene expression, heterochromatin, POF
Introduction
The dosage compensation system of Drosophila melanogaster is a well-known example of whole-chromosome regulation. Dosage compensation is the mechanism used to equalize the transcriptional activities of the two X chromosomes in the homogametic sex with that of the single X chromosome in the heterogametic sex. Dosage compensation strategies vary widely between species. In female mammals, one X chromosome is inactivated and forms the Barr body, whereas in Caenorhabditis elegans, transcription from both X chromosomes in hermaphrodites is downregulated. In D. melanogaster, dosage compensation is achieved by doubling transcription of the single male X chromosome (reviewed in Baker et al, 1994; Lucchesi et al, 2005; Larsson and Meller, 2006). In D. melanogaster, two non-coding RNAs, roX1 and roX2, have been shown to be essential components of the dosage compensation system. Together with the five MSL (male-specific lethal) proteins, roX1 and roX2 ‘paint' the dosage compensated male X chromosome. The MSL complex mediates acetylation of H4 at lysine 16 on the male X chromosome, which in part explains the subsequent hypertranscription (reviewed in Akhtar, 2003; Larsson and Meller, 2006). A number of recent reports have suggested that components generally associated with heterochromatin are important for dosage compensation. A knock-down of Su(var)2-5 (HP1, heterochromatin-associated protein 1) was shown to result in significantly more lethality in males than in females (Liu et al, 2005). A low amount of HP1 along the entire male X chromosome has been found in genome-wide mapping of HP1 using the DamID technique (de Wit et al, 2005). The Su(var)3-7 protein is another component associated with pericentric heterochromatin. Mutation in Su(var)3-7 causes sex-biased lethality and bloating of the male X chromosome. This bloating is dependent on a functional dosage compensation complex (Spierer et al, 2005). Mild overexpression of Su(var)3-7 causes hypercompaction of the male X chromosome (Delattre et al, 2004). These results suggest that Su(var)3-7 is also involved in dosage compensation.
All the above-mentioned mechanisms for regulating entire chromosomes affect the expression of sex chromosomes, but in Drosophila, an additional example of whole-chromosome targeting is provided by the specific binding of POF to the 4th chromosome (Larsson et al, 2001). This suggests that chromosome-specific regulation may be a more general process. POF is a 495-aa protein with a predicted RNA-binding domain (RRM1) in its central part (Larsson et al, 2001) that binds throughout the entire euchromatic portion of the 4th chromosome. The association with the 4th chromosome appears to nucleate in the basal region of this chromosome and spread in cis or trans to coat the length of the chromosome (Larsson et al, 2001). In several species within the genus Drosophila, for example D. virilis and D. pseudoobscura, POF binds specifically to the F-element (the chromosome corresponding to chromosome 4 in D. melanogaster) (Larsson et al, 2004), suggesting that there is a functional association between POF and chromatin. POF also shows a strong association with the MSL dosage compensation complex. In D. busckii, POF binds specifically to the male X chromosome and colocalizes with H4K16Ac. In D. ananassae and D. malerkotliana, POF again binds specifically to the male X chromosome and colocalizes with the MSL complex protein MSL3 (Larsson et al, 2004). These results support the notion that the 4th chromosome in D. melanogaster has a strong relationship with the X chromosome. Indeed it has been argued that the 4th chromosome originates from the X (reviewed in Hochman, 1976; Larsson and Meller, 2006; Riddle and Elgin, 2006).
The 4th chromosome is in many ways an atypical chromosome. The sequenced part of the chromosome is 1.28-Mb long and corresponds roughly to the banded region seen in polytene chromosomes, that is cytogenetic bands 101E–102F. The remaining 3–4 Mb proximal heterochromatic part of the chromosome consists of simple satellite repeats (Locke and McDermid, 1993). The sequenced and banded part of the chromosome includes 92 genes leading to a gene density similar to that of the major chromosome arms. However, this banded part of the chromosome shares properties typical of heterochromatin. Under normal conditions, chromosome 4 does not undergo meiotic recombination (Hochman, 1976; Sandler and Szauter, 1978; Ashburner et al, 2005) and is late replicating (Barigozzi et al, 1966). The banded region appears as a mosaic of unique sequences interspersed with repetitive DNA with a high content of transposable elements (Barigozzi et al, 1966; Miklos et al, 1988; Pimpinelli et al, 1995; Locke et al, 1999a, 1999b; Kaminker et al, 2002; Stenberg et al, 2005). The heterochromatin protein HP1 is abundantly present on the 4th chromosome as are certain histone modifications used to identify heterochromatin, for example, methylated H3K9 (Eissenberg et al, 1992; Czermin et al, 2002; Schotta et al, 2002). Reporter genes inserted in this chromosome often display a partially silenced, variegated expression (Wallrath and Elgin, 1995; Wallrath et al, 1996). Therefore, how expression of the genes on this highly heterochromatic chromosome is controlled remains an intriguing question.
Here we present evidence that POF and HP1 are involved in regulation of the entire 4th chromosome. Pof is not essential for survival of flies with a diplo-4th karyotype, but is necessary for survival of haplo-4th individuals. We show that genes along the entire length of chromosome 4 are downregulated in the absence of Pof and upregulated in the absence of HP1, and that POF binds preferentially within genes. This binding is dependent on heterochromatin, and HP1 and POF colocalize at cytological level and their binding is interdependent.
Results
Characterization of the genomic Pof locus
In our previous study of Pof, we induced and analyzed short deletions in the promoter regions of the Pof gene. These deletions, which include PofD31A and PofD2, result in no detectable Pof transcription, cause female sterility and are homozygous lethal. The lethality could be rescued by a transgenic construct P[w+ Pof] (Figure 1A; Larsson et al, 2004). On the basis of these findings, we proposed that the sterility and lethality were caused by disruption of the Pof gene. However, in later releases of the genome annotation, a novel gene was predicted in the promoter region of Pof, namely CG33228 (Figure 1A). As this predicted gene would be affected by PofD31A and PofD2 deletions and would be included in the transgenic construct P[w+ Pof], we decided to characterize further CG33228. To verify the existence and extent of the CG33228 gene, we performed 3′RACE and 5′RACE experiments using RNA templates from embryos, third instar larvae and adult females and males. We found CG33228 transcript in all these stages and one single 3′ end and one 5′ end were verified (Figure 1A). We cloned the gene using RT–PCR and confirmed its exon/intron structure. In contrast to the predicted gene structure, we found the 5′ end upstream of the transcription start point of Pof and a different exon/intron organization (Figure 1A). We could not detect a transcript with the first exon as predicted in the genome annotation (results not shown). Two P-element insertions, CB-6312-3 and l(2)SH0542, are located within the ORF of CG33228 (Figure 1A). Both these insertions cause recessive lethality, and complementation analysis shows that all trans-heterozygous combinations of PofD31A, PofD2, CB-6312-3 and l(2)SH0542 are lethal. The lethality of these trans-heterozygotes is fully rescued by the P[w+ Pof] transgene, indicating that the lethality lies within the Pof genomic region. These findings indicated that lethality could be because of disruption of CG33228 rather than Pof. To test this possibility, we induced new deletions in the Pof gene, selectively searching for deletions in the transcribed region. Thirteen unique deletions were isolated by imprecise excisions of the EP element in line with EP(2)2285. All of these deletions were homozygous-viable including PofD140, which uncovers the entire transcribed gene. PofD140 (null mutation), PofD119 and PofD114 (probable null mutations) were used for further experiments. Staining of polytene chromosomes from PofD119 homozygotes or PofD119 in combination with PofD2, CB-6312-3 and l(2)SH0542 confirmed that PofD119 and PofD2 do not produce any detectable POF in salivary gland cells, whereas POF expression is unaffected in CB-6312-3 and l(2)SH0542 (Figure 1B). RT–PCR analysis further confirmed that PofD119 and PofD2 have no detectable Pof transcript, whereas CB-6312-3 and l(2)SH0542 produce roughly normal amounts of Pof transcript. The level of transcription from the neighboring gene CG4806 is not affected by these mutations (Figure 1C). We conclude that the lethality and the female sterility caused by PofD2 and PofD31A are the result of disruption of the CG33228 gene and that Pof is not required for viability. The CG33228 gene encodes a novel protein of 215 amino acids with no clear similarity to any protein or domain of known function.
