Abstract
The expression of oncofetal H19 RNA and its localization/cellular source was analyzed in synovial tissue (ST) and isolated synovial macrophages (Mφ) or synovial fibroblasts (SFBs) by reverse transcriptase-polymerase chain reaction (RT-PCR), in situ hybridization, and immunohistochemistry. RT-PCR showed significantly higher H19 expression in ST from patients with rheumatoid arthritis (RA) (P = 0.000) and osteoarthritis (OA) (P = 0.009) than in normal/joint trauma controls (N/JT), but comparable levels in reactive arthritis. In situ hybridization demonstrated strong signals in all RA-ST samples (n = 8), with ≥85% positive cells in the lining layer, diffuse infiltrates, and stroma regions. In lymphoid aggregates and endothelial cells only 20% were positive. RA-ST contained a significantly higher percentage of strongly positive lining cells than OA-ST and N/JT-ST. H19 RNA was expressed in both Mφ and SFBs, as confirmed by RT-PCR in isolated RA Mφ and SFBs (n = 3). In RA-SFBs, low constitutive H19 RNA expression in culture (10% fetal calf serum) was strongly increased on starvation (3.5-fold, 1% fetal calf serum), with or without the addition of interleukin-1β (10 to 100 U/ml), tumor necrosis factor-α (1 to 25 ng/ml), or platelet-derived growth factor-BB (2.5 to 10 U/ml). In OA-SFBs, this starvation-induced increase was lower (twofold), reaching significant differences compared with RA-SFBs after stimulation with interleukin-1β and platelet-derived growth factor-BB. In both RA- and OA-SFBs, the MAP-kinase ERK-1/2 pathway and the phosphatidylinositol-3 kinase pathway influenced H19 RNA expression, as shown by inhibitor studies. Significant overexpression of H19 RNA and its increased sensitivity to starvation/cytokine regulation in RA suggests a pathogenetic role of this oncofetal gene, possibly reflecting embryonal dedifferentiation of the adult ST and/or ongoing inflammatory/oxidative stress.
The hyperplastic synovial tissue (ST) in rheumatoid arthritis (RA) displays several features of a semitransformed tissue, 1-3 such as invasive growth into cartilage and bone, expression of proto-oncogenes and IGF-2, and mutations of the tumor suppressor gene p53. 4 It is presently unclear whether these alterations are because of endogenous genetic alterations (eg, mutations of key regulatory genes) 5,6 or to exogenous alterations induced by (pro)-inflammatory cytokines or oxidative stress. 7,8
The oncofetal H19 gene belongs to a special subgroup of imprinted genes coding for untranslated RNA molecules; 9-11 these genes act as functional RNA rather than protein and their role in physiology/pathophysiology is incompletely understood. However, high expression in fetal tissues, 11 as well as high evolutionary conservation of its exon sequences 12 and its possible secondary structure, 13 are indicative of a biological function of H19 RNA.
H19 RNA is abundantly expressed in embryonal tissue of endodermal and mesodermal origin 14 and is repressed after birth in all tissues except skeletal muscle. 15 Re-expression only occurs in a number of different tumors, 14,16,17 including testicular germ cell tumors and bladder carcinoma. 18 In the latter case, H19 RNA expression appears to reflect the degree of invasiveness. 18 Therefore, H19 RNA has been proposed as a tumor marker or a marker of embryonal dedifferentiation of adult tissues.
On the other hand, H19 RNA expression is suppressed in fibrosarcoma, 19 rhabdomyosarcoma with embryonal histology, 20 Wilms’ tumor, 21 and hormonally active adrenocortical carcinoma, 22 and is variably increased/decreased in breast tumor, 23 in the latter case under the influence of modulation by steroids. 24 Together with the capacity of H19 RNA to inhibit proliferation, clonogenicity, and tumorigenicity on transfection of tumor cell lines, 13,25 these findings suggest that H19 RNA may exhibit tumor-suppressor activity.
Very recently, the potential physiological role of H19 has been defined in more detail. 26-28 Using H19-transfected breast cancer cells, cancerous mammary epithelial cells, or the human bladder carcinoma cell line T24P, a number of H19-regulated target genes have been identified, including thioredoxin (a key protein of cellular redox metabolism), 28 MAP-kinase kinase 1 (MKK1), nuclear factor-κB, c-jun N-terminal kinase 2 (JNK2), tumor necrosis factor (TNF)-α, and interleukin (IL)-6. 26 Because a similar pattern of genes is up-regulated during oxidative stress/hypoxia, and because H19 RNA expression is induced by growth arrest, for example in confluent cell cultures or on serum starvation, 29 H19 may therefore be an indicator of physiological/pathological stress situations.
To verify whether H19 RNA is expressed in RA and bears any specificity for this disease, transcription of H19 RNA was comparatively analyzed in RA-ST and ST from osteoarthritis (OA), reactive arthritis (ReA), and normals/joint trauma (N/JT).
