Abstract
Using a large set of orthologous human and mouse gene pairs, we have characterized genes that have been retained in duplicate in human over timescales comparable to the time of speciation of human and mouse. Orthologous gene pairs for which a paralogous gene has been present for much or all of the time since speciation show an increased rate of nonsynonymous substitution. We have related rate of divergence to functional classification using the Gene Ontology terms. Protein function was found, in some cases, to have a larger impact on rate of evolution than the presence or absence of a paralog. No evidence was found that genes that have been retained in duplicate are weighted toward any functional categories. An increase in the ratio of nonsynonymous to synonymous changes following duplication has previously been reported. However, because amino acid sequences include conservative as well as more freely evolving sites, the ratio of nonsynonymous to synonymous changes tends to be higher for closely related pairs. By measuring the divergence of orthologs only and comparing between genes for which a paralogous gene is either present or absent, we have compared gene pairs that share a common divergence time. We have also found that shorter genes have a higher probability of being found duplicated in the human genome, possibly reflecting a mutational effect.
Genes that have been duplicated through mutation, even if they rise to fixation, are normally lost over long evolutionary timescales because of lower-than-usual purifying selective pressure. More rarely, genes may be retained in duplicate over longer times. In some such cases, the functions and expression patterns of the gene pair may diverge substantially—giving rise to novel functions or specializations in the organism. The decrease in purifying selective pressure that may accompany duplication increases the probability of gene loss, while at the same time, gives rise to the possibility that a novel functionality may evolve. Several examples of the evolution of novel functions through gene duplication have been cited (Li 1997). Hughes and Hughes (1993) performed an early study of the pattern of nucleotide substitution after duplication by comparing a set of duplicated genes from Xenopus laevis to their human orthologs. Although a classic model of functional diversification after duplication indicates that one copy of the gene maintains the original function, whereas the other copy is free to accumulate substitutions and possibly develop a novel function, they found no evidence that one copy of their duplicated genes had evolved more rapidly than the other. More recently, Force et al. (1999) have suggested that gene duplication may allow subfunctionalization to take place if a gene performing more than one function is duplicated. In such a case, the duplicated gene pair might adapt to perform the individual functions separately and more efficiently than the bifunctional parent gene. Many previous studies have attempted to estimate the relative probability of functional adaptation or gene loss through the fixation of null alleles (Walsh 1995; Wagner 1998; Force et al. 1999; Taverna and Goldstein 2000). The likelihood of long-term fixation of novel functions or functional adaptations in a duplicated gene pair has been shown to be influenced by population size (Wagner 2000), as well as numerous additional factors that are difficult to model (Nowak et al. 1997). Redundancy may also be maintained in the genome over long-scales in the absence of adaptation (Nowak et al. 1997; Taverna and Goldstein 2000; Wagner 2000). Factors influencing the initial fixation of duplicated genes have also recently become the subject of research (Lynch et al. 2001).
Lynch and Conery (2000) have suggested that the duplication of genes is a relatively frequent event in evolution—∼0.007 per gene per million years in human (Long and Thornton 2001; Zhang et al. 2001). Typically, these duplicated genes are lost because of a lack of selective pressure to maintain both copies. Lynch and Conery (2000) have estimated a half-life of ∼8 to 16 Myr for the set of genes that do not acquire novel functions and are not retained in duplicate indefinitely (Long and Thornton 2001; Zhang et al. 2001). In their original paper, Lynch and Conery applied a logarithmic decay function to the rate of duplicate gene loss. Using the parameters they have estimated and the comparatively recent date of 65 Myr estimated by them for a genome duplication event in Arabidopsis, no genes originating in the genome duplication event would be expected to be retained in duplicate. Clearly, this logarithmic function would apply only to genes that do not go on to diversify and acquire novel functions. Nonetheless, a surprisingly large number of genes appear to have been retained in duplicate following genome duplication and given the estimate of the rate of gene loss produced by Lynch and Conery. The proportion of duplicated genes retained after genome duplication is highly variable (Massingham et al. 2001; Wolfe 2001), but in a range of organisms, it is higher and, in some cases, markedly higher than would be expected from the application of a decay function with parameters in the ranges estimated by Lynch and Conery (2000), for example, 72% in maize after 11 Myr (Ahn and Tanksley 1993; Gaut and Doebley 1997), 47% in catostomid fish after 53 Myr (Ferris and Whitt 1979), 28% in Arabidopsis after 65 to 200 Myr, and 16% in yeast after 100 Myr (Seoighe and Wolfe 1998).
