Abstract
Supramolecular chemistry at the single-molecule level. An α-hemolysin transmembrane pore can be threaded in a desired orientation by DNA-PEG hybrid strands to yield functional rotaxanes. The single-molecule rotaxanes display stable and reversible two-state switching capacity depending on the applied potential, orientation, and the method of threading and capture used.
Keywords: Single-molecule DNA sequencing, transmembrane pore, rotaxanes, supramolecular chemistry, channel conductance
Recent advances in stochastic DNA sensing technologies,[1] especially those that exploit the transmembrane protein pore α-hemolysin (α-HL), have led to the realization that rapid single-molecule DNA sequencing may be feasible.[2] In an attempt to better ascertain the nucleobase recognition capacity of α-HL and its potential utility in DNA sequencing, we report here methods for capturing single stranded DNA-PEG hybrid molecules inside the pore of the α-HL pore. The DNA-α-HL rotaxanes have been characterized at the single-species level by electrophysiological techniques and display stable and reversible two-state switching capacity depending on the applied potential, orientation, and the method of threading and capture used.
Staphyloccocus aureus α-hemolysin (α-HL) forms large heptameric protein pores in lipid bilayers. Its crystal structure shows a mushroom shaped assembly with a central channel approximately 10 nm long, with a diameter of 1.5 nm at the narrowest point.[3] Macrocyclic adapters have been shown to lodge inside and modify the conductance characteristics the α-HL pore. This approach has been used to devise stochastic sensors for a range of small organic molecules.[4] In addition, the pore structure of α-HL can accommodate polyethyleneglycols (PEG), used to either measure the pore size[5] or in α-HL based biosensors for protein recognition.[6] Moreover, nucleic acids can also be detected during transit transport through the α-HL pore.[2, 7] Homopolymers and block copolymers of DNA such as poly(A) and poly(C), can be distinguished based on differences in the reduction of the α-HL channel conductance.[2b] These elegant studies provide the basis for the designs and studies reported here.
Rotaxanes are among the most commonly used classes of functional supramolecular structures. They have promising applications in nanotechnology as molecular switches, when the thread unit contains at least two dynamic binding sites for the macrocyclic component, each defined by a distinct signal.[8] In the present study, we sought to form rotaxanes by threading ss-DNA-PEG copolymers of defined lengths through the α-HL transmembrane pore. The pore has a dual function in our design. Its protein structure plays a role equivalent to the macrocyclic subunit of the typical small molecule rotaxanes and the perturbations of its native ion conductance provides the sensing signature for evaluating the functional characteristics of the rotaxanes.
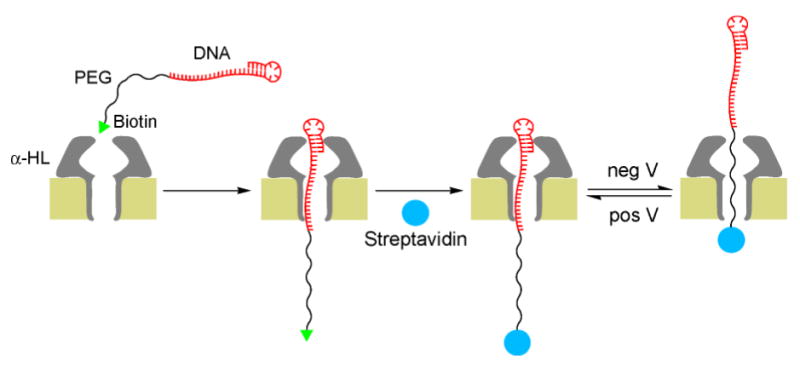
The thread component of the rotaxane is an appropriately functionalized ss-DNA-PEG hybrid molecule designed to afford two different conductance states depending on the portion of the strand (DNA or PEG) that is occupying the channel. DNA Hybrid 1 was constructed as a continuous linear arrangement of the following four segments. The 3’ end bears a 20-base region, designed to bind to a specific oligonucleotide of complementary sequence, followed by a run of poly d(A) (40mer). This segment is extended with 10 units of hexaethylene glycol phosphate and capped with a biotinyl group at the 5’ terminus. This design provides DNA and PEG regions of the appropriate length to allow full threading of the α-HL pore by either of the two polymeric segments. It also provides a mechanism to lock thread 1 at both ends after its insertion into the pore. Formation of a DNA duplex [2a] at the 3’ end of the polymer and binding of streptavidin to the 5’ biotin [6,7b,7d] create structures that are wider than the α-HL entrances and thus can be used to lock-in the rotaxane complex (Figure 1a). Similarly, DNA Hybrid 2 having a double-stranded 3’-DNA stem-loop structure[2c] was designed to control threading orientation and rotaxane formation (Figure 1b). We exploit the anionic character of the ss-DNA-PEG chains to drive the threading process by an applied transmembrane potential. In addition, once a rotaxane is formed, the relative position of the thread inside the pore can be switched by simply adjusting the sign and magnitude of the applied transmembrane potential.