Materials and Methods
Patient Population
ST specimens from patients with RA (n = 26), OA (n = 25), or ReA (n = 2) were obtained during synovectomy or joint replacement surgery from the Immanuel-Krankenhaus (Berlin, Germany) and from the Clinic of Orthopedics [Eisenberg, Friedrich Schiller University (FSU) Jena, Germany] under approval of the responsible ethics committees (Table 1) ▶ . All RA and OA patients fulfilled the respective American College of Rheumatology (ACR) classification criteria. 30,31 ST from patients with either no joint disease (postmortem samples, n = 8) or recent joint trauma (JT) (n = 7), derived from the Tissue Bank of the Institute for Transfusion Medicine (Charité, Berlin) and the Department of Traumatology (FSU), was used as control (Table 1) ▶ . Specimens were embedded in Tissue-Tek OCT Compound (Lab-Tek Products, Elkhart, IN), immediately frozen in isopentane (Merck, Darmstadt, Germany) cooled in liquid nitrogen, and stored at −70°C for immunohistochemistry (IHC) and/or in situ hybridization. Alternatively, samples were homogenized in 4 mol/L of guanidinium isothiocyanate (GuSCN) for reverse transcriptase-polymerase chain reactions (RT-PCR) using a rotor strator homogenizer (DIAX-100; Heidolph, Schwabach, Germany).
Table 1.
Clinical Characteristics of the Patients at the Time of Synovectomy/Sampling
| Patient/diagnosis | Gender/age (yrs) | Disease dur./yrs | RF | ESR (1 h) | CRP (mg/l) | # of ARA-criteria (RA) | Concomitant medication |
|---|---|---|---|---|---|---|---|
| Rheumatoid arthritis | |||||||
| EB31 | F/78 | 2 | n.d. | 52 | 8.5 | 4 | Prednis., Leflunomide |
| EB34 | M/58 | 3 | + | 22 | 10.1 | 4 | NSAIDs, Leflunomide, Prednis. |
| EB42 | F/71 | 44 | ??? | 50 | 35.8 | 4 | NSAIDs, Prednis. |
| EB46 | M/73 | 19 | ??? | 24 | 15.0 | 4 | NSAIDs |
| EB58 | F/74 | 5 | + | 12 | 12.2 | 5 | NSAIDs, MTX, Prednis. |
| Osteoarthritis | |||||||
| EB11 | F/46 | n.d. | n.d. | 9 | 8.2 | 0 | none |
| EB16 | F/69 | n.d. | n.d. | n.d. | 9.6 | 1 | none |
| EB19 | F/70 | n.d. | n.d. | 6 | <5 | 1 | none |
| EB29 | F/76 | 2 | n.d. | 13 | <5 | 1 | NSAIDs |
| EB54 | |||||||
| Normals | |||||||
| NST1 | M/68 | ||||||
| NST2 | F/69 | ||||||
| NST3 | F/74 | ||||||
| NST4 | F/19 | ||||||
| NST5 | M/60 |
Abbreviations are defined as follows: yrs, years; Disease dur., disease duration; RF, rheumatoid factor; ESR, erythrocyte sedimentation rate; CRP, C-reactive protein, *, normal range <5 mg/l; ARA, American Rheumatism Association (now American College of Rheumatology); −, negative; n.d., not determined; RA, rheumatoid arthritis; OA, osteoarthritis; AS, ankylosing spondylitis; PsA, psoriatic arthritis; JRA, juvenile rheumatoid arthritis; SLE, systemic lupus erythematodes; VNS, villonodular synovitis; UA, undifferentiated monoarthritis; MTX, Methotrexate; Prednis., prednisolone; NSAIDs, non-steroidal anti-inflammatory drugs; Sulfas., sulfasalazine; Chlor., chloroquine.
Cell Culture and Cytokine/Growth Factor Stimulation
Isolation of primary culture synovial fibroblasts (SFBs) and Mφ was performed as described, 32 resulting in strong enrichment of SFBs (contamination of <2% leukocytes or endothelial cells) and synovial Mφ [95% purity as assessed by fluorescence-activated cell sorting analysis with anti-CD14 monoclonal antibodies (mAbs)]. Early-passage SFBs (second passage) obtained from two RA patients were either grown in complete Dulbecco’s modified Eagle’s medium (12.5 mmol/L HEPES, 100 U/ml penicillin, 100 μg/ml streptomycin, 2.5 ng/ml amphotericin B; Gibco-BRL, Eggenstein, Germany) supplemented with 10% fetal calf serum (FCS), or serum-starved (1% FCS) for 72 hours and subsequently either left in 1% FCS or stimulated for 24 hours with 10, 50, and 100 U/ml IL-1β (Genzyme, Rüsselsheim, Germany), 2.5, 5, and 10 U/ml platelet-derived growth factor-BB (PDGF-BB) (R&D Systems, Wiesbaden, Germany) or 1, 10, and 25 ng/ml TNF-α (Chemicon, Hofheim, Germany). Inhibitors of the ERK1/2 pathway (U0126, 1 μmol/L; Alexis, Grünberg, Germany); 33 and the phosphatidylinositol-3 kinase pathway (Wortmannin, 1 μmol/L; Alexis); 34 were used in the cases of stimulation with either IL-1β or PDGF-BB. For this purpose, SFBs were preincubated with the inhibitors for 30 minutes at 37°C in complete medium supplemented with 10% FCS. Subsequently, the cytokines/growth factors were added for 24 hours without further change of medium. The cells were then lysed in RNA lysis buffer (4 mol/L GuSCN) supplemented with mercaptoethanol (7 μl/ml).