By measuring the relative rates of synonymous and nonsynonymous substitutions in pairs of paralogous genes Lynch and Conery have suggested a time-dependent impact of duplication on selective pressure—with gene pairs evolving in a nearly neutral way immediately after duplication but becoming more constrained as they become more divergent from their copies. Lynch and Conery have compared the ratio of nonsynonymous substitutions per nonsynonymous site (Ka) to synonymous substitutions per synonymous site (KS) in gene pairs with very different divergence times, and have interpreted their results as a measure of the increase in selection pressure with time in duplicated genes. However, the time since divergence has a significant effect on Ka/KS because amino acid sequence positions evolve at very different rates, whereas synonymous sites probably evolve at much more similar rates. Therefore, Ka/KS should only be compared between sets of gene pairs sharing a divergence time. The difficulty with Lynch and Conery's result is illustrated here by comparing a set of human and chimpanzee orthologs.
In the current work, we have calculated the evolutionary rates of a large set of human and mouse orthologous gene pairs. Using the ENSEMBL (Hubbard et al. 2002) database (v 1.0) of confirmed proteins, we have searched for paralogs of the human proteins for which mouse orthologs were available. We have classified gene functions using the Gene Ontology (GO) Consortium classifications (Ashburner et al. 2000). The aims of the present work are threefold: (1) to characterize the set of genes that are retained in duplicate over different timescales, (2) to measure the effect of gene duplication on rate of sequence evolution, and (3) to look for differences in rate of sequence evolution in different gene-functional classes.
Previous studies of gene families have provided examples of adaptive evolution following gene duplication (Hughes et al. 1998). Adaptive evolution is inferred from rates of nonsynonymous substitution that are higher than synonymous substitution rates. Hughes et al. (2000) have pointed to positive selection in the variable regions of a family of placentally expressed genes. Hughes and co-workers identified variable regions from amino acid alignments and compared Ka with KS in these regions. Because these regions were identified on the basis of variability in amino acid sequence, the comparisons are biased in favor of higher values of KS. Despite this, the very high levels of nonsynonymous substitution appear to reflect functional adaptation. Zhang et al. (1998) found evidence of adaptive evolution in primate ribonuclease genes. The gene families studied are involved in host defense from pathogen attack (Zhang et al. 1998). Although the branch with the highest level of positive selection is close to a duplication event, it is not clear whether the positive selection is a result of gene duplication or pressure for diversification frequently associated with host-pathogen interaction. The focus of the current work is not on discovering instances of adaptive evolution in specific cases of gene duplication but rather on determining the mean effect of the presence of a paralogous gene on the rate of evolution in a set of orthologous genes. This question has recently been approached from a slightly different angle by Kondrashov et al. (2002). They have compared rates of evolution in paralogs and orthologs from a wide range of organisms but have not compared between orthologs and paralogs that share a divergence time, as we have performed here. Kondrashov et al. (2002) have found a larger difference between the rates of divergence of paralogs and orthologs than the one we report here (Ka/KS = 0.451 for mammalian paralog pairs and Ka/KS = 0.131 for mammalian ortholog pairs). We argue that this large difference between orthologs and paralogs is likely to result from more recent average divergence times in the paralog pairs that were used, compared to the ortholog pairs.
RESULTS
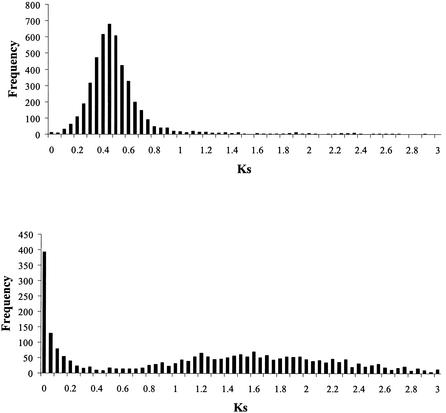
Synonymous (KS) and nonsynonymous (Ka) distances were calculated for the set of 5341 human-mouse orthologs that was extracted from the HomoloGene database and for the set of 2663 human paralog pairs constructed as described above (see Methods). Figure 1a shows histograms of KS for the orthologous data set. The mean and standard deviations of KS in the ortholog data set were 0.54 and 0.34, respectively.