Figure 1.
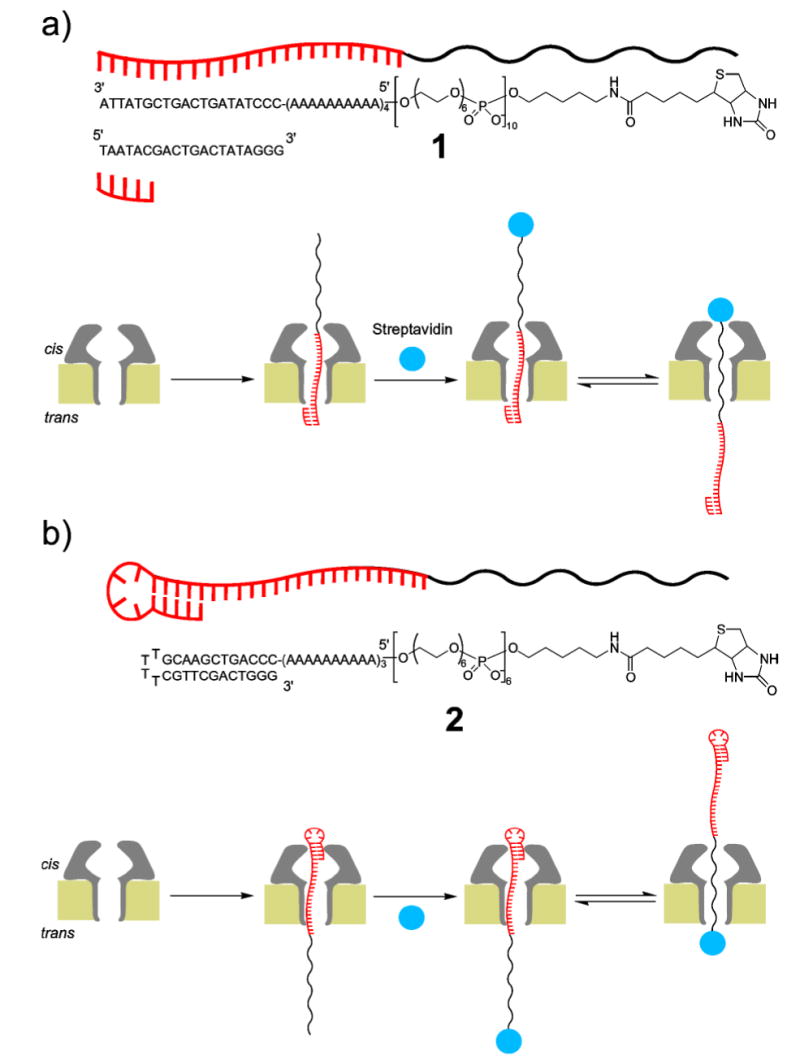
Schematic representation of the systems employed for transmembrane protein rotaxane. a) System employed for rotaxane formation by capture on the cis side by streptavidin and, b) thread molecule with a DNA hairpin for rotaxane formation by capture on the trans side with streptavidin.
Initial experiments were performed with the uncapped ss-DNA-PEG thread 1 added either to the cis or the trans side of the bilayer.[9] At positive transmembrane holding potentials (negative when 1 is added in trans chamber) blocking events displaying two distinct levels were detected suggesting that PEG and DNA subunits can be differentiated (Figure 2a,b). The events can be grouped in two classes, depending on which level is detected first. We have established (vide infra) that the highest blocking step (level 2) results from the DNA segment traversing the pore, while the lower blocking level (level 1) corresponds to the passage of the PEG portion of the strand. Moreover, events that first display the highest blocking level are more numerous indicating that the favored orientation for strand threading is with the 3’-DNA tail first.
Figure 2.