Purification of RNA, Reverse Transcription, and RT-PCR
Total cellular RNA was extracted from 2 to 3 g of ST or cultured synovial Mφ or SFBs (0.5 to 2 × 106 cells) according to Chirgwin and colleagues. 35 In the case of ST, total RNA was purified using a commercially available RNA extraction kit (RNeasy mini kit; Qiagen, Hilden, Germany). In the case of synovial cells, RNA was isolated with the NucleoSpin RNA kit (Macherey-Nagel, Düren, Germany). In both cases, between 2 and 5 μg of total RNA were primed with oligo(d)T and reverse-transcribed with SuperScript-II reverse transcriptase according to the manufacturer’s instructions (Gibco-BRL).
The transcription level of H19 RNA expression was quantified by 32 to 40 cycles of RT-PCR (downstream primer: CCTCATCAGCCCAACATCAA; upstream primer: GGCCGTCT CCACAACTCCAA; resulting fragment length, 313 bp), denaturation at 93°C for 1 minute, followed by annealing at 59°C for 1 minute and extension at 72°C for 35 seconds. To normalize the samples for equal amounts of cDNA, the housekeeping gene glycerol-3-aldehyde phosphatase (GADPH) (downstream primer: GTCGTTGAGGGCAATGCC; upstream primer: GGTCATC CATGACAACTTTG; resulting fragment length, 428 bp) was used as an internal standard (conditions: denaturation at 93°C for 1 minute, followed by annealing at 62°C for 1 minute and extension at 72°C for 45 seconds; total of 28 to 32 cycles). Alternatively, the housekeeping gene β-actin (downstream primer: gtgcgacgaagacgagacc; upstream primer: cagagccg agacaccacg; resulting fragment length, 230 bp) was used as an internal standard (conditions: initial denaturation at 93°C for 3 minutes, followed by a total of 28 to 34 cycles; denaturation at 93°C for 45 seconds, annealing at 62°C for 45 seconds, and extension at 72°C for 30 seconds).
Amplified products were separated and visualized in a 1% agarose gel containing 0.1 μg/ml of ethidium bromide at 280 nm. The intensity of each band was analyzed using an integration image software (Scion Corp., Frederick, MD).
Cloning of PCR Fragments and Nucleic Acid Sequencing
The 313-bp cDNA fragment specific for H19 RNA, transcribed and amplified from RA-ST using the above-mentioned primer pair, was inserted into the pCR-TOP-2.1 vector (Invitrogen, Leek, The Netherlands) by T/A cloning according to the manufacturer’s instructions. The sequence and orientation of the insert was verified by cycle sequencing using an automated laser fluorescence sequencer (A.L.F.; Pharmacia-Biotech, Uppsala, Sweden) using fluorescein-labeled M13 + 40 and M13 reverse standard primers (data not shown).
In Vitro Transcription of the H19 Riboprobe and in Situ Hybridization
After linearization of the pCR-TOPO-2.1 vector harboring the H19 insert (5 μg/25 μl; gene bank accession no. AF125183; position 2896 to 3209) with 10 U XbaI and BamHI (Gibco-BRL) for 1.5 hours at 37°C, in vitro transcription for sense and anti-sense digoxigenin-labeled probes was performed using SP6 (New England Biolabs, Bad Homburg, Germany) and T7 RNA polymerase (Stratagene, Heidelberg, Germany) according to the manufacturer’s instructions.
In situ hybridization was performed as published 36,37 with modifications. Briefly, 6-μm cryostat ST sections were fixed in 3% paraformaldehyde for 1 hour at room temperature and subsequently washed twice each for 5 minutes at room temperature with: 2× standard saline citrate (SSC) (1× SSC: 150 mmol/L NaCl, 15 mmol/L Na-citrate, pH 7.0)/diethyl pyrocarbonate; and 0.1 mol/L triethanolamine-HCl (pH 8.0). After acetylation of the sections for 30 minutes at room temperature with 0.25% acetic anhydride in 0.1 mol/L triethanolamine-HCl (pH 8.0), the sections were again washed twice in 0.1 mol/L triethanolamine-HCl (pH 8.0), prehybridized for 1 hour at room temperature with hybridization buffer containing 50% formamide, 1× Denhardt’s solution, 20% dextran sulfate (in 8× SSC), 500 μg/ml salmon sperm (Gibco-BRL), and 250 μg/ml yeast tRNA (Sigma), and subsequently hybridized with sense or anti-sense digoxigenin-labeled riboprobes (semiquantification by ethidium bromide/agarose gel electrophoresis) in hybridization buffer for 16 hours at 50°C. To exclude that negative results were because of mRNA degradation, sense and anti-sense riboprobes for the constitutively expressed housekeeping gene GAPDH (kindly provided by P. Ruschpler, Institute of Pathology, University of Leipzig, Leipzig, Germany) were applied, with the anti-sense probe yielding positive results in all cases (identical conditions for in situ hybridization; data not shown). After hybridization, washing steps with decreasing stringency were performed at 50°C using the following protocol: 50% formamide/1× SSC (5 minutes), 1× SSC plus 0.1% sodium dodecyl sulfate (15 minutes), 0.25× SSC plus 0.1% sodium dodecyl sulfate (15 minutes), 0.1× SSC plus 0.1% sodium dodecyl sulfate (15 minutes). For detection of the probes, an anti-digoxigenin alkaline phosphatase-conjugated Fab fragment (diluted 1: 500 with 100 mmol/L Tris-HCl, 150 mmol/L NaCl containing 1% horse serum; pH 7.5; Boehringer, Mannheim, Germany) was added for 1 hour at 20°C. After repeated washing steps with 100 mmol/L Tris-NaCl, 150 mmol/L NaCl (pH 7.5), and 100 mmol/L Tris-NaCl/MgCl2 (pH 9.5), staining was performed with 100 mmol/L Tris-NaCl/NaCl, 50 mmol/L MgCl2 staining buffer containing 0.2% (w/v) 5-bromo-4-chloro-3-indolyl-phosphate, 0.22% (w/v) 4-nitro blue tetrazolium chloride, and 1 mmol/L levamisole to block endogenous alkaline phosphatase (Sigma).
