Abstract
Renal tumor classification is important because histopathological subtypes are associated with distinct clinical behavior. However, diagnosis is difficult because tumor subtypes have overlapping microscopic characteristics. Therefore, ancillary methods are needed to optimize classification. We used oligonucleotide microarrays to analyze 31 adult renal tumors, including clear cell renal cell carcinoma (RCC), papillary RCC, chromophobe RCC, oncocytoma, and angiomyolipoma. Expression profiles correlated with histopathology; unsupervised algorithms clustered 30 of 31 tumors according to appropriate diagnostic subtypes while supervised analyses identified significant, subtype-specific expression markers. Clear cell RCC overexpressed proximal nephron, angiogenic, and immune response genes, chromophobe RCC oncocytoma overexpressed distal nephron and oxidative phosphorylation genes, papillary RCC overexpressed serine protease inhibitors, and extracellular matrix products, and angiomyolipoma overexpressed muscle developmental, lipid biosynthetic, melanocytic, and distinct angiogenic factors. Quantitative reverse transcriptase-polymerase chain reaction and immunohistochemistry of formalin-fixed renal tumors confirmed overexpression of proximal nephron markers (megalin/low-density lipoprotein-related protein 2, α-methylacyl CoA racemase) in clear cell and papillary RCC and distal nephron markers (β-defensin 1, claudin 7) in chromophobe RCC/oncocytoma. In summary, renal tumor subtypes were classified by distinct gene expression profiles, illustrating tumor pathobiology and translating into novel molecular bioassays using fixed tissue.
Renal cell carcinoma (RCC) is the most common malignancy of the adult kidney, comprising 3% of all human cancers.1 Localized tumors can be detected by abdominal imaging and cured by surgery.2 However, 25 to 40% of cases present with extrarenal growth or metastases,3 and one-third of apparently localized lesions develop metastases during the postoperative course.4,5 Advanced RCC responds poorly to systemic therapy and has a 5-year survival rate of less than 10%.6,7
Important predictors of outcome for RCC include tumor stage, Fuhrman nuclear grade, histopathological classification, and perioperative thrombocytosis.3,8,9,10 The current renal tumor classification system is based on morphology, as well as underlying genetic differences.3,11,12 More than 90% of clinically significant lesions can be diagnosed as one of the common subtypes of renal epithelial tumor: clear cell (conventional) RCC, papillary RCC, chromophobe RCC, and renal oncocytoma. Mesenchymal renal tumors are more rare and include angiomyolipoma. Recent clinicopathological surveys have indicated that clear cell RCC has the highest rate of metastasis and poorest survival among common renal malignancies.3 Papillary and chromophobe carcinomas are relatively indolent but exhibit potential for metastasis and transformation to high-grade, sarcomatoid tumors. In contrast, typical oncocytomas and angiomyolipomas are considered benign neoplasms.
Renal tumor subtypes exhibit several common morphological characteristics, making diagnosis difficult and subjective in many cases. For instance, eosinophilic variants of clear cell and chromophobe RCC can closely resemble the benign oncocytoma histologically. Therefore, ancillary molecular methods are needed for optimal diagnosis and clinical management. In several types of human cancer, gene expression microarrays have proved to be effective tools for classifying tumors and identifying novel molecular biomarkers.13,14,15,16 Previously, we used microarrays to profile gene expression in clear cell RCC, papillary RCC, chromophobe RCC, and oncocytoma; based on distinct expression patterns, we developed a novel immunohistochemical panel for renal tumor subtyping.17,18 Other groups have confirmed these findings in larger studies.5,19,20,21 We extended our observations in the current experiments, using oligonucleotide microarrays to profile the expression of several thousand genes in a cohort of clear cell RCC, papillary RCC, chromophobe RCC, oncocytoma, and angiomyolipoma. We validated the microarray data for selected, differentially expressed gene products in an independent cohort of fixed tissues, using quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) and immunohistochemistry, and thereby confirmed these gene products as potential expression markers for renal tumor classification.
Materials and Methods
Experimental Specimens
Microarray experiments were performed on frozen specimens from 13 clear cell RCC, 5 papillary RCC, 4 chromophobe RCC, 3 oncocytoma, and 6 angiomyolipoma. Clinicopathological characteristics of this tumor cohort are described in Supplementary Table 1 at http://jmd.amjpathol.org. Quantitative RT-PCR experiments were performed on formalin-fixed paraffin-embedded tissue from an independent cohort of 10 clear cell RCC, 6 papillary RCC, 5 chromophobe RCC, and 7 renal oncocytoma. Immunohistochemistry was performed on a fixed tissue microarray that included 33 clear cell RCC, 19 papillary RCC, 6 chromophobe RCC, and 6 oncocytoma. The Emory University and Atlanta Veterans Affairs Medical Center Departments of Pathology and Laboratory Medicine diagnosed tumors using published criteria:3 clear cell RCC– neoplastic clear cells with an anastomosing vascular network; papillary RCC– circumscription with a fibrous capsule, papillary growth pattern, foam cells, and necrosis; chromophobe RCC– broad alveolar or nested growth pattern, neoplastic cells with irregular nuclei and perinuclear halos, clear flocculent or granular eosinophilic cytoplasm; renal oncocytoma– circumscription with a central scar, nested or tubulocystic growth pattern, and neoplastic oncocytes with small round nuclei and granular eosinophilic cytoplasm; angiomyolipoma– mesenchymal tumor containing mature adipose, spindle, or epithelioid smooth muscle cells and thick-walled blood vessels. Carcinoma grading and staging were based on the standard Fuhrman nuclear grading system and the Tumor-Node-Metastasis staging system (TNM, International Union Against Cancer), respectively.3,8,11 The Emory University Institutional Review Board approved this research under protocol 255-2002.
Microarray Hybridization
Frozen tumor specimens were homogenized in 10 vol of TRIzol (Invitrogen, Carlsbad, CA) per g of tissue. Total RNA was isolated using the standard TRIzol protocol and purified further with the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s recommendations. RNA was quantified and assessed for integrity using a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Expression profiles of all specimens were compared to a commercial universal reference RNA (Clontech, Palo Alto, CA). Probe synthesis and microarray hybridization were performed according to standard Affymetrix protocols, described in detail at the Internet site http://www.affymetrix.com/community/academic/grant.affx. Briefly, total RNA (5 μg per specimen) was reverse-transcribed into double-stranded cDNA, and biotin-labeled cRNA was produced by in vitro transcription. Labeled cRNA was fragmented and digested by DNase I before hybridization. Hybridization cocktails were prepared by combining fragmented targets, probe array controls, bovine serum albumin, and herring sperm DNA. Cocktails were applied to HG Focus oligonucleotide microarrays (Affymetrix, Santa Clara, CA) for 16 hours, followed by automated washing and staining on an Affymetrix workstation. After staining, microarrays were scanned and analyzed with Affymetrix Microarray Suite 5.0 software, to define probe cells, compute signal intensities in each cell, and calculate signal log2 expression ratios for each gene in tumor versus reference specimens. The HG Focus arrays produced data for 8746 genes. All hybridization experiments met the following quality control criteria: average background, <100 U; noise, <5 U; 3′:5′ ratio of control genes, <3; and RNA spikes present with appropriate signal intensities. Scaling factors and transcript presence rates varied less than 20% among tumor samples.
Data Analysis
Affymetrix data sets were normalized with a robust multiarray algorithm22 accounting for GC sequence information (GCRMA algorithm), encoded in GeneTraffic software (Iobion Informatics, La Jolla, CA). Expression profiles were filtered to exclude genes with fewer than two observations of absolute value log2 ratio >2, or with mean log2 ratio range (maximum − minimum) < 2. This procedure selected 4030 differentially expressed genes from the total of 8746. To compare global expression patterns among renal tumor subtypes, the filtered expression profiles were analyzed by unsupervised hierarchical average linkage clustering, using Pearson correlation as the similarity metric.23 To identify genes that correlated significantly with renal tumor subtypes, the unfiltered expression profiles were analyzed with a supervised significance analysis of microarrays (SAM) algorithm,24 using the following parameters: data type = two-class unpaired (ie, one tumor subtype versus all other tumors); imputer engine = 10-nearest neighbor; fold change cutoff = 2.0; permutation number = 500; random number generator seed = 1234567; and median false discovery rate <1%, corresponding to Δ = 1.12, 1.79, 1.44, and 1.53 for clear cell RCC, chromophobe RCC/renal oncocytoma, papillary RCC, and angiomyolipoma, respectively. To determine whether biological processes were highly represented in renal tumor expression profiles, the significant expression markers identified by SAM were analyzed with the GOstat program (http://gostat.wehi.edu.au/).25 This program compiled functional annotations from the Gene Ontology Consortium database26 that were associated with gene lists identified by SAM. Frequencies of specific gene ontology associations were compared in the differentially expressed gene lists versus the entire list of genes featured on HG Focus microarrays. The Fisher’s exact test identified gene ontology terms overrepresented among renal tumor differential expression profiles, using a Benjamini false discovery rate correction for multiple testing.
Quantitative RT-PCR
Experiments were performed according to published protocols with minor modifications.27 Briefly, histological sections were deparaffinized with ethanol and xylene, and cells of interest were microdissected with a sterile scalpel. Tissue was digested in buffer containing proteinase K at 55°C overnight, and total RNA was isolated by phenol/chloroform extraction. The sample was treated with DNase to minimize contamination with genomic DNA. Fluorogenic quantitative RT-PCR assays were performed in triplicate with standard SYBR Green methodology on the I-cycler system (Bio-Rad, Hercules, CA). Reaction specificity was assessed by melting point analyses, in which single melting point peaks were required at temperatures predicted by amplicon sequence. Reactions without reverse transcription and template served as controls for DNA contamination and specimen carry-over. The following test genes were analyzed: megalin/low-density lipoprotein-related protein 2 (LRP2: forward primer, 5′-gctgataaaacgagacgcacagta; reverse primer, 5′-aggacggaaccaatcagtgaag); β-defensin 1 (DEFB1: forward primer, 5′-tttactctctgcttacttttgtctgagatg; reverse primer, 5′-tgctgacgcaattgtaatgatca); and α-methylacyl CoA racemase (AMACR: forward primer, 5′-gggtcaggtcattgatgcaaa; reverse primer, 5′-ttcccacagactcaatttctgagtt). All primer pairs were intron-spanning, and were developed by analysis of intron/exon structures in the Ensembl sequence database (www.ensembl.org), followed by entry of appropriate sequences in Primer Express software (Applied Biosystems, Foster City, CA). Test gene expression was normalized to 28S ribosomal RNA and referenced to a standard RNA specimen. Relative normalized gene expression was compared in renal tumor subtypes, with statistical significance assessed by analysis of variance.