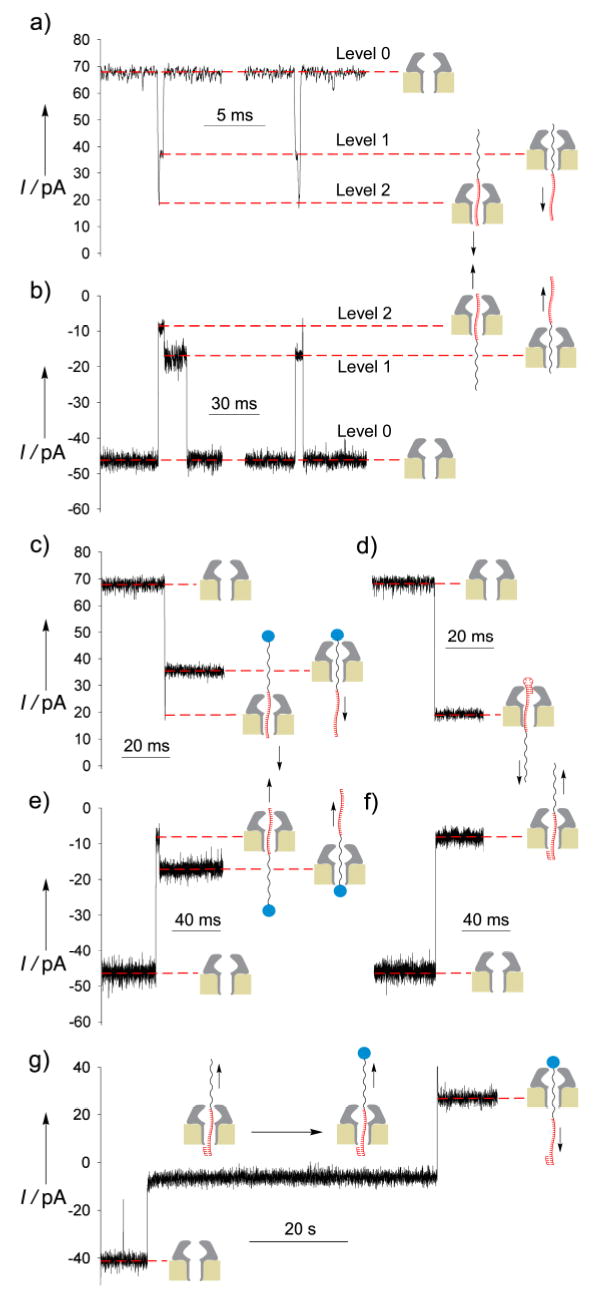
Blocking events caused by thread molecules. Typical events are shown from traces recorded at +140 mV when the following thread molecules were added to the cis chamber: a) 1 μM 1, c) 1μM 1 and 10 μM streptavidin and, d) hairpin thread 1 μM 2. Events were also observed at −140 mV with thread molecules added to the trans side: b) 1μM 1, e) 1μM 1 and 10μM streptavidin, and f) 1μM 1 and 5μM complementary oligonucleotide strand. g) Rotaxane formation at −120/+120 mV transmembrane potential with 1μM 1 and 5μM complementary oligonucleotide strand on the trans side and 10 μM streptavidin on the cis side. Traces were recorded in symmetric KCl 500mM, MOPS 5mM pH 7.5, bessel filtered at 5 kHz and sampled at 6 μs (a), and 100 μs (b-f) and bessel filtered at 2 kHz and sampled at 200 μs (g).
Topoisomeric pseudo-rotaxanes of α-HL, which differ only in the orientation of the threading polymer 1, were used to unequivocally assign the two observed conductance states. The conductance levels attributed to the PEG and DNA can be established when the 3’- or the 5’-termini of 1 is sterically blocked from entering the pore by either DNA duplex formation at the 3’-tail or streptavidin complexation to the 5’-biotin. In experiments where the thread and streptavidin were both present in the same chamber, the 5’-tail was blocked and thus only the DNA end was available to go through the α-HL pore. As expected, the observed blocking pattern showed an initial high level of block (passage of DNA) followed by a recovery in the channel conductance to a lower level assigned to the PEG segment (Figure 2c,e). As expected for a pseudo-rotaxane, the conductance level was stable as long as the transmembrane voltage was applied, indicating that both the steric bulk and strength of the biotin-streptavidin complex prohibits the strand from translocating through the channel pore to the other side of the bilayer.