Immunohistochemical (IHC) Double Staining
To characterize H19 RNA-expressing cells, either double staining with an anti-CD68 monoclonal antibody (peroxidase technique, mAb PG-M1; DAKO, Hamburg, Germany) was performed after completion of in situ hybridization for H19 RNA or single staining with an anti-prolyl-4-hydroxylase mAb (alkaline phosphatase technique, mAb 3-2B12; DAKO) was used in serial sections.
In both cases, all incubations were performed at 20°C in a humid chamber. For double staining, sections were incubated with 0.03% H2O2/PBS (PBS: phosphate-buffered saline; 137 mmol/L NaCl, 2.68 mmol/L KCl, 8.1 mmol/L Na2HPO4, 1.76 mmol/L KH2PO4, pH 7.4) for 20 minutes to inactivate endogenous peroxidase followed by a blocking step for 20 minutes with 5% horse serum/PBS. The anti-CD68 mAb, diluted 1:10 in PBS/5% horse serum, was added for 45 minutes. For detection, a peroxidase-coupled rabbit anti-mouse antibody (1:30 dilution in PBS/5% horse serum; DAKO) was added for 45 minutes. The peroxidase was revealed using diaminobenzidine (0.7 mg/ml in 60 mmol/L Tris-HCl buffer, pH 7.5; Sigma).
For single staining of prolyl-4-hydroxylase, serial sections were fixed with acetone for 10 minutes at 20°C and air-dried. Then the specific antibody, diluted in PBS/5% horse serum, was added for 30 minutes. For detection, sections were incubated with an alkaline phosphatase-coupled goat anti-mouse antibody (DAKO) for 50 minutes (1:30 in buffer-1 containing 100 mmol/L Tris-HCl, pH 7.4, 150 mmol/L NaCl, and 1% horse serum). The alkaline phosphatase was revealed in buffer containing 100 mmol/L Tris-HCl buffer (pH 9.5), 100 mmol/L NaCl, 50 mmol/L MgCl2, 1.0 mg/ml FAST-Red TR, and 0.4 mg/ml Naphtol AS-MX (Sigma). Endogenous alkaline phosphatase was blocked with 0.15 mg/ml levamisole (Sigma). Sections were counterstained with hematoxylin. For single staining of CD68, the peroxidase technique was used as above except that the incubation with the anti-CD68 mAb was performed for 30 minutes only.
Statistical Analysis
Data were analyzed by the multigroup Kruskal-Wallis test, the nonparametric Mann-Whitney U-test, and, in the case of paired synovial Mφ and SFBs, the Wilcoxon test (P < 0.05; SPSS 9.0; SPSS Inc., Chicago, IL). As postmortem samples showed no indication for mRNA degradation [ie, no statistically significant differences were observed by Mann-Whitney U-test between the expression of c-jun, junB, junD, and (short-lived) c-fos in joint trauma and postmortem samples; data not shown], results from joint trauma and normal, postmortem samples were pooled as normal, noninflammatory controls. Correlations were assessed using Spearman’s rank correlation.
Results
RT-PCR of H19 RNA in ST
Whereas GAPDH showed comparable mRNA levels in all RA-ST, H19 RNA expression was highly variable, with clear positivity in 11 of 11 samples and strong expression in 6 of 11 samples (Figure 1A) ▶ . In OA-ST, expression of H19 RNA was generally weaker than in RA-ST, with 7 of 10 weakly positive samples (Figure 1A) ▶ . In controls, ie, ReA (n = 2) and N/JT (n = 8), H19 RNA was weakly expressed in all samples (Figure 1A) ▶ .
Figure 1.