Figure 1.
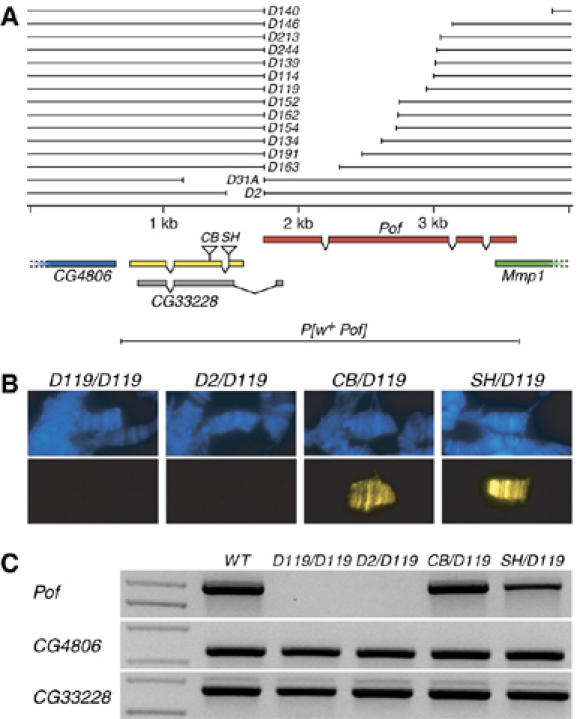
Map of the Pof gene region. (A) The exon–intron structure of the Pof and CG33228 genes are shown below the genomic DNA line. CG4806 and Mmp1 genes extend outside the region shown. Pof is transcribed from left to right, all other genes shown are transcribed in the opposite direction. The confirmed exon–intron structure of CG33228 is shown in yellow and the predicted gene structure from the genome annotation (release 4) is shown in gray. The positions of the P-element insertions in CB-6312-3 and l(2)SH0542 lines are indicated (CB and SH). The deletions PofD31A and PofD2 have been previously described (Larsson et al, 2004). The extent of induced deletions and the transgenic construct P[w+Pof] are shown. (B) Localization of POF on polytene chromosomes from PofD119/PofD119, PofD2/PofD119, CB-6312-3/PofD119 and l(2)SH0542/PofD119 third instar larvae salivary gland cells. DNA is stained with DAPI (blue) and POF is recognized by an anti-POF antibody (yellow). (C) RT–PCR analysis on wild-type, PofD119/ PofD119, PofD2/ PofD119, CB-6312-3/PofD119 and l(2)SH0542/PofD119female poly(A)+ RNA. The primer pairs amplify Pof (top panel), CG4806 (middle) and CG33228 (bottom).
Pof is essential for survival of haplo-4 flies
What are the consequences of lacking a functional Pof gene? It has been proposed that the 4th chromosome also possesses a dosage compensation function, which allows haplo-4th individuals to survive (Hochman, 1976). Crossing males with a compound 4th chromosome (C(4)RM) to females with a normal diploid set of chromosomes will give two classes of offspring, one with three 4th chromosomes and one with a single 4th chromosome. Crosses between PofD140/PofD140; ci57g/ci57 g females and PofD140/CyO; C(4)RM svspa-pol/0 males showed that no haplo-4th PofD140/PofD140 individuals survived (Figure 2A). Other allelic combinations, that is PofD31A/PofD114, PofD119/PofD114 and PofD2/PofD119, also caused haplo-4th-specific lethality (Figure 2B–D). The haplo-4th lethality can be rescued by a single copy of the P[w+ Pof] transgene (Figure 2B). Reciprocal crosses gave similar results (not shown). The trans-heterozygotes with genotypes CB-6312-3/PofD140 and l(2)SH0542/PofD140 did not cause haplo-4th lethality (Figure 2E and F), excluding an influence of CG33228 on haplo-4th lethality. Although most of the PofD140/PofD140; ci57g/0 offspring died as embryos or in early larval stages, a small proportion reached the pupae stage (Supplementary Figure 1). We conclude that Pof is essential for survival of haplo-4th individuals.
Figure 2.
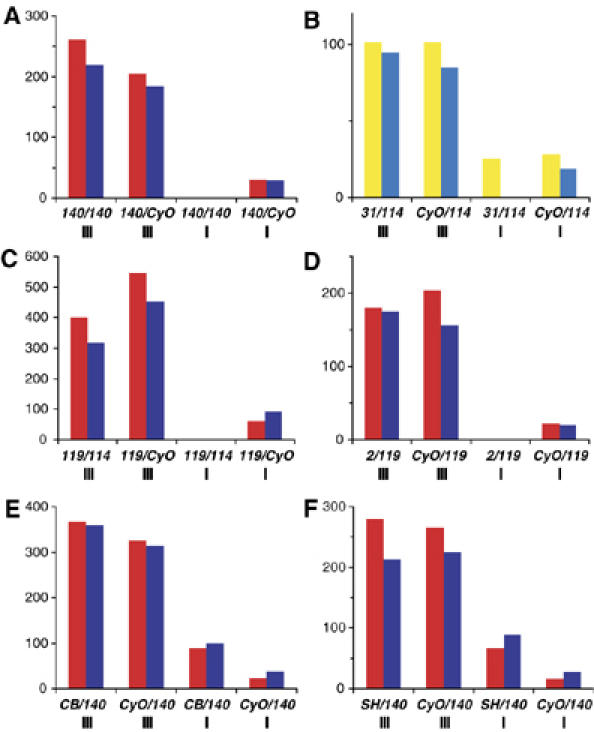
Mutations in Pof cause haplo-4th lethality. The y-axis shows the number of hatched adult flies. The x-axis shows the offspring genotype and the number of 4th chromosomes in roman numerals. Females are shown in red and males in blue (A, C–F). In (B), flies with the P[w+Pof] rescue construct are shown in yellow and their siblings lacking the transgenic construct are shown in light blue. (A) Offspring from PofD140/PofD140; ci57g/ci57 g females × PofD140/CyO; C(4)RM svspa-pol/0 males. No PofD119/PofD119; ci57g/0 (haplo-4th) offspring survive. (B) The haplo-4th lethality is rescued by a transgenic P[w+Pof] construct. The PofD119/PofD114 allelic combination (C) and the PofD2/PofD119 allelic combination (D) causes haplo-4th lethality. In CB-6312-3/PofD140 (E) and l(2)SH0542/PofD140 trans-heterozygotes (F) the haplo-4th individuals survive. The haplo-4th individuals are delayed by 2–4 days and have lower viability than their triplo-4th siblings. This caused the haplo-4th classes to be smaller than theoretically expected in all experiments.