Experiments performed under asymmetrical salt conditions allowed the determination of charge selectivity for the threaded channel.[10] While free α-HL displays a slight anion selectivity (PK+/PCl− = 0.6 in bilayers separating 300 and 500 mM KCl on the cis and trans sides, respectively), when it is threaded from the trans side by the streptavidin-biotin strand complex, it shows PK+/PCl− = 2.0 suggesting that the negatively charged phosphate groups in the PEG segment of the strand change the charge density inside the pore transforming α-HL into a somewhat cation selective pore (Figure 3a).
Figure 3.
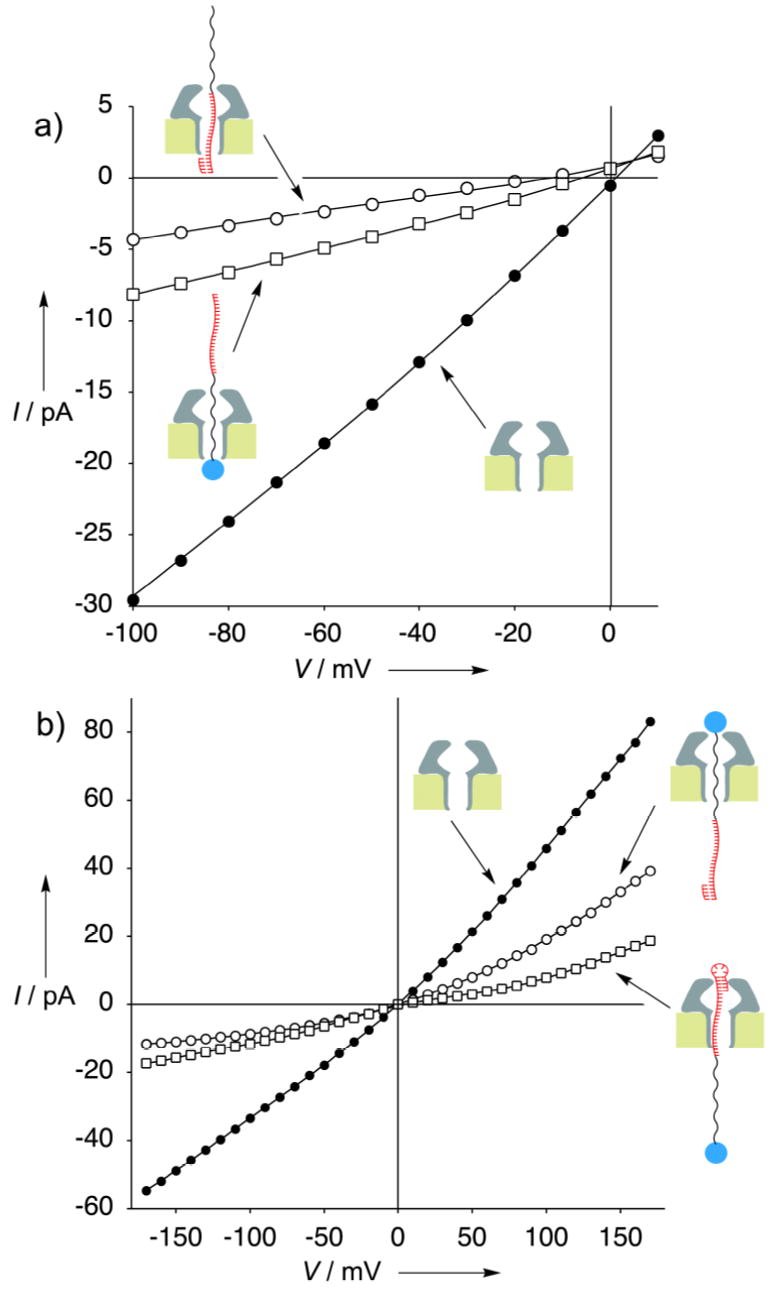
a) Current versus voltage relationships obtained under asymmetric conditions (300 and 500 mM KCl in the cis and trans chambers, respectively, both buffered with 5mM MOPS to pH 7.5) for α-HL (•),α-HL threaded with the streptavidin complex of 1 added to the trans side (▪) and thread 1 hybridized with the complementary oligonucleotide added to the trans side (○). b) Current versus voltage representations for α-HL (•),α-HL rotaxane with thread 1 (○) and rotaxane with strand 2 (□) in KCl 500mM, MOPS 5mM pH 7.5.