Immunohistochemistry
Tissue microarrays were constructed from fixed renal tumor core biopsies, 0.6 mm in diameter and 3 to 4 mm in height, which were placed in recipient paraffin blocks (45.0 × 20.0 mm) with a tissue arrayer (Beecher Instruments, Silver Spring, MD). Three to four tissue cores were arrayed per case. Tissue microarray sections (5 μm) were dewaxed, and steam antigen retrieval was performed at pH 6.5 in a pressure cooker for 20 minutes.28 Tissue sections were incubated with mouse monoclonal antibody against the claudin-7 gene product (CLDN7) for 25 minutes at room temperature (1:400 dilution; Zymed Corporation, South San Francisco, CA). After washing unbound antibody, sections were treated with goat anti-mouse immunoglobulin conjugated to a peroxidase-labeled polymer, according to the manufacturer’s instructions (Envision kit; DAKO Corp., Carpinteria, CA). Immunohistochemical reactions were developed with diaminobenzidine as the chromogenic peroxidase substrate. Sections were counterstained with hematoxylin after immunohistochemistry. Specificity was verified by negative control reactions without primary antibody, as well as appropriate membranous staining reactions in positive control colon tissues. A positive reaction was defined as membranous staining on ≥10% tumor cells in one or more tissue cores. Frequency of reactive cases was compared among renal tumor subtypes, using χ2 analysis to assess for statistical significance.
Results
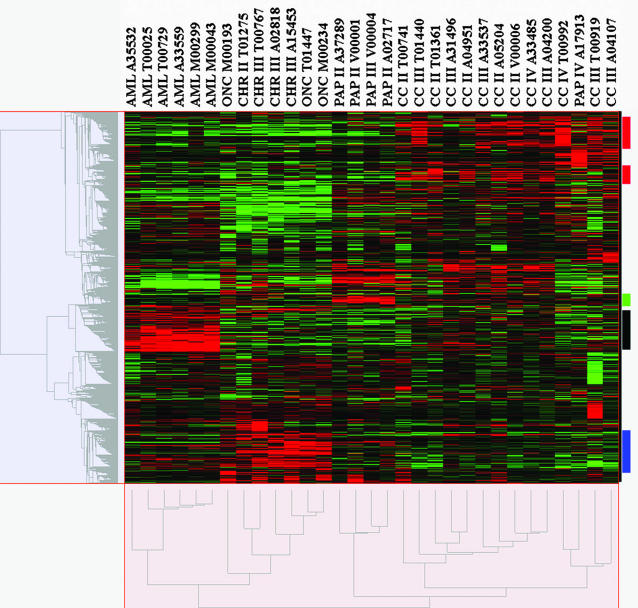
Expression patterns of 8746 genes were measured in 13 clear cell RCCs, 5 papillary RCCs, 4 chromophobe RCCs, 3 renal oncocytomas, and 6 angiomyolipomas using Affymetrix oligonucleotide microarrays. To characterize the unique expression profiles of renal tumor subtypes, unsupervised hierarchical average linkage clustering was used to group tumors and genes by similarity in expression profiles. The resulting molecular classification correlated strongly with histopathology; 30 of 31 tumors were clustered according to appropriate diagnostic subtypes (Figure 1, x axis dendrogram). The sole outlier was a high-grade papillary carcinoma with sarcomatoid transformation, which was clustered with clear cell RCC. The SAM procedure identified genes with the strongest correlation to specific tumor subtypes, and the GOstat program determined the statistical representation of specific gene ontology functional annotation terms in the gene lists identified by SAM. Clear cell RCC overexpressed 402 and underexpressed 220 genes, at a median false discovery rate of 0.99%. As shown in Table 1, clear cell tumors overexpressed a significant number of immune response genes (P = 1.0 × 10−36) and angiogenic factors (P = 8.5 × 10−3). Chromophobe RCC and oncocytoma overexpressed 510 and underexpressed 479 genes, at a median false discovery rate of 0.92%. As shown in Table 2, the overexpressed sequences included a significant number related to electron transport (P = 4.0 × 10−14), oxidative phosphorylation (P = 5.9 × 10−9), and energy pathways (P = 5.9 × 10−9), whereas the underexpressed sequences included I kappa B kinase/nuclear factor-κB signaling activators (P = 1.3 × 10−3). Papillary RCC significantly overexpressed 95 genes and underexpressed only 1 gene, at a median false discovery rate of 0.80%. Although no gene ontology terms were statistically overrepresented in the papillary RCC expression profile, the list of significantly overexpressed genes included several encoding serine-type endopeptidase inhibitors and extracellular matrix products (Table 3). Angiomyolipoma overexpressed 409 and underexpressed 108 genes, at a median false discovery rate of 0.76%. As shown in Table 4, genes related to muscle development (P = 3.6 × 10−4), lipid biosynthesis (P = 4.5 × 10−3), and pigmentation (P > 0.05) were overexpressed in angiomyolipoma. In addition, these tumors expressed vascular endothelial growth factors B and D (VEGFB, VEGFD) at high levels. The complete microarray data are submitted on the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). Detailed results of the SAM and GOstat analyses are presented in Supplementary Tables 2 and 3 at http://jmd.amjpathol.org.
Figure 1.
Unsupervised hierarchical clustering of 31 renal neoplasms. Similarity measurements are based on Pearson correlation. Median-centered differential gene expression is shown in the color-coded grid, with columns representing individual tumors and rows representing individual genes. Red, green, and black grid blocks indicate expression above, below, and at the median of all tumors, respectively. Based on profiles of 4030 differentially expressed genes, tumors were clustered into subgroups corresponding to clear cell RCC, papillary RCC, chromophobe RCC/renal oncocytoma, and angiomyolipoma. The sole outlier (PAP IV A17913) was a high-grade papillary RCC with sarcomatoid transformation, which was clustered with clear cell RCC. The same tumor classification was obtained when all 8746 genes on the microarray were analyzed (data not shown). Red, green, black, and blue bars to the right of the color-coded grid indicate genes overexpressed in clear cell RCC, papillary RCC, angiomyolipoma, and chromophobe RCC/oncocytoma, respectively. Tumor names above the grid are interpreted as follows: CC, clear cell RCC; PAP, papillary RCC; CHR, chromophobe RCC; ONC, renal oncocytoma; AML, angiomyolipoma. Roman numerals indicate Fuhrman nuclear grade. Identification codes with one letter followed by five digits are for internal tracking only. Of the carcinomas in this analysis, only one case (CC IV T00992) was documented to be metastatic.
Table 1.