Figure 1.
Histogram of KS for human and mouse orthologous gene pairs (A) and for human closest paralog pairs (B).
Two sets of human and mouse orthologous pairs were constructed on the basis of the presence or absence of a human paralog within a specified synonymous distance of the human gene. First a set of 180 human-mouse orthologs for which a very close paralog (KS≤0.05) could be identified in human was identified. Second, the mean value of KS for human/mouse orthologs was used to identify human genes from our ortholog set that may have been duplicated at approximately the same time as the divergence of human and mouse. This set contained 70 unique members and was considered to represent the set of human genes with intermediate paralogs. It included all orthologs for which an intermediate paralog (0.34 ≤ KS ≤ 0.74) could be identified (Fig. 1B). Many of these genes are likely to have been present in duplicate in human for much of the time since speciation of human and mouse.
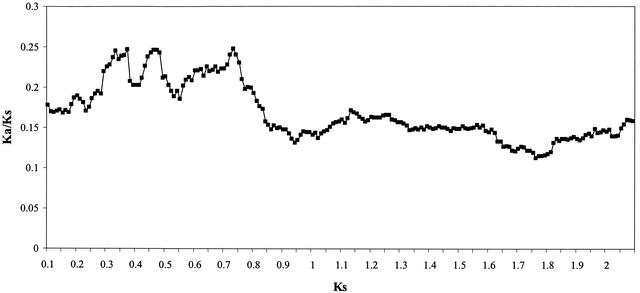
The divergence of human genes for which an intermediate paralog was identified from their mouse orthologs was significantly greater than the average divergence for the whole set. This increased rate of evolution was not found for genes with close paralogs. The effect of the presence of a paralog on the sequence divergence of an ortholog pair depends on how closely related the paralogs are (Fig. 2). Recent duplication does not appear to have had an effect on the divergence of mouse/human orthologs while the presence of older duplicates causes an increase in ortholog divergence. The increased divergence is most apparent in nonsynonymous sites, leading to an increase in the ratio of nonsynonymous to synonymous substitution rates (Table 1). The increase in the synonymous rate can be explained either by the choice of genes in this set or by a correlation between synonymous and nonsynonymous substitution rates (see Discussion). Using the bootstrap resampling procedure described in Methods, the weighted-mean values of KS, Ka, nucleotide divergence, and protein divergence were significantly greater on the set of orthologs for which an intermediate paralog was identified in human (the values were exceeded 1, 0, 1, 11, and 4 times, respectively, from 1000 replicates of the data set).
Figure 2.
Sliding window representation of Ka/KS in the human-mouse orthologs against KS of the closest human paralog. A window size of 0.2 and step size of 0.01 were used.
Table 1.
Sequence Divergence of Ortholog Datasets
| Average length of coding sequencesa (bp) | Numberb | Nucleotide distance | Protein distance | Ka | KS | Ka/KSc | Ka/Ks | |
|---|---|---|---|---|---|---|---|---|
| All orthologues | 1245 | 4218 | 0.201 | 0.202 | 0.108 | 0.535 | 0.171 | 0.202 |
| Orthologues with intermediate paralogue present in human | 997 | 70 | 0.265 | 0.328 | 0.172 | 0.603 | 0.232 | 0.285 |
| Orthologues with close paralogue present in human | 909 | 180 | 0.194 | 0.190 | 0.100 | 0.526 | 0.168 | 0.190 |
| Chimpanzee human orthologues | 835 | 116 | 0.052 | 0.0824 | 0.028 | 0.052 | 0.640 | 0.538 |
aGenes for which only partial coding sequence was available are included.
bOnly gene pairs for which neither Ka nor KS is saturated have been included.
cAverage calculated over gene pairs for which KS > 0.