Pof mutations cause decreased levels of chromosome 4 gene expression
The haplo-4th lethality in Pof mutants shows that Pof is essential for survival of haplo-4th individuals. However, under normal conditions flies are diploid for the 4th chromosome. We therefore tested if Pof mutations have a general effect on chromosome 4 gene expression. We prepared four biological replicates of mRNA from PofD2/PofD2 first instar larvae and compared them to three biological wild-type replicates. An Affymetrix microarray analysis showed that the lack of Pof caused a low but significant reduction of mRNA from a majority of the genes on the 4th chromosome (Supplementary Figure 2A). Although the reduction is too small to be significant at the level of individual genes, the overall reduction on chromosome 4 compared with any of the other chromosome arms is significant (Supplementary Figure 2B and C). However, PofD2/PofD2 larvae were selected from heterozygous parents with maternal POF present and the PofD2 allele also affects the neighboring gene CG33228. We, therefore, performed an additional microarray and compared three biological replicates of mRNA from PofD119/PofD119 first instar larvae with three new biological wild-type replicates. The results confirm and strengthen the conclusion that lack of Pof causes a significant reduction of gene expression from the 4th chromosome (Figure 3A and B; Supplementary Figure 2D). As chromosome 4 only represents 105 expression units on the microarray, we also performed a simulation to verify our results. One-hundred and five genes were randomly selected from the other chromosome arms and the mean reduction was compared with chromosome 4 mean reduction. After 100 million simulations no group with a reduction equal to or stronger than the 4th chromosome was found.
Figure 3.
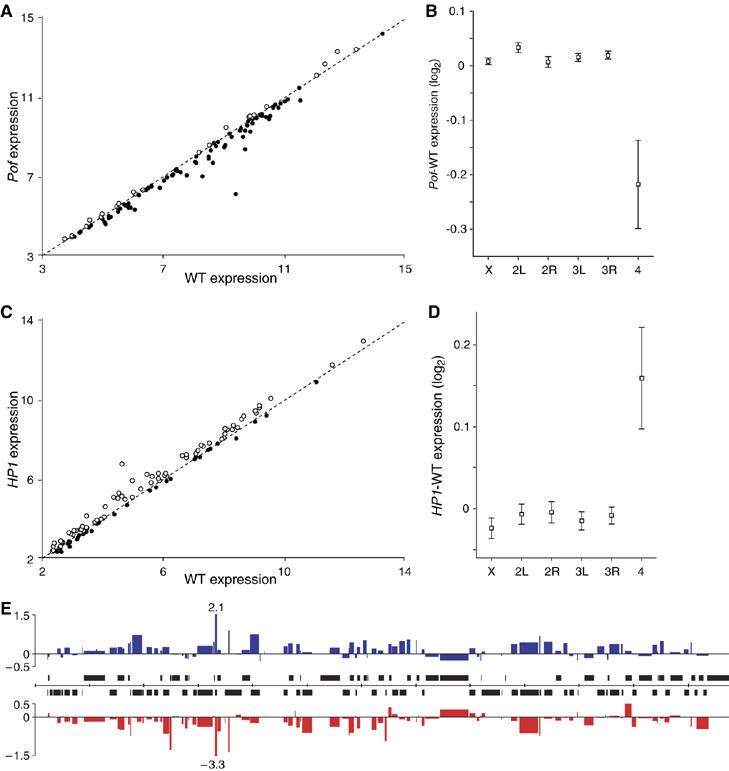
Levels of mRNA from genes located on the 4th chromosome are decreased in a Pof mutant background and increased upon HP1 RNAi when compared with wild type. (A, C) Microarray analysis showing the median level of mRNA from genes on the 4th chromosome in PofD119/PofD119 mutant (A) and HP1 RNAi (C) as a function of the median levels in controls (log2 scale). Genes for which there were reduced amounts of mRNA (below dotted line) are marked with filled circles and genes for which there were increased amounts with open circles. (B, D) Mean change in gene expression for each chromosome arm, PofD119 – wild-type expression (B) and HP1 RNAi – control expression (D) (log2 scale). Squares indicate the mean value and whiskers indicate 95% confidence interval. (E) The microarray results plotted with respect to the position of the genes along the 4th chromosome (annotation release 4.3). The widths of the columns indicate the length of corresponding genes. Expression change (log2 scale) after HP1 RNAi is shown in blue and expression change in PofD119 mutants in red. Tick marks on the scale bar (middle) indicate 100 kb and genes transcribed from left to right are shown above the line and genes transcribed in opposite direction below.
The 4th chromosome is known to be highly heterochromatic and targeted also by the heterochromatin-associated protein HP1. We therefore decided to test also HP1 for general effects on chromosome 4 gene expression. We reanalyzed the microarray data from Liu et al (2005) to see if HP1 RNAi causes chromosome 4-specific expression changes. RNAi-mediated knock-down of HP1 caused a significant general increase in gene expression on chromosome 4 genes (Figure 3C and D; Supplementary Figure 2E). After 100 million simulations (as above), the frequency of gene groups with an increase equal to or stronger than the 4th chromosome was 8.6 × 10−6. Both Pof and HP1 microarray data were reanalyzed after removal of low expressed genes. The results were consistent (results not shown). We next tested if the reduction in gene expression in PofD119 or the increase in gene expression after HP1 RNAi is more pronounced at certain parts of the 4th chromosome, by plotting the relative change against the chromosomal position (Figure 3E). Genes that changed in gene expression are spread along the length of the chromosome. This result suggested a negative correlation between the change caused by PofD119 and HP1 RNAi. Indeed, when tested, the correlation was significant (P<<0.001) with an r-value of −0.60 (Supplementary Figure 2F). This means that removing Pof gives an inverse but proportional effect compared with HP1 RNAi at an individual gene level.
We chose nine genes with different expression levels for confirmatory tests using real-time PCR from RNA templates. Four new biological replicates were prepared from both PofD119/PofD119 and wild-type first instar larvae. All nine genes showed decreased mRNA levels in PofD119/PofD119 mutants when compared with wild-type (Figure 4A).
Figure 4.