Immune Response and Angiogenesis Genes Overexpressed in Clear Cell Renal Cell Carcinoma
| Symbol | UniGene | GenBank | Name |
|---|---|---|---|
| Gene ontology: immune response (GO:0006955; P = 1.0 × 10−56) | |||
| AOAH | Hs.82542 | NM_001637 | Acyloxyacyl hydrolase precursor |
| APOL3 | Hs.241535 | NM_014349 | Apolipoprotein L3 isoform 2 |
| ARHGDIB | Hs.83656 | NM_001175 | Rho GDP dissociation inhibitor (GDI) β |
| BST2 | Hs.118110 | NM_004335 | Bone marrow stromal cell antigen 2 |
| C1QA | Hs.9641 | NM_015991 | Complement component 1, q, α |
| C1QB | Hs.8986 | NM_000491 | Complement component 1, q, β |
| C1QR1 | Hs.97199 | NM_012072 | Complement component 1, q, receptor 1 |
| C1R | Hs.1279 | AL573058 | Complement component 1, r |
| C1S | Hs.434029 | M18767 | Complement component 1, s |
| C3 | Hs.284394 | NM_000064 | Complement component 3 precursor |
| CCL19 | Hs.50002 | U88321 | Small inducible cytokine A19 precursor |
| CCL2 | Hs.303649 | S69738 | Small inducible cytokine A2 precursor |
| CCL20 | Hs.75498 | NM_004591 | Chemokine (C-C motif) ligand 20 |
| CCL4 | Hs.75703 | NM_002984 | Chemokine (C-C motif) ligand 4 precursor |
| CCR1 | Hs.301921 | AI421071 | Chemokine (C-C motif) receptor 1 |
| CCR2 | Hs.395 | NM_000647 | Chemokine (C-C motif) receptor 2 isoform A |
| CCR5 | Hs.54443 | NM_000579 | Chemokine (C-C motif) receptor 5 |
| CCR7 | Hs.1652 | NM_001838 | Chemokine (C-C motif) receptor 7 precursor |
| CD163 | Hs.74076 | NM_004244 | CD163 antigen |
| CD2 | Hs.89476 | NM_001767 | CD2 antigen (p50), sheep red blood cell receptor |
| CD3D | Hs.95327 | NM_000732 | CD3D antigen, δ polypeptide (TiT3 complex) |
| CD53 | Hs.82212 | NM_000560 | CD53 antigen |
| CD74 | Hs.84298 | K01144 | Invariant γ chain |
| CD8A | Hs.85258 | AW006735 | CD8 antigen alpha polypeptide isoform 1 precursor |
| CSF1R | Hs.174142 | NM_005211 | Colony-stimulating factor 1 receptor precursor |
| CSF2RB | Hs.285401 | AV756141 | Colony-stimulating factor 2 receptor, β, low-affinity |
| CST7 | Hs.143212 | AF031824 | Cystatin F |
| CXCL10 | Hs.2248 | NM_001565 | Small inducible cytokine B10 precursor |
| CXCL12 | Hs.237356 | NM_000609 | Chemokine (C-X-C motif) ligand 12 |
| CXCL14 | Hs.24395 | NM_004887 | Small inducible cytokine B14 precursor |
| CXCL9 | Hs.77367 | NM_002416 | Small inducible cytokine B9 precursor |
| ENTPD1 | Hs.205353 | AV717590 | Ectonucleoside triphosphate diphosphohydrolase 1 |
| F8 | Hs.79345 | NM_000132 | Coagulation factor VIII isoform a precursor |
| FCER1G | Hs.433300 | NM_004106 | Fc fragment of IgE, high affinity I, receptor for, γ |
| FCGR2A | Hs.78864 | NM_021642 | Fc fragment of IgG, low affinity IIa, receptor for (CD32) |
| FCGR3A | Hs.176663 | J04162 | Fc fragment of IgG, low affinity IIIa, receptor for (CD16) |
| FOS | Hs.25647 | BC004490 | c-fos |
| FPR1 | Hs.753 | NM_002029 | Formyl peptide receptor 1 |
| G1P2 | Hs.432233 | NM_005101 | Interferon, α-inducible protein (clone IFI-15K) |
| GZMA | Hs.90708 | NM_006144 | Granzyme A precursor |
| HF1 | Hs.250651 | X04697 | H factor 1 (complement) |
| HLA-DMB | Hs.1162 | NM_002118 | Major histocompatibility complex, class II, DM β |
| HLA-DPB1 | Hs.814 | NM_002121 | Major histocompatibility complex, class II, DP β 1 |
| HLA-DQA1 | Hs.198253 | BG397856 | Major histocompatibility complex, class II, DQ α 1 |
| ICSBP1 | Hs.14453 | AI073984 | Interferon consensus sequence-binding protein 1 |
| IFITM1 | Hs.458414 | MGC27165 | Interferon-induced transmembrane protein 1 (9–27) |
| IGJ | Hs.76325 | AV733266 | Immunoglobulin J polypeptide |
| IGSF6 | Hs.135194 | NM_005849 | Immunoglobulin superfamily, member 6 |
| IL10RB | Hs.173936 | BC001903 | Interleukin 10 receptor, β precursor |
| IL1R2 | Hs.25333 | NM_004633 | Interleukin 1 receptor, type II precursor |
| IL2RB | Hs.75596 | NM_000878 | Interleukin 2 receptor β precursor |
| IL2RG | Hs.84 | NM_000206 | Interleukin 2 receptor, γ chain, precursor |
| IL6 | Hs.93913 | NM_000600 | Interleukin 6 (interferon, β 2) |
| IL7R | Hs.362807 | NM_002185 | Interleukin 7 receptor precursor |
| INHBB | Hs.1735 | NM_002193 | Inhibin β B subunit precursor |
| IRF1 | Hs.80645 | NM_002198 | Interferon regulatory factor 1 |
| IRF7 | Hs.166120 | NM_004030 | Interferon regulatory factor 7 isoform a |
| ITGB2 | Hs.83968 | NM_000211 | Integrin β chain, β 2 precursor |
| ITK | Hs.211576 | D13720 | IL2-inducible T-cell kinase |
| JAG2 | Hs.166154 | AF029778 | Jagged 2 isoform a precursor |
| LCP2 | Hs.2488 | AI123251 | Lymphocyte cytosolic protein 2 |
| LIF | Hs.2250 | NM_002309 | Leukemia inhibitory factor |
| LTB | Hs.890 | NM_002341 | Lymphotoxin-β isoform a |
| MNDA | Hs.153837 | NM_002432 | Myeloid cell nuclear differentiation antigen |
| MX1 | Hs.76391 | NM_002462 | Myxovirus resistance protein 1 |
| MX2 | Hs.926 | NM_002463 | Myxovirus resistance protein 2 |
| NK4 | Hs.943 | NM_004221 | Natural killer cell transcript 4 |
| PSMB10 | Hs.9661 | NM_002801 | Proteasome β 10 subunit proprotein |
| PSMB9 | Hs.381081 | NM_002800 | Proteasome β 9 subunit isoform 1 proprotein |
| TCIRG1 | Hs.46465 | NM_006019 | T cell, immune regulator 1, isoform a |
| TLR2 | Hs.63668 | NM_003264 | Toll-like receptor 2 |
| TLR3 | Hs.29499 | NM_003265 | Toll-like receptor 3 |
| TLR7 | Hs.179152 | NM_016562 | Toll-like receptor 7 |
| TNFSF7 | Hs.99899 | NM_001252 | Tumor necrosis factor ligand superfamily, member 7 |
| TYROBP | Hs.9963 | NM_003332 | TYRO protein tyrosine kinase binding protein |
| UBD | Hs.44532 | NM_006398 | Diubiquitin |
| Gene ontology: angiogenesis (GO:0001525; P = 8.5 × 10−3) | |||
| ANGPT2 | Hs.115181 | AF187858 | Angiopoietin 2 |
| ANGPTL4 | Hs.9613 | NM_016109 | Angiopoietin-like 4 protein |
| FLT1 | Hs.381093 | AA058828 | Vascular endothelial growth factor receptor 1 |
| KDR | Hs.12337 | NM_002253 | Vascular endothelial growth factor receptor 2 |
| VEGF | Hs.73793 | AF022375 | Vascular endothelial growth factor |
| VEGFC | Hs.79141 | U58111 | Vascular endothelial growth factor C |
The significance analysis of microarrays (SAM) identified genes overexpressed in clear cell RCC versus all other tumors. The GOstat program identified gene ontology functional annotation terms that were statistically overrepresented in the clear cell RCC expression profile.
Table 2.
Oxidative Phosphorylation and Energy Pathway Genes Overexpressed in Chromophobe Renal Cell Carcinoma and Renal Oncocytoma
| Symbol | UniGene | GenBank | Name |
|---|---|---|---|
| Gene ontology: electron transport (GO:0006118; P = 4.0 × 10−14) | |||
| ACAD8 | Hs.14791 | BC001964 | Acyl-coenzyme A dehydrogenase family, member 8 |
| ACADM | Hs.79158 | NM_000016 | Acyl-coenzyme A dehydrogenase, C-4 to C-12 straight chain |
| ACADSB | Hs.81934 | NM_001609 | Acyl-coenzyme A dehydrogenase, short |
| COVA1 | Hs.155185 | S72904 | Cytosolic ovarian carcinoma antigen 1 |
| COX5A | Hs.323834 | NM_004255 | Cytochrome c oxidase subunit Va precursor |
| COX5B | Hs.1342 | AI557312 | Cytochrome c oxidase subunit Vb precursor |
| COX7A2L | Hs.30888 | NM_004718 | Cytochrome c oxidase subunit VIIa polypeptide 2 like |
| COX7B | Hs.432170 | NM_001866 | Cytochrome c oxidase subunit VIIb precursor |
| COX8A | Hs.433901 | NM_004074 | Cytochrome c oxidase subunit VIII |
| CYC1 | Hs.289271 | NM_001916 | Cytochrome c-1 |
| CYP11B2 | Hs.184927 | X54741 | Cytochrome P450, subfamily XIB polypeptide 2 precursor |
| CYP26A1 | Hs.150595 | NM_000783 | Cytochrome P450, family 26, subfamily A, polypeptide 1 isoform 1 |
| CYP2D6 | Hs.333497 | NM_000106 | Cytochrome P450, subfamily IID, polypeptide 6 |
| DLD | Hs.74635 | J03620 | Dihydrolipoamide dehydrogenase precursor |
| ETFDH | Hs.323468 | NM_004453 | Electron-transferring-flavoprotein dehydrogenase |
| FMO4 | Hs.2664 | NM_002022 | Flavin-containing monooxygenase 4 |
| GSR | Hs.193974 | NM_000637 | Glutathione reductase |
| HCCS | Hs.211571 | AI801013 | Holocytochrome c synthase (cytochrome c heme-lyase) |
| HSPC051 | Hs.284292 | NM_013387 | Ubiquinol-cytochrome c reductase complex 7.2 kd |
| IVD | Hs.374536 | AK022777 | Isovaleryl coenzyme A dehydrogenase |
| MRPS30 | Hs.28555 | NM_016640 | Mitochondrial ribosomal protein S30 |