On average, the sequences for which a close paralog could be identified in human were significantly shorter than the other sequences (P < 0.001; Table 1). For the case of gene-lengths, a two-tailed test of significance was applied because there was no prior expectation that the duplicated genes should be shorter than unduplicated genes. The P value shown above, derived from a nonparametric bootstrap procedure, indicates that this difference is robust to sampling error. Some of the sequences used were fragments of genes. To determine whether the difference in sequence lengths reflected a difference in the lengths of genes that are duplicated intact, we selected only sequences that were annotated as containing the complete coding sequence of the corresponding gene. When full genes only were compared, genes with close paralogs remain significantly shorter than the remainder (P < 0.001). The sequences of the genes with intermediate paralogs were also shorter than average; however, there were insufficient full-length sequences to establish whether this difference is significant or not.
To compare Ka/KS between sets of genes with different divergence times, we constructed a set of human/chimp orthologs. Evolutionary divergences could be calculated for 119 reciprocal best hits between chimp CDSs and ENSEMBL cDNAs. The average value of Ka/KS for this set was 0.64 (Table 1). This high value for closely related orthologs corresponds to the situation immediately following divergence. At this point, the amino acids that are under low selective constraint (or in some cases under positive selection) are unsaturated, and the rate of nonsynonymous substitution is nearly equal to that of synonymous substitution. This illustrates the danger of comparing Ka/KS between gene pairs with very different divergence times, as Lynch and Conery (2000) did. Interestingly the average length of the chimpanzee sequences in our set is significantly less than the average length of the human sequences (Table 1). This is consistent with the general trend in the database in which the average length of human full-length CDSs in the European Molecular Biology Laboratory (EMBL) database is 1461 bp, whereas the equivalent value for chimpanzee sequences was 1108 bp. This difference must reflect differences in the kinds of genes that have been sequenced in chimpanzee rather than real differences in lengths of coding sequences between human and chimpanzee.
Short Sequences
We found that the shortest sequences had higher values of Ka/KS, on average, than did longer sequences. The value of Ka/KS for the shortest 10% of sequences was 0.214. For the shortest 5% of sequences, Ka/KS was equal to the value obtained for the genes with paralogs (0.230). In both cases, KS was close to the value obtained on the entire data set, and the increased value of Ka/KS was brought about by increases in Ka. To test whether the increase in Ka/KS was caused by a greater proportion of shorter sequences among the duplicates, we calculated the average value of Ka/KS in the data set of genes with intermediate paralogs, considering only genes that were longer than average length of genes in the entire orthologous data set. The value of Ka/KS for 22 genes with intermediate paralogs and longer than 1245 bp was also 0.230. Although shorter sequences appear to be evolving faster than longer genes, on average, this is not the sole cause of the apparent increase in the rate of evolution of duplicated genes.
Gene Function and Evolutionary Rate
Classification of genes was performed using the highest GO functional categories, as described in Methods. No significant differences in functional classifications were found when genes with paralogs were compared to the whole set (Table 2). The 31 genes classified as defense related evolved at nearly twice the average rate (Ka/KS = 0.36). The effect of function on rate of evolution is therefore far more substantial than the average impact of gene duplication in the case of defense genes. The number of proteins in a functional category was frequently too small to detect a difference in evolutionary rate. Using a two-tailed test of significance based on random resampling of the data, the only other functional category for which Ka/KS was significantly increased was the set of proteins classified as “molecular function unknown” (Ka/KS = 0.28). The “enzyme,” “motor,” and “ligand binding or carrier” categories showed significant decreases in divergence (Ka/KS = 0.19, 0.13, and 0.19, respectively).
Table 2.
Gene Ontology Functional Classification
| GO functional classification | Number of human orthologs | Number in intermediate paralog set | Number in close paralog set |
|---|---|---|---|
| Total for which classification was possible | 2525 | 49 | 94 |
| Anticoagulant | 2 | 0 | 0 |
| Antioxidant | 1 | 0 | 0 |
| Apoptosis regulator | 20 | 1 | 1 |
| Cell adhesion molecule | 133 | 2 | 5 |
| Cell cycle regulator | 38 | 1 | 3 |
| Chaperone | 160 | 1 | 5 |
| Chaperone regulator | 17 | 0 | 2 |
| Defense/immunity protein | 76 | 1 | 4 |
| Enzyme | 1395 | 29 | 56 |
| Enzyme regulator | 107 | 1 | 6 |
| Ligand binding or carrier | 1121 | 25 | 38 |
| Lysine | 10 | 0 | 0 |
| Molecular function unknown | 113 | 1 | 4 |
| Motor | 33 | 1 | 1 |
| Nucleic acid binding | 595 | 17 | 32 (0.02*) |
| Obsolete | 88 | 0 | 4 |
| Protein stabilization | 1 | 0 | 0 |
| Protein tagging | 1 | 0 | 1 |
| Signal transducer | 992 | 14 | 39 |
| Storage protein | 2 | 0 | 0 |
| Structural protein | 168 | 3 | 6 |
| Toxin | 9 | 0 | 0 |
| Transporter | 486 | 8 | 15 |
P values have been calculated from a two-tailed Fisher's exact test and are shown for values <0.05. Correction for multiple tests has not been performed.