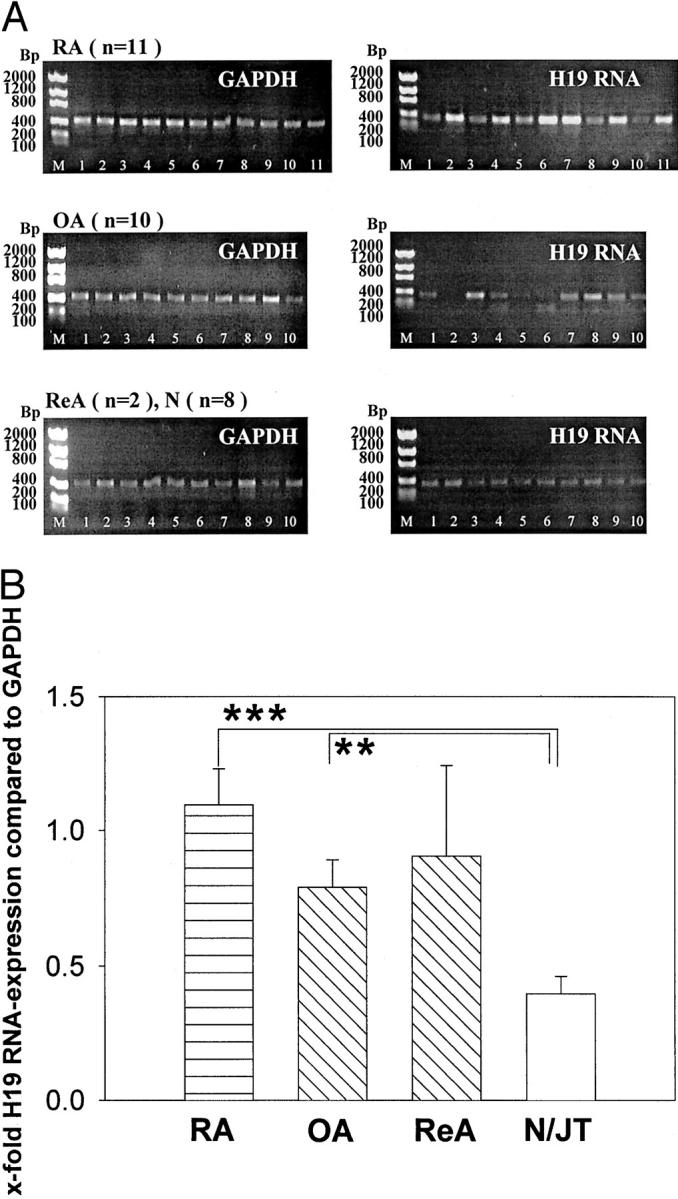
H19 RNA expression in ST (RT-PCR). Representative analysis of ST from patients with RA (n = 23), OA (n = 21), ReA (n = 2; lanes 1 and 2 in the two bottom gel panels), and normals/joint trauma (N/JT; n = 14; lanes 3 to 10 in the two bottom gel panels; A). Semiquantification and normalization to mRNA expression of GAPDH resulted in significantly higher H19 RNA expression in RA-ST versus N/JT-ST (***, P = 0.000) and in OA-ST versus N/JT-ST (**, P = 0.009; B). M, molecular size standard given in bp.
Image analysis quantification and normalization to GAPDH expression resulted in significantly higher H19 RNA expression in RA-ST (n = 23) versus N/JT-ST (n = 14, P = 0.000). However, H19 RNA expression in OA-ST (n = 21) was also significantly higher than in N/JT-ST (P = 0.01, Figure 1B ▶ ). No significant differences were observed among the other groups. There was a weak correlation between the expression level of H19 in the SM and rheumatoid factor positivity in patients with RA, OA, and ReA (r = 0.316, P = 0.036, n = 44). No correlations with other clinical parameters were observed.
Detection of H19 RNA Expression in RA-, OA-, and Normal/JT-ST by in Situ Hybridization
Regional distribution/cellular sources of H19 RNA expression in ST were assessed by in situ hybridization, either separately or in combination with IHC for the fibroblast marker prolyl-4-hydroxylase and the Mφ marker CD68 (mAb PG-M1).
In Situ Hybridization and Double-in Situ Hybridization/IHC
Although the sense-probe for H19 RNA did not bind to RA-ST (Figure 2A) ▶ and OA-ST (Figure 2B) ▶ , the anti-sense probe showed a positive reaction in lining layer, diffuse infiltrates, stroma, and endothelial cells of OA-ST (Figure 2C) ▶ and RA-ST (Figure 2E) ▶ . Using double-in situ hybridization/IHC, H19 RNA was detected in both CD68-positive Mφ and CD68-negative cells of OA-ST (Figure 2D) ▶ . In serial sections, H19 expression in RA-ST could be assigned to prolyl-4-hydroxylase-positive SFBs (Figure 2, F and G) ▶ . In double-in situ hybridization/IHC, ∼50% of the CD68-positive Mφ in lining layer and diffuse infiltrates also showed a positive reaction for H19 RNA (Figure 2, H and I) ▶ .
Figure 2.