| NDUFB2 | Hs.198272 | NM_004546 | NADH dehydrogenase (ubiquinone) 1β subcomplex, 2, 8 kd |
| NDUFB5 | Hs.19236 | NM_002492 | NADH dehydrogenase (ubiquinone) 1β subcomplex, 5, 16 kd |
| NDUFS1 | Hs.8248 | NM_005006 | NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75 kd |
| NDUFS4 | Hs.10758 | BC005270 | NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18 kd |
| NDUFV2 | Hs.51299 | NM_021074 | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24 kd |
| NOX5 | Hs.160199 | NM_024505 | NADPH oxidase, EF hand calcium-binding domain 5 |
| OSBP2 | Hs.7740 | NM_030758 | Oxysterol binding protein 2 |
| PDCD8 | Hs.18720 | NM_004208 | Programmed cell death 8 isoform 1 |
| QF-C | Hs.3709 | NM_014402 | Low molecular mass ubiquinone-binding protein |
| SDHB | Hs.64 | NM_003000 | Succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| SDHD | Hs.168289 | NM_003002 | Succinate dehydrogenase complex, subunit D precursor |
| UQCRC1 | Hs.119251 | NM_003365 | Ubiquinol-cytochrome c reductase core protein I |
| UQCRFS1 | Hs.3712 | BC000649 | Ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| Gene ontology: oxidative phosphorylation (GO:0006119; P = 5.9 × 10−9) | |||
| ATP5B | Hs.406510 | NM_001686 | ATP synthase, H+ transporting, mitochondrial F1 complex, β |
| ATP5G1 | Hs.80986 | AL080089 | ATP synthase, H+ transporting, mitochondrial F0, subunit c, isoform 1 |
| ATP5G3 | Hs.429 | NM_001689 | ATP synthase, H+ transporting, mitochondrial F0, subunit c, isoform 3 |
| ATP5O | Hs.433960 | NM_001697 | ATP synthase, H+ transporting, mitochondrial F1, O subunit |
| ATP6V0B | Hs.7476 | BC005876 | ATPase, H+ transporting, lysosomal 21kD, V0 subunit c″ |
| ATP6V1A1 | Hs.281866 | AF113129 | ATPase, H+ transporting, lysosomal 70 kd, V1 subunit A, isoform 1 |
| ATP6V1B1 | Hs.64173 | NM_001692 | ATPase, H+ transporting, lysosomal 56 |
| ATP6V1C1 | Hs.86905 | NM_001695 | ATPase, H+ transporting, lysosomal 42 kd, V1 subunit C, isoform 1 |
| ATP6V1D | Hs.272630 | AF077614 | ATPase, H+ transporting, lysosomal 34 kd, V1 subunit D |
| ATP6V1E1 | Hs.77805 | BC004443 | ATPase, H+ transporting, lysosomal 31 kd, V1 subunit E isoform 1 |
| Gene ontology: energy pathways (GO:0006091; P = 5.9 × 10−9) | |||
| ACO2 | Hs.300463 | NM_001098 | Aconitase 2 |
| BPGM | Hs.198365 | NM_001724 | 2,3-Bisphosphoglycerate mutase |
| CKMT2 | Hs.80691 | NM_001825 | Sarcomeric mitochondrial creatine kinase precursor |
| ENO3 | Hs.118804 | NM_001976 | Enolase 3 |
| GCGR | Hs.208 | U03469 | Glucagon receptor |
| IDH3A | Hs.250616 | AI826060 | Isocitrate dehydrogenase 3 (NAD+) α precursor |
| NDUFA10 | Hs.198271 | NM_004544 | NADH dehydrogenase (ubiquinone) 1α subcomplex, 10, 42kDa |
| OGDH | Hs.168669 | NM_002541 | Oxoglutarate (α-ketoglutarate) dehydrogenase (lipoamide) |
| OXCT1 | Hs.177584 | NM_000436 | 3-Oxoacid CoA transferase precursor |
| PDHA1 | Hs.1023 | NM_000284 | Pyruvate dehydrogenase (lipoamide) α1 |
| PDHB | Hs.979 | AL117618 | Pyruvate dehydrogenase (lipoamide) β |
| PFKM | Hs.75160 | U24183 | Phosphofructokinase, muscle |
| PHKA1 | Hs.2393 | NM_002637 | Phosphorylase kinase, α1 (muscle) |
| PPARA | Hs.998 | BC000052 | Peroxisome proliferative activated receptor, α |
| SLC25A4 | Hs.2043 | NM_001151 | Solute carrier family 25, member 4 |
| SUCLA2 | Hs.182217 | NM_003850 | Succinate-CoA ligase, ADP-forming, β subunit |
| SUCLG1 | Hs.7043 | NM_003849 | Succinate-CoA ligase, GDP-forming, α subunit |
The significance analysis of microarrays (SAM) identified genes overexpressed in chromophobe RCC/oncocytoma versus all other tumors. The GOstat program identified gene ontology functional annotation terms that were statistically overrepresented in the chromophobe RCC/oncocytoma expression profile.
Table 3.
Protease Inhibitor and Extracellular Matrix Genes Overexpressed in Papillary Renal Cell Carcinoma
| Symbol | UniGene | GenBank | Name |
|---|---|---|---|
| Gene ontology: enzyme inhibitor activity (GO:0004857; P > 0.05) | |||
| ANXA3 | Hs.1378 | M63310 | Annexin A3 |
| GNAI1 | Hs.203862 | AL049933 | G protein, α inhibiting activity polypeptide 1 |
| SERPINE2 | Hs.21858 | AL541302 | Plasminogen activator inhibitor type 1, member 2 |
| SLPI | Hs.251754 | NM_003064 | Secretory leukocyte protease inhibitor precursor |
| TFPI | Hs.170279 | J03225 | Tissue factor pathway inhibitor 1 |
| TFPI2 | Hs.295944 | AL574096 | Tissue factor pathway inhibitor 2 |
| Gene ontology: extracellular matrix (GO:0006119; P > 0.05) | |||
| C6 | Hs.1282 | J05064 | Complement component 6 precursor |
| FLRT3 | Hs.41296 | NM_013281 | Fibronectin leucine rich transmembrane protein 3 |
| GLRB | Hs.32973 | AF094754 | Glycine receptor β |
| LAMB1 | Hs.82124 | NM_002291 | Laminin, β1 precursor |
| LAMC2 | Hs.54451 | NM_005562 | Laminin, γ2 isoform a precursor |
| MMP15 | Hs.80343 | NM_002428 | Matrix metalloproteinase 15 preproprotein |
The significance analysis of microarrays (SAM) identified genes overexpressed in papillary RCC versus all other tumors. The GOstat program did not identify gene ontology functional annotation terms that were statistically overrepresented in the papillary RCC expression profile.
Table 4.
Muscle Development, Lipid Biosynthesis, and Pigmentation Genes Overexpressed in Angiomyolipoma
| Symbol | UniGene | GenBank | Name |
|---|---|---|---|
| Gene ontology: muscle development (GO:00075177; P = 3.6 × 10−4) | |||
| ACTA1 | Hs.1288 | NM_001100 | α1 actin precursor |
| ACTA2 | Hs.195851 | NM_001613 | α2 actin |
| ACTG2 | Hs.378774 | NM_001615 | Actin, γ2 propeptide |
| AEBP1 | Hs.439463 | NM_001129 | Adipocyte enhancer binding protein 1 precursor |
| APEG1 | Hs.21639 | NM_005876 | Aortic preferentially expressed gene 1 |
| CALD1 | Hs.325474 | AL577531 | Caldesmon 1 isoform 3 |
| COL6A3 | Hs.80988 | NM_004369 | α3 type VI collagen isoform 1 precursor |
| DMD | Hs.169470 | NM_004010 | Dystrophin Dp427c isoform |
| GATA6 | Hs.50924 | D87811 | GATA binding protein 6 |
| HDAC9 | Hs.116753 | NM_014707 | Histone deacetylase 9 isoform 3 |
| ITGB1BP2 | Hs.109999 | NM_012278 | Melusin |
| LAMA2 | Hs.75279 | NM_000426 | Laminin α2 subunit precursor |
| MYH11 | Hs.78344 | AI889739 | Smooth muscle myosin heavy chain 11 isoform SM1 |
| MYL9 | Hs.9615 | NM_006097 | Myosin regulatory light polypeptide 9 isoform a |
| TAGLN | Hs.433399 | NM_003186 | Transgelin |
| TPM1 | Hs.77899 | Z24727 | Tropomyosin 1 α |
| TPM2 | Hs.300772 | NM_003289 | Tropomyosin 2 β |
| TPM4 | Hs.250641 | AI214061 | Tropomyosin 4 |
| Gene ontology: lipid biosynthesis (GO:0008610; P = 4.5 × 10−3) | |||
| ACACA | Hs.7232 | BE855983 | Acetyl-coenzyme A carboxylase α |
| CYP51A1 | Hs.226213 | NM_000786 | Cytochrome P450, family 51 |
| FADS1 | Hs.132898 | AL512760 | Fatty acid desaturase 1 |
| FADS2 | Hs.184641 | NM_004265 | Fatty acid desaturase 2 |
| FDFT1 | Hs.48876 | BC003573 | Farnesyl-diphosphate farnesyltransferase 1 |
| ISYNA1 | Hs.405873 | AL137749 | Myo-inositol 1-phosphate synthase A1 |
| LSS | Hs.93199 | AW084510 | Lanosterol synthase |
| SR-BP1 | Hs.24447 | NM_005866 | Type I σ receptor isoform 1 |
| PBX1 | Hs.155691 | NM_002585 | Pre-B-cell leukemia transcription factor 1 |
| PC | Hs.89890 | NM_022172 | Pyruvate carboxylase precursor |
| PTGDS | Hs.430637 | NM_000954 | Prostaglandin D2 synthase 21 kd (brain) |
| RODH | Hs.11958 | U89281 | 3-Hydroxysteroid epimerase |
| SC4MOL | Hs.239926 | AV704962 | Sterol-C4-methyl oxidase-like |
| SC5DL | Hs.288031 | D85181 | Sterol-C5-desaturase-like |
| SCD | Hs.119597 | AB032261 | Stearoyl-CoA desaturase (δ-9-desaturase) |
| SIAT8A | Hs.82527 | L32867 | Sialyltransferase 8A |
| Gene ontology: pigmentation (GO:0048066; P > 0.05) | |||
| OA1 | Hs.74124 | NM_000273 | Ocular albinism 1 (Nettleship-Falls) protein |
| SILV | Hs.95972 | U01874 | Silver homolog |
| TYRP1 | Hs.75219 | NM_000550 | Tyrosinase-related protein 1 |
| UROD | Hs.78601 | M14016 | Uroporphyrinogen decarboxylase |
The significance analysis of microarrays (SAM) identified genes overexpressed in angiomyolipoma versus all other tumors. The GOstat program identified gene ontology functional annotation terms that were statistically overrepresented in the angiomyolipoma expression profile.