DISCUSSION
Starting from a relatively large data set of orthologs and making use of a human protein set that is coming closer to being complete has made it possible to characterize the genes that are retained in duplicate and to measure the effect of the presence of paralogs on the rate of sequence evolution. Because the ENSEMBL protein set that was used is likely not to include all human proteins, some of the genes for which paralogs genes are present in the genome will have been missed. This will mean that the identification of the set of genes for which paralogs are available will be less than perfectly efficient. The effect will be to reduce the magnitude of the observed differences between the sets with paralogs and the rest of the ortholog data set. As a result, the estimate of the effect on rate of evolution of the presence of a duplicated gene should be considered to be a conservative estimate. Synonymous distances between human paralogs were calculated for only the highest nonself BLAST hits of the human gene against the ENSEMBL data set. In some cases, a human gene will have close and more distant paralogs. No additional category was established for such genes, and they will have been included in the set of genes for which close paralogs were available. As a result, the distinction between these two categories of genes is blurred. Despite this, there is a significant difference between the two groups. The rate of acceleration that was observed in the set of human genes with intermediate paralogs was not found in the genes with close paralogs.
Increase in the Rate of Synonymous Substitution in the Duplicated Gene Set
All of the measures of ortholog distance, including KS, were greater in the set of genes with intermediate paralogs. This increased value of KS can be explained by a divergence time for genes with intermediate paralogs and their mouse counterparts that is older than the time of speciation of human and mouse. It is easy to see how this could be the case. Genes that duplicated in human some time before the speciation of human and mouse could easily have been wrongly assigned to an orthologous pair in the HomoloGene data set, such that the mouse gene was associated with the wrong human gene from the duplicated pair. This would result in a divergence time equal to the time, before speciation, of the duplication of the human gene. Although this could account for some of the increased divergence between these human and mouse orthologs, it cannot account for the fact that the increase in Ka is higher than the increase in KS, resulting in a significantly higher value of Ka/KS. The increased value of KS can be explained without reference to the difficulty in determining orthology in the presence of paralogous pairs by noting the correlation between synonymous and nonsynonymous substitutions that has been observed in mammals (Wolfe and Sharp 1993). A high frequency of double-nucleotide substitutions could link synonymous and nonsynonymous rates, although it is not clear whether the observed rate of double-nucleotide substitutions (Averof et al. 2000) is high enough to produce a significant effect.
Increased Rate of Evolution After Duplication
Our observation of an increase of 36% in the average value of Ka/KS for the case of genes with an intermediate paralog is at odds with a recent study of 19 duplicated gene sets in fish, in which no evidence was found for an increased rate of evolution following gene duplication (Robinson-Rechavi and Laudet 2001). In another recent study, Ka/KS was compared between sets of paralogous and orthologous genes (Kondrashov et al. 2002). The difference observed was much greater than that observed in the current work. However, because the ortholog and paralog sets compared would have had different divergence times, the observed difference in Ka/KS may be a significant overestimate. By studying a set of orthologous genes, we have attempted to compare only gene pairs that share a divergence time.