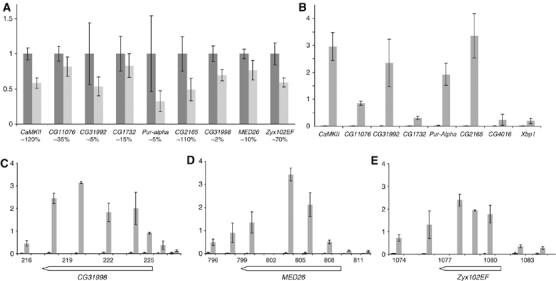
POF regulates and binds within chromosome 4 genes. (A) Comparison of the mean levels of mRNA from nine genes by real-time RT–PCR in wild type (dark gray) and PofD119/PofD119 mutants (light gray). mRNA levels were normalized against actin gene mRNA in each replicate and 100% was set as the mean value of expression in wild type. Error bars represent the standard deviations of four biological replicates. The y-axis shows the relative amount of transcript and the x-axis shows the gene name with an estimate of the relative amount of mRNA (expression level) as a percentage of actin mRNA below. (B) The binding of POF to six chromosome 4 genes and two genes from chromosome 2R (CG4016 and Xbp1) were analyzed using ChIP. The y-axis indicates enrichment plotted as percent of input. Mean POF enrichments (gray bars) and MOCK (black bars) with standard deviations from three independent chromatin preparations are shown. The binding of POF to three additional genes CG31998, region 220 kb (C), MED26, 800 kb (D) and Zyx102EF, 1080 kb (E) were analyzed using ChIP. The extent of the genes is shown below the x-axis.
POF binds preferentially within genes
The above results indicate that POF is necessary to assist transcription of genes on chromosome 4. We, therefore, decided to examine in more detail the distribution of POF on the chromosome in Schneider 2 cells by chromatin immunoprecipitation (ChIP), followed by real-time PCR. To get a rough idea of POF binding, we analyzed approximately half of the sequenced chromosome at 10 kb resolution (Supplementary Figure 3). The results suggest that POF is highly enriched on this part of the chromosome, with two regions showing less enrichment (230–320 and 730–790 kb regions). During the course of this study, high-resolution ChIP results were published showing that the MSL proteins bind within genes on the X chromosome (Alekseyenko et al, 2006; Gilfillan et al, 2006; Legube et al, 2006). As the 10 kb spacing was not suitable to draw conclusion on binding of POF at the gene level, we chose three well isolated genes with POF binding according to ChIP results for more detailed mapping: CG31998, MED26 and Zyx102EF. We analyzed these three genes at approximately 2 kb resolution using ChIP and the results are shown in Figure 4C–E. In these three genes POF clearly binds within the transcribed region rather than outside the genes. In addition, the amount of transcript from these three genes is decreased in a Pof mutant background (Figure 4A). We also tested the remaining six genes in Figure 4A for POF binding by ChIP. The results show that all chromosome 4 genes tested except CG1732 represent targets for POF binding. It should be noted that CG1732 shows the smallest reduction in gene expression in PofD119 mutants in Figure 4A.
Binding of POF to the 4th chromosome depends on heterochromatin
To identify POF requirements for binding to the 4th chromosome, we immunostained polytene chromosomes from larval strains with translocated 4th chromosomes. Six different T(1;4)s with karyotype pXd4; p4dX; 4 were stained to detect POF distal-4th chromosome (d4) binding and scored as no binding, single-band binding, partial binding (a few bands) and entire d4 binding (Supplementary Figure 4). In two of these translocations, we detected nuclei with single-band binding (T(1;4)wm5 and T(1;4)wm258−18) or partial binding (T(1;4)wm258−18). We decided to study these two translocations in more detail. By growing the larvae at different temperatures it became clear that at 18°C a majority of the nuclei in T(1;4)wm5 showed no binding on d4, although the normal 4th chromosome was still decorated with POF as usual. However, a fraction of the nuclei showed single-band binding (Figure 5 and Supplementary Figure 4). In contrast, in T(1;4)wm258−18, which includes more of the centromere proximal heterochromatin, the majority of the nuclei showed single-band binding and a fraction showed partial binding. If these larvae were grown at 25°C, no binding was detected in T(1;4)wm5, the fraction of single-band binding in T(1;4)wm258−18 was decreased and no partial binding was found. Because an increase in temperature is known to reduce the compaction of heterochromatin, we decided to also analyze chromosomes from T(1;4)/0 males, that is males lacking the Y chromosome. It is also known that partial or total removal of the Y chromosome increases the compaction of heterochromatin (Gowen and Gay, 1934; Spofford, 1976; Dimitri and Pisano, 1989). Removal of the Y chromosome caused a similar effect to lowering the temperature and, strikingly, in T(1;4)wm258−18/0 males grown at 18°C, a fraction of the d4 showed staining of the entire polytenized region (Figure 5). We conclude that the binding of POF depends on the amount and compaction of nearby heterochromatin. It should be stressed that POF is not detected within the centromere proximal heterochromatic region of the 4th chromosome. Whenever a single-band is seen in nuclei it is located at 101F–102B2.
Figure 5.
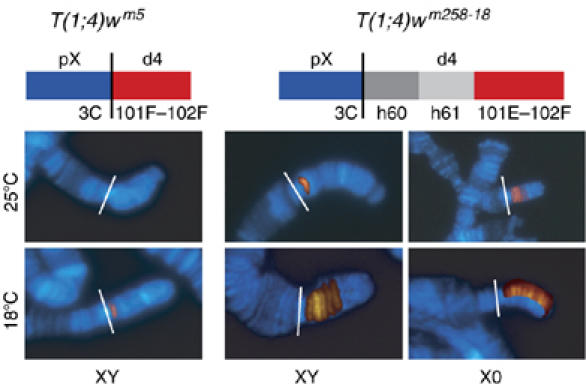
POF localization on translocated chromosomes. T(1;4) males stained with DAPI (blue) anti-POF (red) are shown as merged images. Original images are shown in Supplementary Figure 4. Schematic illustrations of the two translocations are shown on top (not to scale). The proximal X region is labeled pX and the distal 4 portion translocated to pX is labeled d4. The vertical black lines indicate the translocation breakpoints and are shown by white lines in the images. 3C indicates the cytological position of the X chromosome breakpoint, and h60 and h61 correspond to the heterochromatic regions 60 and 61. The temperature for growing the larvae is shown to the left of the images and the presence or absence of the Y chromosome is indicated below the images. The T(1;4)wm258−18/0 males grown at 18°C (bottom right) is a composite image in which half the POF-stained d4 has been collated on the DAPI-stained 4th chromosome.