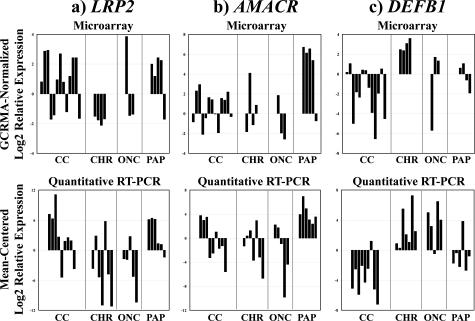
By microarray, clear cell and papillary RCC overexpressed markers of proximal nephron epithelium, such as cubilin (CBLN) and megalin/low-density lipoprotein-related protein 2 (LRP2). Papillary RCC specifically overexpressed the proximal nephron marker AMACR. In contrast, chromophobe RCC and oncocytoma overexpressed markers of distal nephron epithelium, such as parvalbumin (PVALB), chloride channels Ka and Kb (CLCNKA, CLCNKB), and DEFB1 (see complete microarray data). To validate these findings, quantitative RT-PCR was performed in an independent cohort of fixed renal tumors to measure mRNA expression of LRP2, AMACR, and DEFB1 (Figure 2). Consistent with the microarray data, DEFB1 was overexpressed in chromophobe RCCs and renal oncocytomas versus clear cell and papillary RCCs (P = 0.024), and AMACR was overexpressed in papillary RCC versus all other tumors (P = 0.0047). LRP2 was expressed at higher levels in clear cell and papillary RCCs versus chromophobe RCC and oncocytoma, although the difference did not reach statistical significance (P = 0.30).
Figure 2.
Differential expression of proximal and distal nephron markers in renal epithelial neoplasms: validation of microarray data by quantitative RT-PCR. Graphs at the top of the figure show normalized log2 gene expression ratios in tumor RNA relative to reference RNA, determined by oligonucleotide microarray. Graphs at the bottom of the figure show mean-centered log2 gene expression ratios in tumor RNA relative to reference RNA, determined by quantitative RT-PCR. In all graphs, bars indicate relative gene expression in individual tumors. Tumor subtypes are indicated below the x axes: CC, clear cell RCC; PAP, papillary RCC; CHR, chromophobe RCC; ONC, renal oncocytoma. a: Megalin/low-density lipoprotein-related protein 2 (LRP2, left). The proximal nephron marker LRP2 was typically expressed at high levels in clear cell and papillary RCC, although the confirmatory data did not reach statistical significance (P = 0.30 by quantitative RT-PCR). b: α-Methylacyl CoA racemase (AMACR, center). The proximal nephron marker AMACR was overexpressed significantly in papillary RCC (P = 0.0047 by quantitative RT-PCR). c: β-Defensin 1 (DEFB1, right). The distal nephron marker DEFB1 was overexpressed significantly in chromophobe RCC and oncocytoma (P = 0.024 by quantitative RT-PCR).
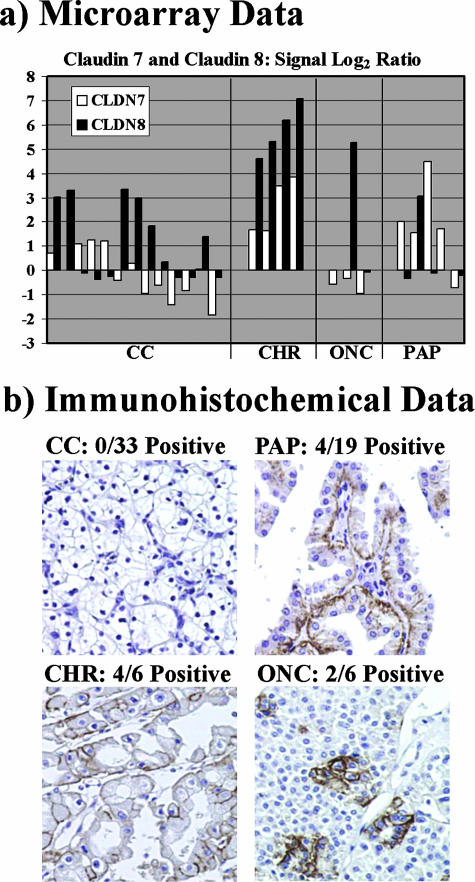
Unsupervised clustering of the microarray data did not resolve chromophobe RCC from oncocytoma, indicating similar overall expression patterns. In addition, the SAM algorithm did not identify any individual genes that were expressed differentially between the two subtypes. Nevertheless, within limits set by the microarray study size, a small number of gene products showed a trend toward differential expression between chromophobe RCC and oncocytoma. For example, claudins 7 and 8 (CLDN7, CLDN8) were relatively overexpressed in chromophobe RCC (Figure 3a). To test the validity of this finding, immunohistochemistry for CLDN7 was performed on a renal epithelial tumor tissue microarray (Figure 3b). Strong membranous staining was produced in the tumor cells of 4 of 6 chromophobe RCCs versus 2 of 6 oncocytomas. In papillary RCC, membranous staining of tumor cells was obtained in 4 of 19 cases, although the predominant staining pattern was cytoplasmic in stromal cells. In contrast, 0 of 33 clear cell RCC cases were positive for CLDN7 gene product in tumor or stromal cells (P ≤ 0.001, chromophobe RCC versus all other tumors).
Figure 3.
Expression of distal nephron claudins in renal epithelial neoplasms. a: Microarray data. Graph shows normalized log2 gene expression ratios in tumor RNA relative to reference RNA. White bars, claudin 7 (CLDN7); black bars, claudin 8 (CLDN8). Bars indicate relative gene expression in individual tumors. Tumor subtypes are indicated below the x axis: CC, clear cell RCC; PAP, papillary RCC; CHR, chromophobe RCC; ONC, renal oncocytoma. CLDN7 and CLDN8 were overexpressed in chromophobe RCC relative to oncocytoma. b: Immunohistochemical data. CLDN7 gene product was detected by immunoperoxidase reactions, using diaminobenzidine (brown) as the chromogenic peroxidase substrate and hematoxylin (blue) as the nuclear counterstain. Representative images are shown for each renal tumor subtype, with tumor subtype and frequency of positive reactions indicated above each panel. CLDN7 gene product was detected in 4 of 6 chromophobe RCCs, 2 of 6 oncocytomas, 4 of 19 papillary RCCs, and 0 of 33 clear cell RCCs. In chromophobe RCC and oncocytoma, the immunohistochemical staining pattern was membranous in tumor cells. In papillary RCC, the predominant staining pattern was cytoplasmic in stromal cells. Original magnifications, ×100.
Discussion
Until quite recently, the classification of adult renal tumors was limited to two major subtypes: clear cell and granular cell. Subsequent morphological and cytogenetic studies led to the recognition of several distinct renal tumor subtypes, culminating in the contemporary World Health Organization classification published in 2004.29 The clinical relevance of this classification system is underscored by distinct prognoses associated with different renal tumor subtypes,3,8,30 a finding that has prompted several proposals to account for histological subtype in the design of new therapies and clinical trials.31,32,33 However, new approaches are needed for diagnosis and clinical management of renal tumors. Light microscopy, the standard method for diagnostic classification, is difficult because renal tumor subtypes share common histopathological features.3,8,11 This problem is likely to grow with the advent of diagnostic procedures that result in small, distorted renal tumor biopsies.34 Gene expression profiling is a promising approach to address this problem, because expression microarrays can resolve certain tumors into diagnostic, prognostic, and therapeutic subclasses that are difficult to distinguish by light microscopy.13,14,15,16 The current experiments, as well as previous studies,5,12,17,18,19,20,21 have confirmed that renal tumor subtypes can be classified diagnostically using microarrays, on the basis of distinct and reproducible gene expression profiles. Of 31 cases analyzed in the current study, the sole outlier was a papillary RCC with extensive sarcomatoid transformation, which was most similar to clear cell RCC in terms of gene expression. Extensive review of this case by light microscopy revealed no clear cell histology, leading us to suspect that misclassification occurred due to gene expression from the spindle cell component. Alternatively, the tumor may have been a transformed, high-grade clear cell RCC with sarcomatoid and papillary features, containing scant residual clear cell histology, which was misdiagnosed by light microscopy.
The current study represents a progression of our previous microarray analysis of renal tumors,17 with several important enhancements. For example, the current study characterized gene expression in a larger number of tumors (31 versus 7 cases), and included samples of papillary RCC and angiomyolipoma, in addition to clear cell RCC, chromophobe RCC, and oncocytoma. Furthermore, the current experiments, which used oligonucleotide microarrays, produced expression data from a greater number of genes than our previous assays, which used cDNA microarrays. These differences in study design could explain why the current experiments identified many more candidate expression markers for renal tumor subtypes. Nevertheless, results of the two studies were consistent, in that the current analysis confirmed many specific markers for clear cell RCC and chromophobe RCC/oncocytoma identified in our previous work.
In the current study, clear cell RCC overexpressed several genes encoding proximal nephron markers, including CBLN and LRP2 (functional partners in receptor-mediated endocytosis),35,36 consistent with histogenetic models that relate this tumor subtype to proximal nephron epithelium.37 Clear cell RCC also overexpressed angiogenic factors and receptors, consistent with the anastomosing vascular network that makes this tumor a promising target for anti-angiogenic therapies.6 Angiogenesis in clear cell RCC is due primarily to loss-of-function mutation of the von Hippel-Lindau (vHL) gene on chromosome 3p25, the most common genetic defect in both hereditary and sporadic lesions.38 In our data, vHL gene expression did not vary significantly among clear cell tumors or between renal tumor subtypes. Although cytogenetic studies were not available to correlate with the expression data, a lack of significant variation in vHL expression has been noted in previous microarray studies of renal tumors.20 Clear cell tumors overexpressed a variety of immune response genes, including several members of the class II MHC. Class II MHC expression has been related to lymphocyte infiltration in RCC,39 possibly indicative of anti-tumor immunity, and clinical studies have shown that immunomodulatory therapies are more effective against advanced clear cell RCC than cases with nonclear cell histology.33 Thus, expression profiles of immune response genes may be therapeutically significant in the characterization of clear cell renal tumors.