Hybridization of thread 1 to its complementary 20-mer DNA sequence prevents the 3’ DNA tail from entering the α-HL lumen; thus, only the 5’-tail (PEG segment) is free to enter the pore. Indeed, when 1 and the complementary short DNA were both added to the trans side (Figure 2f) the blocking events observed by applying negative transmembrane potentials were stable with time and showed the high level of block associated with the DNA segment of the thread being inside the α-HL. When the same experiment was performed under asymmetric conditions (Figure 3a) high charge selectivity was observed (PK+/PCl− > 100) accounted for by the high density of phosphate groups from the DNA inside the pore.
Formation of a single-molecule α-HL/DNA-PEG rotaxane is a two-step process. The first step as described above involves formation of a pseudo-rotaxane. This process is driven by the applied transmembrane potential to force threading of the free tail of a one-end capped (ds-DNA or streptavidin) polymer through the transmembrane pore and holding it stably in that configuration. The second step (locking) requires efficient capture of the protruding end of the thread on the other side of the bilayer. Practical considerations precluded the use of a streptavidin-capped strand as the starting material for rotaxane formation since the locking step would require capture of the 3’ end by the complementary DNA oligonucleotide. We found that the best approach to achieve rotaxane formation was to employ the pre-hybridized strand on the trans side of the pore with streptavidin present in the cis side (Figure 1a). Our studies show that once the pseudo-rotaxane was captured (typically in <1 minute under conditions employed), α-HL conductance could no longer be returned to its native conductance state, even when the sign of the applied transmembrane potential was changed (Figure 2g). Instead the α-HL/DNA-PEG rotaxane displayed the conductance level at positive potentials corresponding to the PEG moiety and at negative potentials conductance level corresponding to the DNA subunit residing inside the pore. The rotaxane structure proved to be stable between the range of transmembrane potential studied (from −170 to +170 mV) and could be switched multiple times between the DNA and PEG states (Figure 2g).
The DNA-PEG polymer 2 with a double-stranded 3’-DNA stem-loop structure was used to construct a corresponding rotaxane structure of opposite strand orientation (Figure 1b). The DNA stem-loop is known to prevent entry into the vestibule of the α-HL channel when applied to the cis side.[2c] Addition of 2 to the cis side of the α-HL pore resulted in the formation of a pseudo-rotaxane as evidenced by the observed permanent block when a high positive transmembrane potential was applied. The residual conductance corresponded to the expected blockade caused by DNA residing inside the pore (Figure 2d). Furthermore, the α-HL native conductance could be fully recovered by switching back and forth between negative and positive transmembrane potentials (threading/dethreading). However, when streptavidin was added to the trans side the thread was permanently trapped in the α-HL channel by the resulting rotaxane structure. This new rotaxane showed a conductance level at negative holding potentials typical of the PEG segment lodged inside the channel signifying formation of a rotaxane of the opposite thread orientation as compared to the one described above. The rotaxane structure was also shown to be stable over a wide range of applied transmembrane potentials (from −170 to +170 mV) (Figure 3b).
In summary, we have described a methodology for controlled formation of individual rotaxanes of desired strand orientation with tunable conformational states. This methodology is expected to open up new possibilities for studying nucleic acids and other polymers at single-molecule level. In addition, this study provides a direct method for trapping of DNA strands of defined lengths and sequences inside the α-HL channel structure in order to define the scope and limitations of the nucleobase recognition capacity of α-HL and establish its utility in DNA sequencing. The work also suggests that DNA-α-HL rotaxanes might provide a means for multiple pass single-molecule DNA sequencing which is likely to have significantly lower sequencing errors than single pass analyses.
Supporting Information
Single Molecule DNA Rotaxanes of a Transmembrane Pore Protein**
Jorge Sánchez-Quesada, Alan Saghatelian, Stephen Cheley, Hagan Bayley, and M. Reza Ghadiri*
DNA-PEG Hybrid Strands Synthesis
Synthesis was accomplished on an Applied Biosystems 391 PCR-MATE synthesizer (Applied Biosystems, Foster City, CA) using standard cyanoethylphosphoramidite chemistry. Nucleobase, hexaethylene glycol and biotin β-cyanoethylphosphoramidites derivatives, solvents and reagents were used as provided (Glen Research, Sterling, VA). DNA-PEG hybrid strands were purified by preparative denaturing polyacrylamide gel electrophoresis, isolated by the crush and soak method, and desalted using C18 Sep-Pak cartridges (Waters, Milford, MA). Quantification was determined from the absorbance at 260 nm, and stored at −20 °C.