The observed increase may reflect both relaxation of selection pressure and selection for novel function following duplication in a subset of the genes and over some fraction of the time interval following duplication. The time since divergence of mouse and human is too great for a high probability of observation of positive selection in the orthologous data set. Few of the orthologous pairs show evidence of positive selection (five pairs of orthologous genes from a data set of 4218 pairs show Ka/KS > 1). Of the gene pairs for which Ka/KS > 1, four have very high values of both Ka and KS and may correspond to incorrectly assigned orthologous pairs and, in at least one case, a pseudogene. The remaining pair, a spliceosomal protein, has an atypically low value of KS. To observe positive selection following gene duplication, sets of orthologous genes from closely related organisms should be compared. Provided nearly complete genome sequence information exists for at least one of the organisms, it should be possible to detect differences in evolutionary rate between genes with and without paralogs. For this approach to be effective, the age of the duplications being studied must be similar to the time of speciation, so that the divergence of the orthologs has been affected significantly by the presence of the paralog. Because Ka/KS tends to decrease with time, the number of sites evolving under positive selection required for a high probability of Ka/KS > 1 increases with time since gene divergence. Observation of Ka/KS > 1 in a more divergent gene pair is likely to reflect stronger selection than in a less diverged pair.
Expression and Subfunctionalization
The rate of evolution in tissue-specific genes and ubiquitously expressed genes has previously been compared (Duret and Mouchiroud 2000; K. Crum and C. Seoighe, unpubl.). In a set of human and rodent genes, Duret and Mouchiroud (2000) found that genes that were expressed in a small number of tissues were evolving up to three times faster than were genes that were more ubiquitously expressed. Force et al. (1999) first put forward the suggestion that subfunctionalization may be an important mode of evolution following duplication. Following duplication, genes that previously performed two or more functions might lose one function and experience a narrowing of expression pattern. If this has taken place frequently in the set of human paralogs studied here, then the observed decrease in selective pressure following duplication may be related to a reduction in the number of tissues in which an individual gene from a duplicated gene pair is expressed. This hypothesis can be tested further by measuring the ubiquity of genes that have been duplicated.
Partial Gene Sequences and Incorrectly Identified Orthologs
The data set used here includes partial sequences of genes. Unlike in some previous work (for example, see Lynch and Conery 2000), the coding sequences used were not required to start with a methionine. All sequences for which at least 50 codons were available were used. The data set may also include some pseudogenes and hypothetical proteins for which experimental evidence was not available. The number of such sequences is not expected to be higher in the set of genes identified as having a paralog, and such genes should therefore not bias the results substantially. Furthermore, misassembles or misannotation in the ENSEMBL database could lead to spurious paralogs being identified. This can weaken the result but should not introduce a bias. The observed increase in the rate of evolution of the duplicated genes is thus a conservative estimate.
The reciprocal BLAST method that was used in the production of the HomoloGene database is not expected to yield perfect assignment of ortholog pairs. In fact, the accuracy with which orthology is determined may be reduced by the presence of paralogs, causing an apparent increase in the divergence of ortholog pairs for which paralogs are present. Although this can cause an apparent increase in divergence on a nucleotide or amino acid sequence level, it cannot account for the observed increase in the ratio of nonsynonymous to synonymous change.
Lengths of Duplicated Genes
The interesting observation that duplicated genes are shorter than average is likely to reflect a mutational phenomenon. The probability of a mutation resulting in the complete duplication of a gene is likely to be higher for shorter genes. If this relationship is caused by a mutational effect, as indicated here, then it would not be expected to hold in the case of genes duplicated during whole genome duplication. Interestingly, analysis of a data set of duplicated genes used to infer genome duplication in Saccharomyces cerevisiae (Wolfe and Shields 1997) shows no relationship between gene length and gene duplication (average length of 812 genes from duplicated pairs, 518 bp; overall average gene length, 488 bp). It has previously been suggested that the probability of intact transfer of genes may be dependent on gene size (Millen et al. 2001), and the same may hold true for the duplication of an intact gene. It may be possible to make use of the size distributions of duplicated genes to help to infer whether sets of duplicated genes, produced at different times, result from large-scale chromosomal or genome duplications or from individual gene duplications.
METHODS
Orthologous Sequences
The HomoloGene database (http://www.ncbi.nlm.nih.gov/HomoloGene/; Zhang et al. 2000) was downloaded from the National Center for Biotechnology Information on September 20, 2001. EMBL accessions for human and mouse orthologous pairs that were described as either reciprocal best hits or manually curated were extracted from the database. The HomoloGene database is based on similarity searches of UniGene clusters. The inclusion of manually curated ortholog pairs and reciprocal best matches introduces some redundancy into the data set. This redundancy was reduced by allowing each EMBL accession to occur just once.