Gene expression was remarkably similar in chromophobe RCC and renal oncocytoma, consistent with other microarray studies.17,20,21 These tumor subtypes share several clinical, morphological, and molecular features,40,41,42,43 and both arise frequently in Birt-Hogg-Dubé syndrome, an autosomal dominant, multiorgan system tumor syndrome mapped to chromosome 17p12-q11.2 (other renal tumor subtypes, including clear cell RCC, arise less frequently in this syndrome).44,45 The Birt-Hogg-Dubé gene, termed folliculin (FLCN), is of unknown function and was not probed by the microarrays used in our study, precluding interpretation of our data in context of this sequence. However, recent studies have shown that the Birt-Hogg-Dubé locus may be inactivated, either by loss of heterozygosity or promoter hypermethylation, in sporadic renal tumors of all histological subtypes.46 Chromophobe RCC and oncocytoma are related to intercalated cells of the cortical collecting duct.47,48 Correspondingly, these tumors overexpressed genes for distal nephron markers, such as DEFB1 (a small cationic antimicrobial peptide),49 PVALB (a calcium-binding protein),50 and others (see complete microarray data). Overexpression of DEFB1 was confirmed by quantitative RT-PCR. These findings were consistent with previous immunohistochemical studies.18,51 Chromophobe RCC and oncocytoma also overexpressed genes related to energy pathways, electron transport, and oxidative phosphorylation, which may reflect the characteristically abundant mitochondria in their neoplastic cells.42 Previous research has correlated high content of oxidative phosphorylation complexes in oncocytoma with a slow-growing, noninvasive phenotype, in comparison to clear cell RCC.52 Chromophobe RCC and oncocytoma underexpressed activators of the nuclear factor-κB signaling cascade and genes related to apoptosis. In several types of cancer, nuclear factor-κB signal transduction has been shown to promote oncogenesis by inhibiting cell death and activating cell proliferation and angiogenesis.53 However, the role of this signaling pathway in renal tumor development has not been studied in detail.
Notwithstanding their many similarities, chromophobe RCC and renal oncocytoma are distinct in their potential for malignant behavior, making accurate classification one of the most important, and difficult, diagnoses in renal tumor pathology. Immunohistochemical or other expression markers would be particularly useful in this setting, but none have attained widespread acceptance.54 Our microarray experiments identified the CLDN7 and CLDN8 gene products (tight junction proteins expressed normally in distal nephron epithelium)55 as candidate expression markers for chromophobe RCC. Immunohistochemistry suggested that CLDN7 was expressed at the protein level in chromophobe RCC, and to a lesser degree in oncocytoma, but not in clear cell RCC, consistent with the mRNA data, as well as the aforementioned histogenetic models for these tumors. Larger studies are needed to validate the utility of CLDN7 for discriminating chromophobe RCC from oncocytoma. Recently, an immunohistochemical study showed that the RON oncogene product (macrophage-stimulating protein receptor) was overexpressed specifically in oncocytoma,56 although this finding was not repeated in an independent study.57 If confirmed in future studies, RON and the distal nephron claudins could be clinically useful as an immunohistochemical panel with differential reactivity toward chromophobe RCC and oncocytoma.
Papillary RCC expressed several proximal nephron markers, which could indicate a relationship between this tumor and proximal nephron epithelium. In particular, papillary carcinoma strongly overexpressed the proximal nephron marker AMACR,58 consistent with previous microarray experiments.20,21 The protein product of this gene can be probed with well-established immunohistochemical assays, emphasizing its potential diagnostic utility.59 In this study, we developed a novel quantitative RT-PCR assay for AMACR applicable to formalin-fixed tissues, which might complement immunohistochemistry in clinical diagnosis. Papillary carcinomas also overexpressed genes encoding serine protease inhibitors and extracellular matrix proteins. Although this expression profile has not been described before in papillary RCC, genes with this activity are known to be important factors in tumor growth and invasion, and specific gene products have been identified as potential targets of cancer therapy.60,61 The rare, autosomal dominant familial papillary RCC syndrome is linked to oncogenic mutations of the c-MET gene on chromosome 7q31-34.62 Mutations of c-MET are uncommon in sporadic tumors, although protein is detected in most cases by immunohistochemistry.63,64 In our study, c-MET mRNA was overexpressed in several papillary carcinomas, although the difference between papillary RCC and other renal tumor subtypes did not attain statistical significance using the SAM algorithm, due to the limited number of tumors and variability in expression among cases.
This study includes one of the largest microarray analyses of renal angiomyolipoma yet performed. Angiomyolipoma is a mesenchymal neoplasm caused by proliferation of perivascular epithelioid cells. Most cases are composed of variable amounts of mature adipose, smooth muscle, and atypical thick-walled blood vessels.65 Angiomyolipoma is associated with the autosomal dominant genetic disorder tuberous sclerosis, caused by mutations in the TSC1 or TSC2 tumor suppressors on chromosomes 9q34 and 16p13.66,67,68 In our study, TSC2 was underexpressed consistently in angiomyolipoma, although the difference did not reach statistical significance using the SAM algorithm. The TSC1 and TSC2 gene products form a heterodimer that antagonizes cell growth and angiogenesis.69,70 Therefore, our finding of TSC2 underexpression, as well as significant overexpression of vascular endothelial growth factors B and D (VEGFB, VEGFD), could be related to the distinctive angiogenesis of angiomyolipoma. Atypical angiogenesis is characteristic of other tuberous sclerosis-related neoplasms,71 and future studies are indicated to determine whether this vascularity is related to differential expression of TSC1/2, VEGFB, or VEGFD. In addition to angiogenic factors, angiomyolipoma overexpressed myoid, adipose, and melanocytic markers, consistent with immunohistochemical profiles of this tumor.65,72 These results could suggest that the grossly dissected specimens used in our study contained a typical range of vascular, myoid, and adipose histology. Laser capture microdissection would be useful to isolate areas with these histological features before microarray hybridization, to determine whether gene expression profiles in angiomyo-lipoma vary with histological pattern.
The RCC cases in our study were not associated with long-term clinical follow-up, preventing discovery of expression profiles that correlate with outcome. However, Takahashi and colleagues73 used high-density cDNA microarrays to define a gene expression profile that predicted cause-specific survival, independent of histological grade and pathological stage, in a well-characterized cohort of clear cell RCC. Similarly, Vasselli and colleagues74 analyzed the primary tumors from a series of metastatic RCC, and identified several expression markers that correlated with survival. The oligonucleotide arrays in our study contained probes for 49 of the prognostic markers described in these two reports. Based on the expression patterns of these genes, clear cell tumors in our study could be clustered into two major categories, one of which consisted entirely of high-grade lesions (Fuhrman grade III/IV; data not shown). Additional clinical follow-up will be necessary to determine whether this classification defines cases with distinct clinical outcomes.
In summary, histopathological subtypes of renal neoplasms expressed distinct, biologically relevant molecular signatures. For example, clear cell RCC was revealed as an immunogenic and angiogenic tumor related to proximal nephron epithelium. Chromophobe RCC and oncocytoma appeared to be closely related neoplasms, overexpressing distal nephron markers and energy pathway genes, and underexpressing IκB kinase/nuclear factor-κB regulators and cell death genes. Papillary RCC expressed a distinct molecular signature, including serine protease inhibitors, extracellular matrix products, and proximal nephron markers such as AMACR. Angiomyolipoma was characterized as a mesenchymal tumor with adipose, smooth muscle, vascular, and melanocytic features. Additional clinical or pathological properties may be revealed by further analysis of the microarray data and the case cohort. Consistent with our previous research, microarray data could be translated into specific quantitative RT-PCR and immunohistochemical assays using formalin-fixed paraffin-embedded tissues, which may be applicable in clinical settings for diagnosis and clinical management of renal tumors.
Footnotes
Supported by a Veterans Affairs merit review grant (to A.N.Y.).
A.N.S. and Q.Y.-G. contributed equally to this study.
Supplementary material for this article can be found at http://jmd.amjpathol.org.