To analyze further the dependence on heterochromatin for POF binding to the 4th chromosome, we analyzed POF binding in H2Av, HP1, Su(var)3-7, Su(var)3-9 and Su(UR) larvae (mutants in which heterochromatin formation is affected). The results are shown in Figure 6A. On the 4th chromosomes from H2Av810 and HP104/HP105, mutant larvae the staining of POF was dramatically decreased although not completely absent. Upon increasing the exposure time, weak staining with the normal distribution of POF on the 4th chromosome was seen (results not shown). No effect was seen in mutants of Su(var)3-7, Su(var)3-9 or Su(UR). We then tested the possible interdependence of binding by POF and HP1 or H2Av by staining chromosomes from PofD119/PofD119 mutant larvae with antibodies raised against H2Av and HP1 (Figure 6B). A PofD119 mutant background has no effect on H2Av staining, but markedly reduces the binding of HP1 to the 4th chromosome. Pericentric heterochromatic regions show no detectable difference in HP1 binding between PofD119 mutants and wild type (Figure 6B). The reduction of HP1 binding to the 4th chromosome in PofD119/PofD119 mutant background is less dramatic than the reduction of POF binding in HP104/HP105 mutants. Notably, immunostaining also showed that POF and HP1 colocalized close to perfect at the level of cytology within the 4th chromosome (Figure 6C). We conclude that POF is dependent on heterochromatin for binding to the 4th chromosome and that POF and HP1 binding to the 4th chromosome is interdependent.
Figure 6.
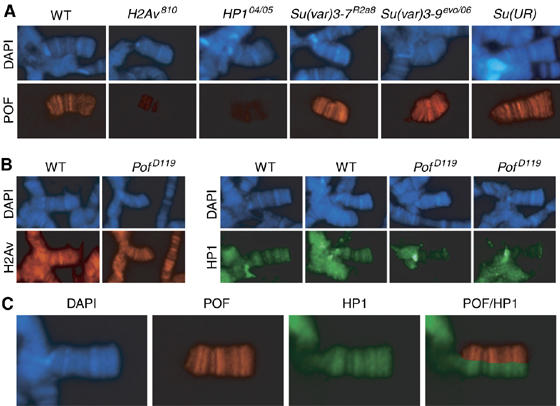
Association of POF and HP1 with the 4th chromosome is interdependent. (A) Polytene chromosomes were prepared from wild-type, H2Av810/H2Av810, HP104/HP105, Su(var)3-7R2a8/ Su(var)3-7R2a8, Su(var)3-9evo/Su(var)3-906 and Su(UR)/Su(UR) third instar larvae and stained with DAPI (blue) and POF (red). The staining of POF is dramatically reduced in H2Av810 and HP1 mutant backgrounds. (B) Polytene chromosomes from wild type and PofD119/PofD119 mutants were stained with antibodies raised against H2Av (left panel) and against HP1 (right panel). Two nuclei from each genotype are shown with HP1 staining. Note the reduction of HP1 staining on the 4th chromosome but not in the chromocenter in PofD119/PofD119 mutants compared with wild type. (C) Localization of POF and HP1 on chromosome 4. The combined POF/HP1 image shows that POF colocalizes perfectly with HP1 on the 4th chromosome.
Discussion
Chromosome-specific regulation has long been known as a mechanism used to maintain correct expression levels of sex chromosomes, that is, dosage compensation. Discovery of the binding of POF to the 4th chromosome in, for example, D. melanogaster and to the male X chromosome in some related species suggests that the 4th chromosome is also regulated by a chromosome-specific system. Further, haplo-4th individuals of D. melanogaster can survive, and two hypotheses have been proposed to explain their survival: (i) a dosage compensation mechanism may compensate for the lack of one 4th chromosome (Hochman, 1976) or (ii) loss of one 4th may be non-lethal simply because of its very small size as, for instance, small regions of chromosome 2 or 3 can be deleted without haplo-lethal consequences (Lindsley et al, 1972); discussed in Larsson et al (2004) and Larsson and Meller (2006). In this study, we present evidence that POF is needed for survival of haplo-4th individuals. Also the expression of chromosome 4 genes in diplo-4th individuals is decreased in the absence of Pof and increased after HP1 RNAi. ChIP on three isolated genes suggested that POF associates with genes. Furthermore, we show that POF binding is dependent on heterochromatin, and that the binding of POF and HP1 on the 4th chromosome is interdependent.
Pof is essential for haplo-4th survival
In our previous work we concluded that Pof is essential for survival as deletions of the promoter region of Pof caused lethality, which could be rescued by a genomic Pof construct (Larsson et al, 2004). More recent updates of the D. melanogaster genome predicted a gene within this deleted region, namely CG33228. We have confirmed the existence of this gene and shown that the lethality is caused by the disruption of CG33228. By inducing a number of deletions in the transcribed region of Pof we have analyzed the consequences of a Pof knockout without including any deleterious effects from CG33228. Results show that Pof is essential for survival of haplo-4th individuals. It should be stressed that our results do not show, nor have we attempted to show, the existence of a chromosome 4 dosage compensation mechanism. The lethality of haplo-4 flies in Pof mutant background may well be the combined effect of a decrease in chromosome gene template with the disruption of a chromosome-wide transcription-facilitating system. The haplo-4th survival would then be a consequence rather than the underlying cause.
Lack of Pof affects chromosome 4 gene expression opposite to HP1
Somatic elimination of the 4th chromosome is relatively frequent in D. melanogaster (Mohr, 1932; Ashburner et al, 2005), but the 4th chromosome is normally present in diploid form in both males and females. Microarray analyses detected significant decreases in chromosome 4 gene expression in diplo-4 flies in Pof mutant backgrounds, but we saw no correlation between the degree of reduction and the expression level of individual genes. The genes affected are located along the entire 4th chromosome and their degree of reduction is not correlated to their distance from the centromere. We have confirmed the decrease in expression for nine genes using quantitative RT–PCR. On the contrary, HP1 knock-down causes a significant increase in chromosome 4 gene expression. Furthermore, the increase in gene expression after HP1 RNAi correlates at the gene by gene level to the decrease seen in PofD119. This suggests that POF and HP1 are functionally related as regulators for genes on the 4th chromosome.
POF binds within genes
Using ChIP we have mapped POF binding along 600 kb of the 1.28 Mb sequenced part of the 4th chromosome at 10 kb resolution, and three specific regions at a 2 kb resolution. The mapping suggests that, like the MSL complex (Alekseyenko et al, 2006; Gilfillan et al, 2006; Legube et al, 2006), POF binds within genes. This strengthens the similarity between POF and the MSL proteins in binding behavior at the gene level and provides further support for the model of POF being a transcription facilitating factor for the 4th chromosome. It will be interesting to investigate the relation between POF binding and transcription in more detail.