Distance Calculations
Protein-coding nucleic acid sequences and protein sequences were extracted from the EMBL database (release 67, http://www.ebi.ac.uk/embl; Stoesser et al. 2002) using CODERET from the EMBOSS suite of programs (Rice et al. 2000). Gene pairs for which less than half of the human sequence could be aligned to the mouse sequence were omitted from the analysis. Only pairs of sequences containing >50 aligned amino acids were retained. If more than one protein was identified for an EMBL accession, the sequence of the first translation was selected from the EMBL entry. Protein sequences were aligned using ClustalW (Thompson et al. 1994) and default parameters. Protein and nucleotide distance matrices were calculated using ClustalW with the option for Kimura correction of multiple hits. The protein-coding sequences were aligned, using the protein alignment as a guide with the program align2aa.pl (http://sunflower.bio.indiana.edu/∼wfischer/PerlScripts). Evolutionary distance parameters were estimated using the Diverge program of GCG (http://www.accelrys.com/products/gcg_wisconsin_package/index.html), which is based on the method of Li (1997). Mean values of evolutionary distance were weighted by the number of sites analyzed in each pair-wise comparison.
Identification of Human Paralogs
Human protein-coding sequences from this data set were compared to confirmed cDNA sequences from the ENSEMBL database (version 1.0) using BLASTN (Altschul et al. 1997). Because it was assumed that the highest BLAST hit would normally correspond to the query sequence itself, the second highest hit corresponding to a different gene and separated physically from the highest hit by at least the length of the query sequence (using ENSEMBL genome co-ordinates) was extracted from the BLAST output file. The incompleteness of the ENSEMBL database and the strictness of the selection criteria is likely to result in some genes that do have paralogs not being identified. Missed paralogs can weaken, but should not bias, the results obtained. Protein coding sequences for ENSEMBL cDNA sequences were obtained by searching the cDNA with the corresponding ENSEMBL protein using computer scripts written in Perl. Only sequences >50 amino acids were retained.
Functional Classification
The GO database (Ashburner et al. 2000) was downloaded from www.geneontology.org on December 5, 2001. A list of GO identifiers was associated with each human EMBL entry by finding cross-references to protein databases in the EMBL database entries and using the GO gene associations database. Using the GO identifiers, a Perl script was written to associate as many highest-level GO terms as possible to each human gene from our data set. At least one highest-level GO term was associated with each of 3526 of the 5400 genes in the data set.
Chimpanzee Sequences
Chimpanzee sequences were extracted from the EMBL database (release 67), and a reciprocal BLASTN search was performed against the ENSEMBL database of human confirmed cDNA sequences. Reciprocal best hits were extracted from the BLAST output file and subjected to the same distance calculation pipeline described above. A list of the EMBL and ENSEMBL accession numbers of the chimpanzee-human gene pairs can be viewed in the supplementary information (http://www.sanbi.ac.za/∼cathal/supp.html).
Significance Testing
Significance testing was performed using a nonparametric bootstrap procedure. For example, to test whether a parameter a had a higher value in a subset, S, of some larger set L, 1000 replicates of S were generated randomly with replacement from the set L. The parameter was evaluated on each replicate, and the number of times the parameter was larger than the value on S was counted. If this number was <10, that is, <1% of replicates, the difference was considered significant.
WEB SITE REFERENCES
http://sunflower.bio.indiana.edu/∼wfischer/PerlScripts; program align2aa.pl.
http://www.accelrys.com/products/gcg_wisconsin_package/index.html; Diverge program of GCG.
http://www.ebi.ac.uk/embl; European Molecular Biology Laboratory database.
http://www.geneontology.org; Gene Ontology database.
http://www.ncbi.nlm.nih.gov/HomoloGene/; HomoloGene database.
http://www.sanbi.ac.za/∼cathal/supp.html; list of the EMBL and ENSEMBL accession numbers of the chimpanzee-human gene pairs.
Acknowledgments
We are grateful for very helpful comments on the manuscript from Ken Wolfe, Denis Shields, and Win Hide, as well as for the financial support to V.N. and J.K. from the South African National Research Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL cathal@sanbi.ac.za; FAX 27-21-959-2512.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.270902.