References
- Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, Ward E, Feuer EJ, Thun MJ. Cancer Statistics, 2004. CA Cancer J Clin. 2004;54:8–29. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- Homma Y, Kawabe K, Kitamura T, Nishimura Y, Shinohara M, Kondo Y, Saito I, Minowada S, Asakage Y. Increased incidental detection and reduced mortality in renal cancer—recent retrospective analysis at eight institutions. Int J Urol. 1995;2:77–80. doi: 10.1111/j.1442-2042.1995.tb00428.x. [DOI] [PubMed] [Google Scholar]
- Amin MaB, Amin MiB, Tamboli P, Javidan J, Stricker H, de-Peralta Venturina M, Deshpande A, Menon M. Prognostic impact of histologic subtyping of adult renal epithelial neoplasms: an experience of 405 cases. Am J Surg Pathol. 2002;26:281–291. doi: 10.1097/00000478-200203000-00001. [DOI] [PubMed] [Google Scholar]
- Zisman A, Pantuck AJ, Wieder J, Chao DH, Dorey F, Said JW, deKernion JB, Figlin RA, Belldegrun AS. Risk group assessment and clinical outcome algorithm to predict the natural history of patients with surgically resected renal cell carcinoma. J Clin Oncol. 2002;20:4559–4566. doi: 10.1200/JCO.2002.05.111. [DOI] [PubMed] [Google Scholar]
- Gieseg MA, Cody T, Man MZ, Madore SJ, Rubin MA, Kaldjian EP. Expression profiling of human renal carcinomas with functional taxonomic analysis. BMC Bioinformatics. 2002;3:26. doi: 10.1186/1471-2105-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantuck AJ, Zeng G, Belldegrun AS, Figlin RA. Pathobiology, prognosis, and targeted therapy for renal cell carcinoma: exploiting the hypoxia-induced pathway. Clin Cancer Res. 2003;9:4641–4652. [PubMed] [Google Scholar]
- Zisman A, Pantuck AJ, Dorey F, Said JW, Shvarts O, Quintana D, Gitlitz BJ, deKernion JB, Figlin RA, Belldegrun AS. Improved prognostication of renal cell carcinoma using an integrated staging system. J Clin Oncol. 2001;19:1649–1657. doi: 10.1200/JCO.2001.19.6.1649. [DOI] [PubMed] [Google Scholar]
- Moch H, Gasser T, Amin MB, Torhorst J, Sauter G, Mihatsch MJ. Prognostic utility of the recently recommended histologic classification and revised TNM staging system of renal cell carcinoma: a Swiss experience with 588 tumors. Cancer. 2000;89:604–614. [PubMed] [Google Scholar]
- O’Keefe SC, Marshall FF, Issa MM, Harmon MP, Petros JA. Thrombocytosis is associated with a significant increase in the cancer specific death rate after radical nephrectomy. J Urol. 2002;168:1378–1380. doi: 10.1016/S0022-5347(05)64453-9. [DOI] [PubMed] [Google Scholar]
- Symbas NP, Townsend MF, El-Galley R, Keane TE, Graham SD, Petros JA. Poor prognosis associated with thrombocytosis in patients with renal cell carcinoma. BJU Int. 2000;86:203–207. doi: 10.1046/j.1464-410x.2000.00792.x. [DOI] [PubMed] [Google Scholar]
- Reuter VE, Presti JC., Jr Contemporary approach to the classification of renal epithelial tumors. Semin Oncol. 2000;27:124–137. [PubMed] [Google Scholar]
- Zambrano NR, Lubensky IA, Merino MJ, Linehan WM, Walther MM. Histopathology and molecular genetics of renal tumors: toward unification of a classification system. J Urol. 1999;162:1246–1258. [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, Gascoyne RD, Grogan TM, Muller-Hermelink HK, Smeland EB, Chiorazzi M, Giltnane JM, Hurt EM, Zhao H, Averett L, Henrickson S, Yang L, Powell J, Wilson WH, Jaffe ES, Simon R, Klausner RD, Montserrat E, Bosch F, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Fisher RI, Miller TP, LeBlanc M, Ott G, Kvaloy S, Holte H, Delabie J, Staudt LM. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3:185–197. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- Khan J, Wei JS, Ringner M, Saal LH, Ladanyi M, Westermann F, Berthold F, Schwab M, Antonescu CR, Peterson C, Meltzer PS. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med. 2001;7:673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Xu J, Stolk JA, Zhang X, Silva SJ, Houghton RL, Matsumura M, Vedvick TS, Leslie KB, Badaro R, Reed SG. Identification of differentially expressed genes in human prostate cancer using subtraction and microarray. Cancer Res. 2000;60:1677–1682. [PubMed] [Google Scholar]
- Young AN, Amin MB, Moreno CS, Lim SD, Cohen C, Petros JA, Marshall FF, Neish AS. Expression profiling of renal epithelial neoplasms: a method for tumor classification and discovery of diagnostic molecular markers. Am J Pathol. 2001;158:1639–1651. doi: 10.1016/S0002-9440(10)64120-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AN, de Oliveira Salles PG, Lim SD, Cohen C, Petros JA, Marshall FF, Neish AS, Amin MB. Beta defensin-1, parvalbumin, and vimentin: a panel of diagnostic immunohistochemical markers for renal tumors derived from gene expression profiling studies using cDNA microarrays. Am J Surg Pathol. 2003;27:199–205. doi: 10.1097/00000478-200302000-00008. [DOI] [PubMed] [Google Scholar]
- Boer JM, Huber WK, Sultmann H, Wilmer F, von Heydebreck A, Haas S, Korn B, Gunawan B, Vente A, Fuzesi L, Vingron M, Poustka A. Identification and classification of differentially expressed genes in renal cell carcinoma by expression profiling on a global human 31,500-element cDNA array. Genome Res. 2001;11:1861–1870. doi: 10.1101/gr.184501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JP, Shinghal R, Gill H, Reese JH, Terris M, Cohen RJ, Fero M, Pollack JR, van de Rijn M, Brooks JD. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am J Pathol. 2003;162:925–932. doi: 10.1016/S0002-9440(10)63887-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Yang XJ, Sugimura J, Backdahl J, Tretiakova M, Qian CN, Gray SG, Knapp R, Anema J, Kahnoski R, Nicol D, Vogelzang NJ, Furge KA, Kanayama H, Kagawa S, Teh BT. Molecular subclassification of kidney tumors and the discovery of new diagnostic markers. Oncogene. 2003;22:6810–6818. doi: 10.1038/sj.onc.1206869. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented gene ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht K, Richter T, Muller U, Walch A, Werner M, Hofler H. Quantitative gene expression analysis in microdissected archival formalin-fixed and paraffin-embedded tumor tissue. Am J Pathol. 2001;158:419–429. doi: 10.1016/S0002-9440(10)63985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton AJ, Jordan S, Yeomans P. Brief, high-temperature heat denaturation (pressure cooking): a simple and effective method of antigen retrieval for routinely processed tissues. J Pathol. 1994;173:371–379. doi: 10.1002/path.1711730413. [DOI] [PubMed] [Google Scholar]
- Eble JN, Sauter G, Epstein JI, Sesterhenn IA. Tumors of the kidney. Lyon: IARC Press; WHO Classification of TumoursTumours of the Urinary System and Male Genital Organs. 2004 [Google Scholar]
- Kattan MW, Reuter V, Motzer RJ, Katz J, Russo P. A postoperative prognostic nomogram for renal cell carcinoma. J Urol. 2001;166:63–67. [PubMed] [Google Scholar]
- Chow GK, Myles J, Novick AC. The Cleveland Clinic experience with papillary (chromophil) renal cell carcinoma: clinical outcome with histopathological correlation. Can J Urol. 2001;8:1223–1228. [PubMed] [Google Scholar]
- Beck SD, Patel MI, Snyder ME, Kattan MW, Motzer RJ, Reuter VE, Russo P. Effect of papillary and chromophobe cell type on disease-free survival after nephrectomy for renal cell carcinoma. Ann Surg Oncol. 2004;11:71–77. doi: 10.1007/BF02524349. [DOI] [PubMed] [Google Scholar]
- Motzer RJ, Bacik J, Mariani T, Russo P, Mazumdar M, Reuter V. Treatment outcome and survival associated with metastatic renal cell carcinoma of non-clear-cell histology. J Clin Oncol. 2002;20:2376–2381. doi: 10.1200/JCO.2002.11.123. [DOI] [PubMed] [Google Scholar]
- Chen RN, Novick AC, Gill IS. Laparoscopic cryoablation of renal masses. Urol Clin N Am. 2000;27:813–820. doi: 10.1016/s0094-0143(05)70128-2. [DOI] [PubMed] [Google Scholar]
- Christensen EI, Verroust PJ. Megalin and cubilin, role in proximal tubule function and during development. Pediatr Nephrol. 2002;17:993–999. doi: 10.1007/s00467-002-0956-5. [DOI] [PubMed] [Google Scholar]
- Hammad SM, Stefansson S, Twal WO, Drake CJ, Fleming P, Remaley A, Brewer HB, Jr, Argraves WS. Cubilin, the endocytic receptor for intrinsic factor-vitamin B(12) complex, mediates high-density lipoprotein holoparticle endocytosis. Proc Natl Acad Sci USA. 1999;96:10158–10163. doi: 10.1073/pnas.96.18.10158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AC, Nairn RC. Renal tubular antigens in kidney tumors. Cancer. 1972;29:977–981. doi: 10.1002/1097-0142(197204)29:4<977::aid-cncr2820290444>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, Latif F, Liu S, Chen F, Duh FM, Lubensky I, Duan DR, Florence C, Pozzatti R, Walther MM, Bander NH, Grossman HB, Brauch H, Pomer S, Brooks JD, Isaacs WB, Lerman MI, Zbar B, Linehan WM. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- Saito T, Kimura M, Kawasaki T, Sato S, Tomita Y. MHC class II antigen-associated invariant chain on renal cell cancer may contribute to the anti-tumor immune response of the host. Cancer Lett. 1997;115:121–127. doi: 10.1016/s0304-3835(97)04754-x. [DOI] [PubMed] [Google Scholar]
- Brown JA, Takahashi S, Alcaraz A, Borell TJ, Anderl KL, Qian J, Persons DL, Bostwick DG, Lieber MM, Jenkins RB. Fluorescence in situ hybridization analysis of renal oncocytoma reveals frequent loss of chromosomes Y and 1. J Urol. 1996;156:31–35. [PubMed] [Google Scholar]