POF binding depends on heterochromatin
Our results from the polytene chromosome staining experiments indicate that binding of POF is dependent on heterochromatin. Reducing the amount of heterochromatin by increasing the temperature or genetic means (i.e. mutations in the heterochromatin formation pathway or presence of the Y chromosome) decreases the amount of POF binding to the 4th chromosome. The temperature and the presence of the Y-chromosome have only been shown to affect the POF binding to translocated 4th chromosomes, but we have shown that the amount of POF on the wild-type 4th chromosome is reduced in HP1 or H2Av mutant backgrounds, where there is a strong reduction in the amount of heterochromatin. POF is released from the 4th chromosome in the absence of HP1 and, likewise, lack of POF releases HP1 binding, though this is less absolute. Indeed, on the 4th chromosome with the exception of the most centromere proximal region, POF and HP1 show close to perfect colocalization at the level of cytology. Whether this is true also at the gene level remains elusive, although the negative correlation in gene expression changes for Pof mutations and HP1 RNAi suggests that it is. The interdependence between POF and HP1 suggests they are involved in a balancing mechanism that helps maintain appropriate levels of expression of genes on the 4th chromosome. The replacement of H2A by H2Av in the nucleosome is proposed to be an early step in the pathway leading to heterochromatin (Swaminathan et al, 2005). A later step is the deacetylation and methylation of H3K9, which serves as a target for HP1 (Lachner et al, 2001; Nakayama et al, 2001). As Su(var)3-9 is known to methylate H3K9, it should be noted that POF binding to the 4th chromosome is not affected by mutations in Su(var)3-9. This is explained by the fact that although Su(var)3-9 mutants lack H3K9 methylation in the chromocentre regions, chromosome 4 is not affected (Czermin et al, 2002; Schotta et al, 2002). The methylation of H3K9 on the 4th chromosome is probably executed by a different protein.
The balance between repressing and activating factors may represent a more general mechanism for regulating whole chromosomes. Although generally accepted, it has only recently been shown that disruption of the MSL complex, using RNAi-mediated knockdown in SL2 tissue culture cells, causes a reduction in the amounts of mRNA from X-linked genes (Hamada et al, 2005; Straub et al, 2005). The problem of showing downregulation of most genes along the X chromosome at the level of the whole organism could be because of the balancing mechanisms between activating and repressing complexes. We speculate that in dosage compensation of X chromosomes, correct gene expression is achieved by a balance between the MSL complex and repressing components such as HP1 and Su(var)3-7. This may explain why clearcut evidence for lack of compensation is difficult to find when studying mutants of components in the MSL complex.
How general are whole chromosome regulatory mechanisms?
One tentative explanation for the regulatory system of the 4th chromosome appears to be that it had an X-chromosomal origin and POF (an ancient component of a dosage compensation complex) was retained on it when it started to segregate as a unique autosome. Chromosomal rearrangements occurring through evolution will occasionally select for new or adapted functions of pre-existing regulatory mechanisms. For example, it has been argued that imprinting originates from X-inactivation centers (Huynh and Lee, 2001, 2005) sharing monoallelic expression of large non-coding RNAs (Sleutels et al, 2002; Thakur et al, 2004). Whether or not Drosophila is unique in having two chromosome-wide targeting and regulatory systems is still unknown. However, the fact that the 4th chromosome is subject to a chromosome-specific regulatory mechanism highlights the importance of this issue. Our results also show the importance of balancing mechanisms in chromosome-specific regulation, in this case exemplified by POF and HP1, providing feedback systems for fine-tuning expression status.
Materials and methods
Fly stocks and genetic crosses
Flies were cultivated and crossed in vials with potato mash-yeast-agar medium at 25°C unless otherwise indicated. The T(1;4) stocks were obtained from the Bloomington Drosophila Stock Center. The His2Av810, Su(var)2-504, Su(var)2-505 (HP1), Su(var)3-9evo and Su(var)3-906 alleles were obtained from Victor Corces (Johns Hopkins University, Baltimore). The Su(var)3-7R2a8 was obtained from Marion Delattre (University of Geneva, Switzerland) and is considered to be a null mutation (Spierer et al, 2005). The Su(UR) stock was obtained from Igor Makunin (Institute of Cytology and Genetics, Russian Academy of Sciences, Novosibirsk). To test for haplo-4th lethality, PofD140/PofD140; ci57g/ci57 g, females were mated to PofD140/CyO; C(4)RM svspa-pol/0 males. All offspring were counted and classified. Other allelic combinations were tested with corresponding crosses. To test for rescue by a transgenic P[w+Pof] construct w1118; PofD31A/CyO; P[w+ Pof]/+; ci57g/ci57 g females were mated to PofD114/CyO; C(4)RM svspa-pol/0 males. Male offspring were counted and classified. Only males with yellow eyes survived as haplo-4th individuals, that is w1118; PofD31A/ PofD114; P[w+ Pof]/+; ci57g/0.
Generation of Pof deletions by P-element excision
To create deletions of the transcribed region of the Pof gene we excised the P-element inserted close to the transcription start of Pof in the fly line EP(2)2285. Single white-eyed, non-Sco males from the cross yw/yw; Sco/CyO × w1118; EP(2)2285/CyO; mus309D3, Δ2-3/mus309D2 were crossed to yw; Sco/CyO females. After 4 days, the single males were removed from the crosses, DNA was prepared and PCR was used to screen for short imprecise excisions. The primer pair used is listed in Supplementary Table I. A total of 269 males were crossed and analyzed and 16 deletions were found. Crosses from males with excisions were continued to establish balanced stocks. The exact breakpoints of the deletions were determined by sequencing. Three of the deletions occurred twice, leading to a total of 13 unique deletions in the transcribed and coding region of Pof.
Characterization of CG33228
To verify the existence of the predicted CG33228 gene and to characterize it, poly(A)+ RNA was isolated from embryos (0–16 h), third instar larvae, adult females and males, as described previously (Svensson et al, 2003). The 5′ and 3′ ends of the gene were determined using the BD SMART™ RACE cDNA amplification kit according to the manufacturer's instructions (Clontech). The transcribed region of CG33228 was amplified using Ready-To-Go RT–PCR beads (Amersham Biosciences) and primers designed from its 5′ and 3′ ends. The PCR products were sequenced to verify the exon/intron structure of the gene.
Immunostaining of polytene chromosomes
Immunostaining of polytene chromosomes were performed essentially as described previously (Larsson et al, 2001). We used primary antibody raised against POF (1:400 dilution), MSL3 (1:2000), H2Av (1:1000) (Leach et al, 2000) or HP1 (C1A9, 1:50; PRB-291C, 1:300). Donkey anti-rabbit, anti-IgY and anti-goat (Jackson Laboratories) conjugated with Cy3 or FITC (diluted 1:400) or goat anti-rabbit, anti-mouse and anti-IgY (MedProbe) conjugated with AlexaFluor555 or AlexaFluor488 (diluted 1:300) were used as secondary antibodies. Preparations were analyzed using a Zeiss Axiophot microscope equipped with a KAPPA DX20C CCD camera. Images were assembled and merged electronically using Adobe Photoshop. For quantitative comparisons of stainings in mutants versus wild type, preparations and stainings were run in parallel. Nuclei with clear cytology were chosen based on DAPI staining and photographed. All settings were identical for each specific antibody. At least 20 nuclei of each genotype were used in the comparisons and at least five slides of each genotype were analyzed. To analyze POF staining in T(1;4) translocations a minimum of five slides were analyzed for each translocation and for each condition. Nuclei with clear cytology were chosen based on DAPI staining, and the POF binding in these nuclei was then determined.