REFERENCES
- Ahn S, Tanksley SD. Comparative linkage maps of the rice and maize genomes. Proc Natl Acad Sci. 1993;90:7980–7984. doi: 10.1073/pnas.90.17.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology: The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averof M, Rokas A, Wolfe KH, Sharp PM. Evidence for a high frequency of simultaneous double-nucleotide substitutions. Science. 2000;287:1283–1286. doi: 10.1126/science.287.5456.1283. [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: Expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 2000;17:68–74. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- Ferris SD, Whitt GS. Evolution of the differential regulation of duplicate genes after polyploidization. J Mol Evol. 1979;12:267–317. doi: 10.1007/BF01732026. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Doebley JF. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci. 1997;94:6809–6814. doi: 10.1073/pnas.94.13.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Green JA, Garbayo JM, Roberts RM. Adaptive diversification within a large family of recently duplicated, placentally expressed genes. Proc Natl Acad Sci. 2000;97:3319–3323. doi: 10.1073/pnas.050002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MK, Hughes AL. Evolution of duplicate genes in a tetraploid animal, Xenopus laevis. Mol Biol Evol. 1993;10:1360–1369. doi: 10.1093/oxfordjournals.molbev.a040080. [DOI] [PubMed] [Google Scholar]
- Kondrashov, F.A., Rogozin, I.B., Wolf, Y.I., and Koonin, E.V. 2002. Selection in the evolution of gene duplications. Genome Biol. 3:RESEARCH0008. [DOI] [PMC free article] [PubMed]
- Li W-H. Molecular evolution. Sunderland, Massachusetts: Sinauer Associates Inc.; 1997. [Google Scholar]
- Long M, Thornton K. Gene duplication and evolution. Science. 2001;293:1551. doi: 10.1126/science.293.5535.1551a. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery J S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Lynch M, O'Hely M, Walsh B, Force A. The probability of preservation of a newly arisen gene duplicate. Genetics. 2001;159:1789–1804. doi: 10.1093/genetics/159.4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massingham T, Davies LJ, Lio P. Analyzing gene function after duplication. Bioessays. 2001;23:873–876. doi: 10.1002/bies.1128. [DOI] [PubMed] [Google Scholar]
- Millen RS, Olmstead RG, Adams KL, Palmer JD, Lao NT, Heggie L, Kavanagh TA, Hibberd JM, Gray JC, Morden CW, et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 2001;13:645–658. doi: 10.1105/tpc.13.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, Boerlijst MC, Cooke J, Smith JM. Evolution of genetic redundancy. Nature. 1997;388:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Laudet V. Evolutionary rates of duplicate genes in fish and mammals. Mol Biol Evol. 2001;18:681–683. doi: 10.1093/oxfordjournals.molbev.a003849. [DOI] [PubMed] [Google Scholar]
- Seoighe C, Wolfe KH. Extent of genomic rearrangement after genome duplication in yeast. Proc Natl Acad Sci. 1998;95:4447–4452. doi: 10.1073/pnas.95.8.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoesser G, Baker W, van den Broek A, Camon E, Garcia-Pastor M, Kanz C, Kulikova T, Leinonen R, Lin Q, Lombard V, et al. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 2002;30:21–26. doi: 10.1093/nar/30.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna, D.M. and Goldstein, R.M. 2000. The evolution of duplicated genes considering protein stability constraints. Pac. Symp. Biocomput. 69–80. [DOI] [PubMed]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. The fate of duplicated genes: loss or new function? Bioessays. 1998;20:785–788. doi: 10.1002/(SICI)1521-1878(199810)20:10<785::AID-BIES2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Wagner A. The role of population size, pleiotropy and fitness effects of mutations in the evolution of overlapping gene functions. Genetics. 2000;154:1389–1401. doi: 10.1093/genetics/154.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh JB. How often do duplicated genes evolve new functions? Genetics. 1995;139:421–428. doi: 10.1093/genetics/139.1.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH. Yesterday's polyploids and the mystery of diploidization. Nat Rev Genet. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Sharp PM. Mammalian gene evolution: Nucleotide sequence divergence between mouse and rat. J Mol Evol. 1993;37:441–456. doi: 10.1007/BF00178874. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Zhang J, Rosenberg HF, Nei M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Gaut B S, Vision TJ. Gene duplication and evolution. Science. 2001;293:1551. [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]