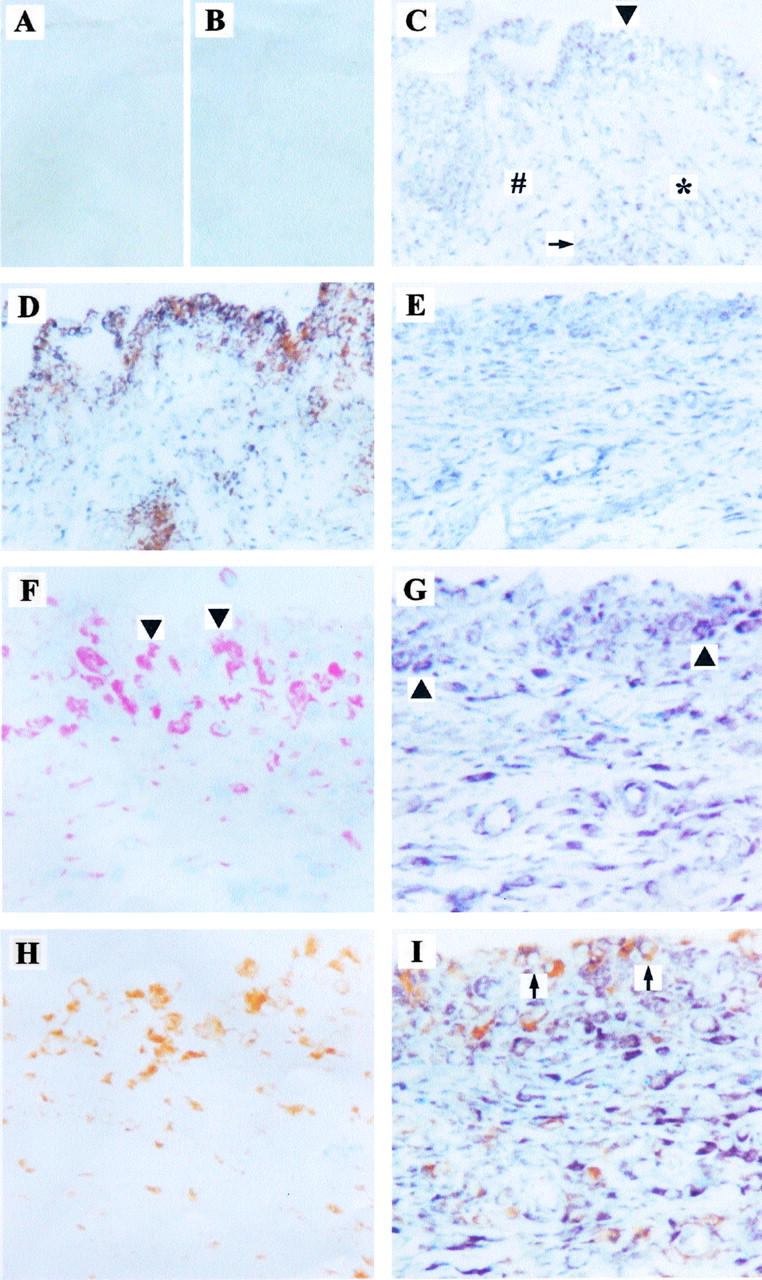
Distribution and cellular sources of H19 RNA expression in ST (in situ hybridization). Six-μm cryostat sections from OA-ST (B, C, D) or RA-ST (A, E–I). The sense probe for H19 (negative control) did not bind to RA-ST (A) and OA-ST (B). The anti-sense probe for H19 showed a positive reaction in lining layer (arrowhead in C), diffuse infiltrates (arrow in C), stroma (# in C), and endothelial cells (* in C) of OA-ST (C) and RA-ST (E). Using double-in situ hybridization/IHC, H19 RNA was detected in CD68-positive Mφ and CD68-negative cells of OA-ST (D) and RA-ST (H and I, arrows); in serial sections, H19 expression in CD68-negative cells of RA-ST was predominantly in prolyl-4-hydroxylase-positive SFBs (F and G, arrowheads). Original magnifications: ×92 (A–E); ×184 (F–I).