- Bugert P, Gaul C, Weber K, Herbers J, Akhtar M, Ljungberg B, Kovacs G. Specific genetic changes of diagnostic importance in chromophobe renal cell carcinomas. Lab Invest. 1997;76:203–208. [PubMed] [Google Scholar]
- Tickoo SK, Lee MW, Eble JN, Amin M, Christopherson T, Zarbo RJ, Amin MB. Ultrastructural observations on mitochondria and microvesicles in renal oncocytoma, chromophobe renal cell carcinoma, and eosinophilic variant of conventional (clear cell) renal cell carcinoma. Am J Surg Pathol. 2000;24:1247–1256. doi: 10.1097/00000478-200009000-00008. [DOI] [PubMed] [Google Scholar]
- Tickoo SK, Reuter VE, Amin MB, Srigley JR, Epstein JI, Min KW, Rubin MA, Ro JY. Renal oncocytosis: a morphologic study of fourteen cases. Am J Surg Pathol. 1999;23:1094–1101. doi: 10.1097/00000478-199909000-00013. [DOI] [PubMed] [Google Scholar]
- Nickerson ML, Warren MB, Toro JR, Matrosova V, Glenn G, Turner ML, Duray P, Merino M, Choyke P, Pavlovich CP, Sharma N, Walther M, Munroe D, Hill R, Maher E, Greenberg C, Lerman MI, Linehan WM, Zbar B, Schmidt LS. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell. 2002;2:157–164. doi: 10.1016/s1535-6108(02)00104-6. [DOI] [PubMed] [Google Scholar]
- Pavlovich CP, Walther MM, Eyler RA, Hewitt SM, Zbar B, Linehan WM, Merino MJ. Renal tumors in the Birt-Hogg-Dube syndrome. Am J Surg Pathol. 2002;26:1542–1552. doi: 10.1097/00000478-200212000-00002. [DOI] [PubMed] [Google Scholar]
- Khoo SK, Kahnoski K, Sugimura J, Petillo D, Chen J, Shockley K, Ludlow J, Knapp R, Giraud S, Richard S, Nordenskjold M, Teh BT. Inactivation of BHD in sporadic renal tumors. Cancer Res. 2003;63:4583–4587. [PubMed] [Google Scholar]
- Ortmann M, Vierbuchen M, Fischer R. Renal oncocytoma. II. Lectin and immunohistochemical features indicating an origin from the collecting duct. Virchows Arch B Cell Pathol Incl Mol Pathol. 1988;56:175–184. [PubMed] [Google Scholar]
- Storkel S, Steart PV, Drenckhahn D, Thoenes W. The human chromophobe cell renal carcinoma: its probable relation to intercalated cells of the collecting duct. Virchows Arch B Cell Pathol Incl Mol Pathol. 1989;56:237–245. doi: 10.1007/BF02890022. [DOI] [PubMed] [Google Scholar]
- Valore EV, Park CH, Quayle AJ, Wiles KR, McCray PB, Jr, Ganz T. Human beta-defensin-1: an antimicrobial peptide of urogenital tissues. J Clin Invest. 1998;101:1633–1642. doi: 10.1172/JCI1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindels RJ, Timmermans JA, Hartog A, Coers W, van Os CH. Calbindin-D9k and parvalbumin are exclusively located along basolateral membranes in rat distal nephron. J Am Soc Nephrol. 1991;2:1122–1129. doi: 10.1681/ASN.V261122. [DOI] [PubMed] [Google Scholar]
- Martignoni G, Pea M, Chilosi M, Brunelli M, Scarpa A, Colato C, Tardanico R, Zamboni G, Bonetti F. Parvalbumin is constantly expressed in chromophobe renal carcinoma. Mod Pathol. 2001;14:760–767. doi: 10.1038/modpathol.3880386. [DOI] [PubMed] [Google Scholar]
- Simonnet H, Alazard N, Pfeiffer K, Gallou C, Beroud C, Demont J, Bouvier R, Schagger H, Godinot C. Low mitochondrial respiratory chain content correlates with tumor aggressiveness in renal cell carcinoma. Carcinogenesis. 2002;23:759–768. doi: 10.1093/carcin/23.5.759. [DOI] [PubMed] [Google Scholar]
- Gilmore T, Gapuzan ME, Kalaitzidis D, Starczynowski D. Rel/NF-kappa B/I kappa B signal transduction in the generation and treatment of human cancer. Cancer Lett. 2002;181:1–9. doi: 10.1016/s0304-3835(01)00795-9. [DOI] [PubMed] [Google Scholar]
- Castren JP, Kamel DE, Nurmi MJ, Collan YU. Cathepsin H expression distinguishes oncocytomas from renal cell carcinomas. Anticancer Res. 2000;20:537–540. [PubMed] [Google Scholar]
- Li WY, Huey CL, Yu AS. Expression of claudin-7 and -8 along the mouse nephron. Am J Physiol. 2004;286:F1063–F1071. doi: 10.1152/ajprenal.00384.2003. [DOI] [PubMed] [Google Scholar]
- Rampino T, Gregorini M, Soccio G, Maggio M, Rosso R, Malvezzi P, Collesi C, Dal Canton A. The Ron proto-oncogene product is a phenotypic marker of renal oncocytoma. Am J Surg Pathol. 2003;27:779–785. doi: 10.1097/00000478-200306000-00008. [DOI] [PubMed] [Google Scholar]
- Patton KT, Tretiakova MS, Yao JL, Papavero V, Huo L, Adley BP, Wu G, Huang J, Pins MR, Teh BT, Yang XJ. Expression of RON Proto-oncogene in Renal oncocytoma and chromophobe renal cell carcinoma. Am J Surg Pathol. 2004;28:1045–1050. doi: 10.1097/01.pas.0000128661.58697.7d. [DOI] [PubMed] [Google Scholar]
- Lin F, Brown RE, Shen T, Yang XJ, Schuerch C. Immunohistochemical detection of P504S in primary and metastatic renal cell carcinomas. Appl Immunohistochem Mol Morphol. 2004;12:153–159. doi: 10.1097/00129039-200406000-00010. [DOI] [PubMed] [Google Scholar]
- Tretiakova MS, Sahoo S, Takahashi M, Turkyilmaz M, Vogelzang NJ, Lin F, Krausz T, Teh BT, Yang XJ. Expression of alpha-methylacyl-CoA racemase in papillary renal cell carcinoma. Am J Surg Pathol. 2004;28:69–76. doi: 10.1097/00000478-200401000-00007. [DOI] [PubMed] [Google Scholar]
- Dong Y, Berners-Price SJ, Thorburn DR, Antalis T, Dickinson J, Hurst T, Qiu L, Khoo SK, Parsons PG. Serine protease inhibition and mitochondrial dysfunction associated with cisplatin resistance in human tumor cell lines: targets for therapy. Biochem Pharmacol. 1997;53:1673–1682. doi: 10.1016/s0006-2952(97)00015-4. [DOI] [PubMed] [Google Scholar]
- Sanz L, Kristensen P, Russell SJ, Ramirez Garcia JR, Alvarez-Vallina L. Generation and characterization of recombinant human antibodies specific for native laminin epitopes: potential application in cancer therapy. Cancer Immunol Immunother. 2001;50:557–565. doi: 10.1007/s00262-001-0235-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt L, Junker K, Weirich G, Glenn G, Choyke P, Lubensky I, Zhuang Z, Jeffers M, Vande Woude G, Neumann H, Walther M, Linehan WM, Zbar B. Two North American families with hereditary papillary renal carcinoma and identical novel mutations in the MET proto-oncogene. Cancer Res. 1998;58:1719–1722. [PubMed] [Google Scholar]
- Sweeney P, El-Naggar AK, Lin SH, Pisters LL. Biological significance of c-met over expression in papillary renal cell carcinoma. J Urol. 2002;168:51–55. [PubMed] [Google Scholar]
- Lindor NM, Dechet CB, Greene MH, Jenkins RB, Zincke MT, Weaver AL, Wilson M, Zincke H, Liu W. Papillary renal cell carcinoma: analysis of germline mutations in the MET proto-oncogene in a clinic-based population. Genet Test. 2001;5:101–106. doi: 10.1089/109065701753145547. [DOI] [PubMed] [Google Scholar]
- Fetsch PA, Fetsch JF, Marincola FM, Travis W, Batts KP, Abati A. Comparison of melanoma antigen recognized by T cells (MART-1) to HMB-45: additional evidence to support a common lineage for angiomyolipoma, lymphangiomyomatosis, and clear cell sugar tumor. Mod Pathol. 1998;11:699–703. [PubMed] [Google Scholar]
- Fryer AE, Chalmers A, Connor JM, Fraser I, Povey S, Yates AD, Yates JR, Osborne JP. Evidence that the gene for tuberous sclerosis is on chromosome 9. Lancet. 1987;1:659–661. doi: 10.1016/s0140-6736(87)90416-8. [DOI] [PubMed] [Google Scholar]
- Kandt RS, Haines JL, Smith M, Northrup H, Gardner RJ, Short MP, Dumars K, Roach ES, Steingold S, Wall S, Blanton SH, Foldman P, Kwiatkowski DJ, Jewell A, Weber JL, Roses AD, PericakVance MA. Linkage of an important gene locus for tuberous sclerosis to a chromosome 16 marker for polycystic kidney disease. Nat Genet. 1992;2:37–41. doi: 10.1038/ng0992-37. [DOI] [PubMed] [Google Scholar]
- Chen CH, Tzeng CC, Cheng TC, Chiu AW. Angiomyolipoma of kidney as a part of tuberous sclerosis complex. J Postgrad Med. 2003;49:278–279. [PubMed] [Google Scholar]
- Li Y, Corradetti MN, Inoki K, Guan KL. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem Sci. 2004;29:32–38. doi: 10.1016/j.tibs.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Brugarolas JB, Vazquez F, Reddy A, Sellers WR, Kaelin WG., Jr TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell. 2003;4:147–158. doi: 10.1016/s1535-6108(03)00187-9. [DOI] [PubMed] [Google Scholar]
- Arbiser JL, Brat D, Hunter S, D’Armiento J, Henske EP, Arbiser ZK, Bai X, Goldberg G, Cohen C, Weiss SW. Tuberous sclerosis-associated lesions of the kidney, brain, and skin are angiogenic neoplasms. J Am Acad Dermatol. 2002;46:376–380. doi: 10.1067/mjd.2002.120530. [DOI] [PubMed] [Google Scholar]
- L’Hostis H, Deminiere C, Ferriere JM, Coindre JM. Renal angiomyolipoma: a clinicopathologic, immunohistochemical, and follow-up study of 46 cases. Am J Surg Pathol. 1999;23:1011–1020. doi: 10.1097/00000478-199909000-00003. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Rhodes DR, Furge KA, Kanayama H, Kagawa S, Haab BB, Teh BT. Gene expression profiling of clear cell renal cell carcinoma: gene identification and prognostic classification. Proc Natl Acad Sci USA. 2001;98:9754–9759. doi: 10.1073/pnas.171209998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasselli JR, Shih JH, Iyengar SR, Maranchie J, Riss J, Worrell R, Torres-Cabala C, Tabios R, Mariotti A, Stearman R, Merino M, Walther MM, Simon R, Klausner RD, Linehan WM. Predicting survival in patients with metastatic kidney cancer by gene-expression profiling in the primary tumor. Proc Natl Acad Sci USA. 2003;100:6958–6963. doi: 10.1073/pnas.1131754100. [DOI] [PMC free article] [PubMed] [Google Scholar]