Microarray analysis
For microarray analysis, PofD2 versus wild type, poly(A)+RNA was isolated using DynabeadsOligo (dT)25 (Dynal). Four hundred first instar larvae were used for each of four biological replicates of y1w67c23; PofD2/PofD2, selected as non-GFP larvae from a y1w67c23; PofD2/CyO GFP balanced stock and three replicates of y1w67c23 as controls. The larvae were frozen at −70°C and homogenized in 0.1 M Tris–HCl (pH 8.0), 0.5 M LiCl, 10 mM EDTA, 1% SDS and 5 mM DTT. Dynal's instructions were then followed. The poly(A)+RNA was used for labeling according to the standard Affymetrix protocol. The seven labeled cDNA probes were hybridized to an Affymetrix gene chip (version 1). For microarray analysis, PofD119 versus wild type, total RNA was isolated using TRIzol reagent (Invitrogen) followed by a purification using RNeasy kit (Qiagen) according to the instruction by the suppliers. Two hundred first instar larvae were used for each of three biological replicates of y1w67c23; PofD119/PofD119 and three replicates of y1w67c23 as controls. The six labeled cDNA probes were hybridized to an Affymetrix gene chip (version 2). For microarray analysis, HP1 RNAi versus control, we analyzed the data from Liu et al (2005). Four replicates of HP1 RNAi (two male and two female arrays) were compared with four control replicates (HP1 RNAi without driver, two male two female arrays). Intensity values were normalized and summarized using robust multiarray analysis (RMA) (Irizarry et al, 2003). Other methods, like MAS5, were tested and all gave similar results. All microarray data analyses were carried out using R (www.R-project.org) and the Bioconductor package (Gentleman et al, 2004). ANOVA tests for all chromosome arms were performed on log2 differences between median wild type and median mutant expression. A significant ANOVA (P<0.05) was followed by Duncan's multiple range test to pairwise compare all chromosome arms. Statistical analysis was carried out using Statistica 7.1 (StatSoft). To exclude that the significant difference in gene expression of the 4th chromosome was a consequence of the small sample size (105 expression units) a simulation was performed. The number of simulations was increased until the P-value was stabilized. Microarray data are available at: http://www.ncbi.nlm.nih.gov/geo/ (accession: GSE5984 and GSE6839).
Chromatin immunoprecipitation
For ChIP experiments we used Schneider's Drosophila line 2 cells (ATCC CRL-1963) grown at 25°C in Erlenmeyer flasks to a density of 0.5−1.5 × 107 cells/ml in Drosophila SFM medium (Invitrogen) supplemented with 100 U/ml of penicillin G, 100 μg/ml of streptomycin sulfate and 2 mM of L-glutamine. The cells were crosslinked, washed and sonicated as described by Schwartz et al (2006). For ChIP, 150 μl of cell lysate was diluted 10-fold in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl, (pH 8.0) and 167 mM NaCl) and protein inhibitors were added. The diluted lysate was precleared by incubation with 30 μl of Dynabeads conjugated to Protein A (Dynal), preblocked by equilibration in 150 μl ChIP buffer containing 12 μg sonicated herring sperm DNA. The cleared lysate was incubated with 3 μl affinity purified POF antibody (∼1 μg) overnight at 4°C. The antibody complexes were precipitated by incubation with DNA-blocked Protein A Dynabeads for 1 h at 4°C. The beads were washed once with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM Tris–HCl, pH 8.0 and 150 mM NaCl), once with high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM Tris–HCl, pH 8.0 and 500 mM NaCl), once with LiCl-containing buffer (250 mM LiCl, 10 mM Tris–HCl pH 8.0, 1 mM EDTA, 1% NP-40 and 1% sodium deoxycholate) and twice with TE buffer (10 mM Tris–HCl, pH 8.0 and 1 mM EDTA). The protein–DNA complex was eluted from the antibody by incubating 2 × 15 min at RT in 250 μl elution buffer (1% SDS and 0.1 M NaHCO3) with rotation. NaCl was added to a final concentration of 200 mM and protein–DNA crosslinks were reversed by heating at 65°C for 4 h. A 10 μl volume of 0.5 M EDTA, 20 μl of 1 M Tris–HCl, pH 6.5, and 1 μl of 20 mg/ml proteinase K were added before an additional incubation at 45°C for 1 h. The DNA was recovered by phenol–chloroform extraction followed by ethanol precipitation. The immunoprecipitated DNA was then dissolved in 100 μl water.
Real-time PCR
DNA from the ChIP reactions was amplified by real-time PCR using SYBR Green PCR Master Mix (Bio-Rad). Primer pairs used are listed in Supplementary Table I. The amounts of specific DNA fragments were derived by comparing the threshold cycle value to a five-point standard curve corresponding to input DNA ranging from 0.02 to 1.92%. All PCR assays were controlled by melt curve analysis for specificity. Six control regions outside chromosome 4 were tested and used as negative controls. All primer pairs were also tested on MOCK precipitations. For quantitative real-time PCR of RNA templates we used the iScript One-Step RT–PCR system with SYBR Green (Bio-Rad). poly(A)+RNA was isolated using DynabeadsOligo (dT)25 as described above. One hundred first instar larvae were used for each biological replicate, with four biological replicates of wild type and four of PofD119/PofD119. The expression levels were normalized to the amount of actin mRNA in each replicate. The mean levels of expression, with standard deviations, were calculated (as presented) and a relative expression level for each gene was determined as a percentage of actin expression.
Supplementary Material
Supplementary Figures and Legends
Supplementary Table 1
Acknowledgments
We thank Fang-Lin Sun for allowing us to analyze their microarray data, Victor Corces, Carole Seum and Igor Makunin for fly lines, David Glaser for H2Av antisera and Mitzi Kuroda for MSL antibodies. We also thank Karin Ekström for technical assistance and Simon Tuck for comments on the manuscript. This work was supported by grants from the Nilsson-Ehle foundation to PS and from the Swedish Research Council, Åke Wiberg, Carl Tryggers and Philip Sörenssen foundations to JL.