Planar Lipid Bilayer Experiments
Lipid bilayers of 1,2-diphytanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids, Alabaster, AL) were formed on a 80-100 μm orifice in a 25 μm thick Teflon film (Goodfellow Corporation, Malvern, PA) separating the two chambers of a Teflon lipid bilayer apparatus following the procedure described by Montal and Mueller. Chambers were filled with 0.5 M KCl solutions containing 5 mM MOPS (3-(N-morpholino)propanesulfonic acid) and titrated to pH 7.5 with KOH. Chambers were named cis and trans, cis chamber was at virtual ground, hence a positive potential indicates a higher potential in the trans side and a positive current is one in which cations flow from the trans to the cis chamber. Monomeric α-hemolysin (α-HL) was added to a final concentration of approximately 50 nM in the cis chamber leading to single channel appearance in a matter of minutes, chamber was then flushed with fresh buffer solution to avoid further incorporation of additional α-HL channels. Thread molecules were added to final concentrations of 0.5 − 1.0 μM, short complementary DNA strand and streptavidin were employed at 5–10 μM final concentration. Transmembrane currents were recorded with an Axopatch-1D amplifier (Axon Instruments, Foster City, CA). The electrodes were 1 mm diameter Ag/AgCl pellets connected to a silver wire surrounded by a wax-sealed Teflon tube (Axon Instruments). For analysis, currents were further low-pass filtered with an eight-pole Bessel filter (model 902, Frequency Devices, Haverhill, PA) with the corner frequency set at 5 or 2 kHZ (-3dB), and sampled by computer at 6–200 μs (Digidata A/D converter and pClamp 8 software, Axon Instruments).
Charge Selectivity Measurements
Experiments were performed in independent experiments under non-symmetrical conditions with planar bilayers separating solutions of 300 and 500mM KCl in cis and trans sides, respectively. Reverse potentials or potentials at zero current were determined from both graphical representations of current versus potential and readings from the amplifier output. Permeability ratios were calculated by substituting reverse potentials values and ionic activities for the solutions employed into the Goldman-Hodgkin-Katz equation:
An additional experiment was performed to prove the assignment of the DNA blocking level. A DNA single strand of d(A)40 was prepared containing a biotin group at the 5’ terminus. When this strand was added to the cis side in the presence of streptavidin, pseudorotaxane formation was observed by the decrease in conductance of the αHL pore to a similar level to the one observed when hybrid 2 was employed. This was an indication that neither the nature of the stopper nor the orientation of the strand (3’ to 5’ and 5’ to 3’) shows any remarkable effect in the blocking current in the system studied.
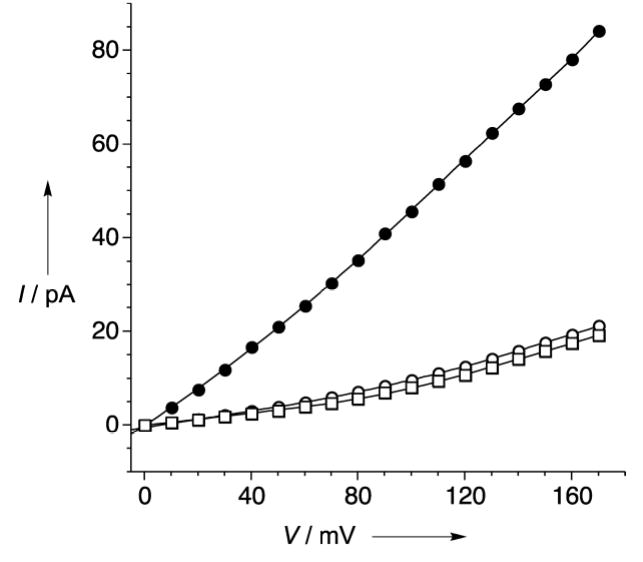
Current versus voltage representations for α-HL (•), thread molecule 2 added to the cis chamber (□) and d(A)40-biotin + streptavidin to the cis side(○). 500mM KCl, 5mM MOPS, pH 7.5.
Footnotes
This work was supported by a grant from the Office of Naval Research (MURI-99, N000149910717).