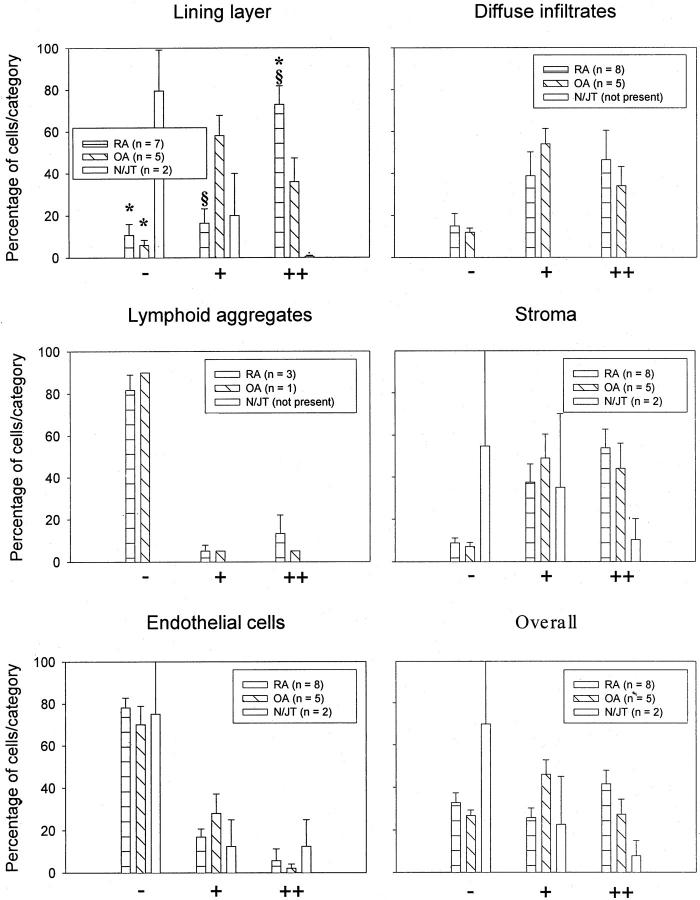
On average (RA-ST, n = 8; OA-ST, n = 5; normal/JT, n = 2), ∼50% of the H19-positive cells in lining layer and stroma of RA-, OA-, and N/JT-ST were CD68-positive Mφ. On the basis of serial IHC staining with prolyl-4-hydroxylase, the remaining H19-positive cells appeared to be predominantly SFBs. The same applied to diffuse infiltrates in RA-ST and OA-ST. Interestingly, ∼80 to 90% of H19-positive cells in lymphoid aggregates of RA-ST (present in three of eight samples) and OA-ST (present in one of five samples) were CD68-positive Mφ.
Semiquantification of H19 RNA Expression in ST by in Situ Hybridization
H19 RNA Expression in RA-ST
Strong signals for H19 RNA were obtained in all RA synovial specimens (n = 8, Figure 3 ▶ ). In the lining layer, ∼90% of the cells showed a positive signal, with 73% showing a strong signal. In diffuse infiltrates and stroma regions, a total of 85% expressed H19 RNA, with approximately equal proportions of weakly and strongly positive cells. In lymphoid aggregates (only present in three of eight samples), only ∼20% of the cells were weakly or strongly positive, comparable to the situation in endothelial cells (Figure 3) ▶ .
Figure 3.
Distribution of H19 RNA expression in ST (semiquantitation of in situ hybridization). Strong signals for H19 RNA were obtained in all RA synovial specimens (n = 8). In lining layer, diffuse infiltrates, and stroma, ≥85% of the cells showed a positive signal, whereas lymphoid aggregates (only present in three of eight samples) and endothelial cells contained only ∼20% of positive cells. The H19 RNA expression in OA-ST (n = 5) was generally comparable to that in RA-ST. Significant differences between RA and OA (§) were restricted to the lining layer. The percentage of H19-positive cells in lining layer and stroma of normal/joint trauma-ST (N/JT) was lower than in RA and OA, with significant differences in comparison with both RA and OA (*) in the lining layer. The percentage of H19-positive endothelial cells was comparable in RA-, OA-, and N/JT-ST.
H19 RNA Expression in OA-ST
The H19 RNA expression in OA-ST (n = 5) was generally comparable to that in RA-ST. Differences between RA and OA were restricted to the lining layer, in which RA-ST contained a significantly higher percentage of strongly positive cells (73% versus 36%) and consequently a significantly lower percentage of weakly positive cells (16% versus 58%, Figure 3 ▶ ).
H19 RNA Expression in N/JT-ST
The percentage of H19-positive cells in lining layer and stroma was lower than in RA and OA, with significant differences in comparison with both RA and OA in the lining layer. The percentage of H19-positive endothelial cells was comparable in RA-, OA-, and N/JT-ST (Figure 3) ▶ .
The average of the individual histological regions, calculated to estimate the overall percentage of H19-positive cells in ST, confirmed the results obtained by semiquantification of H19 RNA in situ hybridization (Figure 3) ▶ and by RT-PCR in whole ST (Figure 1) ▶ .