References
- Akhtar A (2003) Dosage compensation: an intertwined world of RNA and chromatin remodelling. Curr Opin Genet Dev 13: 161–169 [DOI] [PubMed] [Google Scholar]
- Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI (2006) High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev 20: 848–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Golic KG, Hawley RS (2005) Drosophila A Laboratory Handbook. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Baker BS, Gorman M, Marín I (1994) Dosage compensation in Drosophila. Annu Rev Genet 28: 491–521 [DOI] [PubMed] [Google Scholar]
- Barigozzi C, Dolfini S, Fraccaro M, Raimondi GR, Tiepolo L (1966) In vitro study of the DNA replication patterns of somatic chromosomes of Drosophila melanogaster. Exp Cell Res 43: 231–234 [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- de Wit E, Greil F, van Steensel B (2005) Genome-wide HP1 binding in Drosophila: developmental plasticity and genomic targeting signals. Genome Res 15: 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M, Spierer A, Jaquet Y, Spierer P (2004) Increased expression of Drosophila Su(var)3-7 triggers Su(var)3-9-dependent heterochromatin formation. J Cell Sci 117: 6239–6247 [DOI] [PubMed] [Google Scholar]
- Dimitri P, Pisano C (1989) Position effect variegation in Drosophila melanogaster: relationship between suppression effect and the amount of Y chromosome. Genetics 122: 793–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Morris GD, Reuter G, Hartnett T (1992) The heterochromatin-associated protein HP1 is an essential protein in Drosophila with dosage-dependent effects on position effect variegation. Genetics 131: 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan GD, Straub T, de Wit E, Greil F, Lamm R, van Steensel B, Becker PB (2006) Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev 20: 858–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen JW, Gay EH (1934) Chromosome constitution and behavior in eversporting and mottling in Drosophila melanogaster. Genetics 19: 189–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada FN, Park PJ, Gordadze PR, Kuroda MI (2005) Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev 19: 2289–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochman B (1976) The fourth chromosome of Drosophila melanogaster. In: Ashburner M, Novitski E (eds) The Genetics and Biology of Drosophila, pp 903–928. New York: Academic Press [Google Scholar]
- Huynh KD, Lee JT (2001) Imprinted X inactivation in eutherians: a model of gametic execution and zygotic relaxation. Curr Opin Cell Biol 13: 690–697 [DOI] [PubMed] [Google Scholar]
- Huynh KD, Lee JT (2005) X-chromosome inactivation: a hypothesis linking ontogeny and phylogeny. Nat Rev Genet 6: 410–418 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, Ashburner M, Celniker SE (2002) The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol 3, RESEARCH0084.1-0084.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- Larsson J, Chen JD, Rasheva V, Rasmuson Lestander A, Pirrotta V (2001) Painting of fourth, a chromosome-specific protein in Drosophila. Proc Natl Acad Sci USA 98: 6273–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J, Meller VH (2006) Dosage compensation, the origin and the afterlife of sex chromosomes. Chromosome Res 14: 417–431 [DOI] [PubMed] [Google Scholar]
- Larsson J, Svensson MJ, Stenberg P, Mäkitalo M (2004) Painting of fourth in genus Drosophila suggests autosome-specific gene regulation. Proc Natl Acad Sci USA 101: 9728–9733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach TJ, Mazzeo M, Chotkowski HL, Madigan JP, Wotring MG, Glaser RL (2000) Histone H2A.Z is widely but nonrandomly distributed in chromosomes of Drosophila melanogaster. J Biol Chem 275: 23267–23272 [DOI] [PubMed] [Google Scholar]
- Legube G, McWeeney SK, Lercher MJ, Akhtar A (2006) X-chromosome-wide profiling of MSL-1 distribution and dosage compensation in Drosophila. Genes Dev 20: 871–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley DL, Sandler L, Baker BS, Carpenter AT, Denell RE, Hall JC, Jacobs PA, Miklos GL, Davis BK, Gethmann RC, Hardy RW, Steven AH, Miller M, Nozawa H, Parry DM, Gould-Somero M, Gould-Somero M (1972) Segmental aneuploidy and the genetic gross structure of the Drosophila genome. Genetics 71: 157–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LP, Ni JQ, Shi YD, Oakeley EJ, Sun FL (2005) Sex-specific role of Drosophila melanogaster HP1 in regulating chromatin structure and gene transcription. Nat Genet 37: 1361–1366 [DOI] [PubMed] [Google Scholar]
- Locke J, Howard LT, Aippersbach N, Podemski L, Hodgetts R B (1999a) The characterization of DINE-1, a short, interspersed repetitive element present on chromosome and in the centric heterochromatin of Drosophila melanogaster. Chromosoma 108: 356–366 [DOI] [PubMed] [Google Scholar]
- Locke J, McDermid H (1993) Analysis of Drosophila chromosome four by pulse field electrophoresis. Chromosoma 102: 718–723 [DOI] [PubMed] [Google Scholar]
- Locke J, Podemski L, Roy K, Pilgrim D, Hodgetts R (1999b) Analysis of two cosmid clones from chromosome 4 of Drosophila melanogaster reveals two new genes amid an unusual arrangement of repeated sequences. Genome Res 9: 137–149 [PMC free article] [PubMed] [Google Scholar]
- Lucchesi JC, Kelly WG, Panning B (2005) Chromatin remodeling in dosage compensation. Annu Rev Genet 39: 615–651 [DOI] [PubMed] [Google Scholar]
- Miklos GLG, Yamamoto MT, Davies J, Pirrotta V (1988) Microcloning reveals a high frequency of repetitive sequences characteristic of chromosome four and the β−heterochromatin of Drosophila melanogaster. Proc Natl Acad Sci USA 85: 2051–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr OL (1932) Genetic and cytological proof of somatic elimination of the fourth chromosome in Drosophila melanogaster. Genetics 17: 60–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- Pimpinelli S, Berloco M, Fanti L, Dimitri P, Bonaccorsi S, Marchetti E, Caizzi R, Caggese C, Gatti M (1995) Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc Natl Acad Sci USA 92: 3804–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle NC, Elgin SC (2006) The dot chromosome of Drosophila: insights into chromatin states and their change over evolutionary time. Chromosome Res 14: 405–416 [DOI] [PubMed] [Google Scholar]
- Sandler L, Szauter P (1978) The effect of recombination-defective meiotic mutants on fourth-chromosome crossing over in Drosophila melanogaster. Genetics 90: 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G (2002) Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J 21: 1121–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V (2006) Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP (2002) The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415: 810–813 [DOI] [PubMed] [Google Scholar]
- Spierer A, Seum C, Delattre M, Spierer P (2005) Loss of the modifiers of variegation Su(var)3-7 or HP1 impacts male X polytene chromosome morphology and dosage compensation. J Cell Sci 118: 5047–5057 [DOI] [PubMed] [Google Scholar]
- Spofford JB (1976) Position-effect varigation in Drosophila. In: Ashburner M, Novitski E (eds) The Genetics and Biology of Drosophila, pp 955–1018. London: Academic Press [Google Scholar]
- Stenberg P, Pettersson F, Saura AO, Berglund A, Larsson J (2005) Sequence analysis of chromosome identity in three Drosophila species. BMC Bioinform 6: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T, Gilfillan GD, Maier VK, Becker PB (2005) The Drosophila MSL complex activates the transcription of target genes. Genes Dev 19: 2284–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson MJ, Chen JD, Pirrotta V, Larsson J (2003) The ThioredoxinT and deadhead gene pair encode testis- and ovary-specific thioredoxins in Drosophila melanogaster. Chromosoma 112: 133–143 [DOI] [PubMed] [Google Scholar]
- Swaminathan J, Baxter EM, Corces VG (2005) The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev 19: 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur N, Tiwari VK, Thomassin H, Pandey RR, Kanduri M, Göndör A, Grange T, Ohlsson R, Kanduri C (2004) An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Mol Cell Biol 24: 7855–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath L, Elgin S (1995) Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev 9: 1263–1277 [DOI] [PubMed] [Google Scholar]
- Wallrath L, Guntur V, Rosman L, Elgin S (1996) DNA representation of variegating heterochromatic P-element inserts in diploid and polytene tissues of Drosophila melanogaster. Chromosoma 104: 519–527 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Legends
Supplementary Table 1