References
- 1.a) Bayley H, Martin CR. Chem Rev. 2000;100:2575. doi: 10.1021/cr980099g. [DOI] [PubMed] [Google Scholar]; b) Bezrukov SM. J Membrane Biol. 2000;174:1. doi: 10.1007/s002320001026. [DOI] [PubMed] [Google Scholar]; c) Bayley H, Cremer PS. Nature. 2001;413:226. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- 2.a) Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proc Natl Acad Sci USA. 1996;93:13770. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Biophys J. 1999;77:3227. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Vercoutere W, Winters-Hilt S, Olsen H, Deamer D, Haussler D, Akeson M. Nat Biotechnol. 2001;19:248. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]; d) Howorka S, Cheley S, Bayley H. Nat Biotechnol. 2001;19:636. doi: 10.1038/90236. [DOI] [PubMed] [Google Scholar]
- 3.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Science. 1996;274:1859. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 4.a) Gu LQ, Braha O, Conlan S, Cheley S, Bayley H. Nature. 1999;398:686. doi: 10.1038/19491. [DOI] [PubMed] [Google Scholar]; b) Sánchez-Quesada J, Ghadiri MR, Bayley H, Braha O. J Am Chem Soc. 2000;122:11756. [Google Scholar]; c) Gu LQ, Cheley S, Bayley H. Science. 2001;291:636. doi: 10.1126/science.291.5504.636. [DOI] [PubMed] [Google Scholar]; d) Gu LQ, Cheley S, Bayley H. J Gen Physiol. 2001;118:481. doi: 10.1085/jgp.118.5.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Korchev YE, Bashford CL, Alder GM, Kasianowicz JJ, Pasternak CA. J Membrane Biol. 1995;147:233. doi: 10.1007/BF00234521. [DOI] [PubMed] [Google Scholar]; b) Bezrukov SM, Vodyanov I, Brutyan RA, Kasianowicz JJ. Macromolecules. 1996;29:8517. [Google Scholar]; c) Howorka S, Movileanu L, Lu X, Magnon M, Cheley S, Braha O, Bayley H. J Am Chem Soc. 2000;122:2411. [Google Scholar]
- 6.a) Movileanu L, Howorka S, Braha O, Bayley H. Nat Biotechnol. 2000;18:1091. doi: 10.1038/80295. [DOI] [PubMed] [Google Scholar]; b) Movileanu L, Cheley S, Howorka S, Braha O, Bayley H. J Gen Physiol. 2001;117:239. doi: 10.1085/jgp.117.3.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Proc Natl Acad Sci USA. 2000;97:1079. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Henrickson SH, Misakian M, Robertson B, Kasianowicz JJ. Phys Rev Lett. 2000;85:3057. doi: 10.1103/PhysRevLett.85.3057. [DOI] [PubMed] [Google Scholar]; c) Meller A, Nivon L, Branton D. Phys Rev Lett. 2001;86:3435. doi: 10.1103/PhysRevLett.86.3435. [DOI] [PubMed] [Google Scholar]; d) Kasianowicz JJ, Henrickson SE, Weethall HH, Robertson B. Anal Chem. 2001;73:2268. doi: 10.1021/ac000958c. [DOI] [PubMed] [Google Scholar]; e) Howorka S, Movileanu L, Braha O, Bayley H. Proc Natl Acad Sci USA. 2001;98:12996. doi: 10.1073/pnas.231434698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raymo FM, Stoddart JF. Chem Rev. 1999;99:1643. doi: 10.1021/cr970081q. [DOI] [PubMed] [Google Scholar]
- 9.Lipid bilayers were formed from 1,2-diphytanoyl-sn-glycero-3-phosphocholine in a Teflon planar bilayer apparatus: Montal M, Mueller P. Proc Natl Acad Sci USA. 1972;69:3561. doi: 10.1073/pnas.69.12.3561. The cis chamber was at virtual ground, and a positive potential refers to a higher potential in the trans chamber. A positive current is produced by cations flowing from the trans to the cis side or aninons flowing in the opposite direction. α-HL is a non-symmetric channel; its orientation in planar lipid bilayers was controlled by adding the preassembled heptamers to the cis chamber. α-HL incorporates into lipid bilayers through the stem domain that ends up spanning the bilayer, while the cap remains on the cis side.
- 10.Permeability ratios were calculated from reverse potential measurements and ionic concentrations, converted into ionic activities, by using the Goldman-Hodgkin-Katz equation: B. Hille, Ionic channels of excitable membranes, 3rd ed. Sinauer, Sunderland, 2001.