RT-PCR of H19 RNA in Isolated SFBs and Macrophages
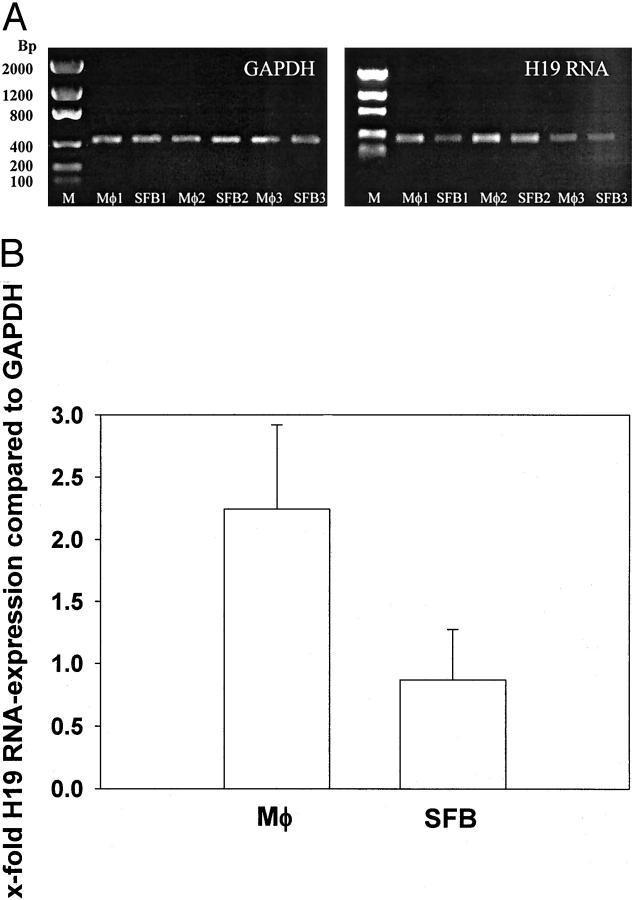
H19-RNA was detected by RT-PCR in paired SFBs and synovial Mφ isolated from primary culture synovial cells of three RA patients (Figure 4A) ▶ , resulting in numerically higher H19 RNA expression in synovial Mφ than in SFBs (2.2-fold versus 0.9-fold in comparison with the expression of GAPDH; not significant; Figure 4B ▶ ). This analysis confirmed the double-in situ hybridization/IHC result that H19 RNA is expressed in both RA synovial Mφ and RA-SFBs.
Figure 4.
H19 RNA expression in synovial Mφ and SFBs isolated from RA (RT-PCR). A: Synovial Mφ and SFBs from patients with RA (n = 3) were analyzed by RT-PCR. B: Image analysis quantification and normalization to mRNA expression of GAPDH showed numerically higher H19 RNA expression in Mφ than in SFBs, but without statistical significance. M, molecular size standard given in bp.
Regulation of H19 RNA Expression in RA SFBs on Starvation and Stimulation with IL-1β, PDGF-BB, and TNF-α
SFBs from one RA patient (Figure 5A) ▶ and SFBs from a total of four representative RA patients (Figure 5B) ▶ showed low constitutive expression of H19 RNA on culture in medium containing 10% FCS. H19 RNA expression was strongly induced by starving the cells for 96 hours (1% FCS, on average 4.5-fold), either with or without stimulation with IL-1β, PDGF-BB, and TNF-α (Figure 5, A and B) ▶ . In comparison to OA-SFBs, this induction was significantly (or borderline significantly) stronger after stimulation of RA-SFBs with 1% FCS, 100 or 50 U/ml of IL-1β, as well as 10 or 5 U/ml of PDGF-BB (Figure 5B) ▶ .
Figure 5.
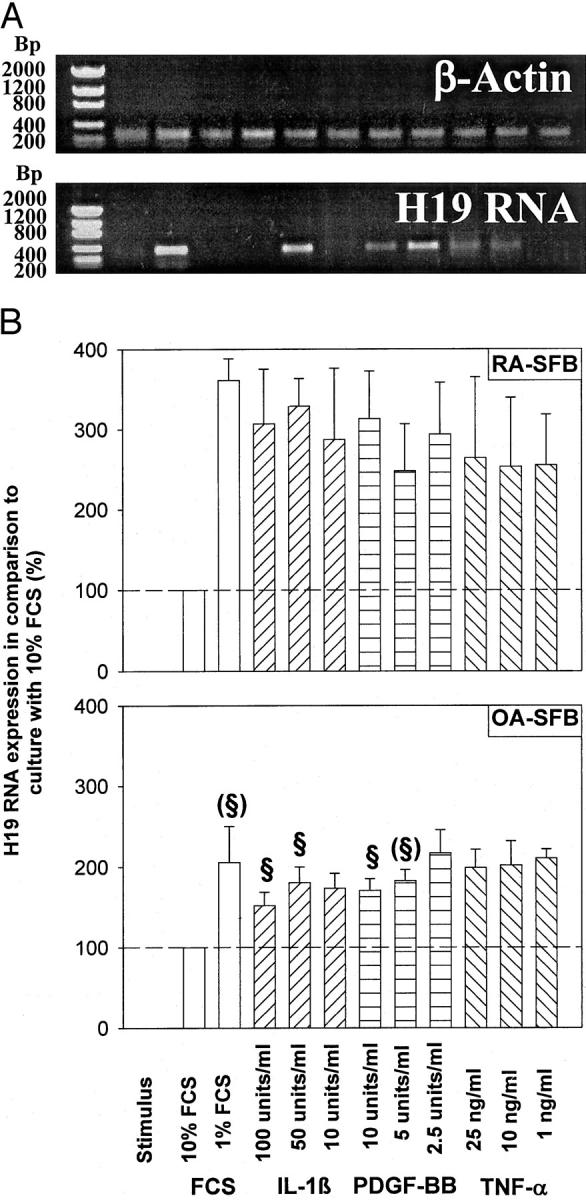
Regulation of H19 RNA expression in RA-SFBs/OA-SFBs on starvation (1% FCS) and stimulation with IL-1β, PDGF-BB, or TNF-α. A: Gel analysis of the H19 and β-actin PCR products from one RA patient. B: The means and standard errors of the mean (SEM) of the gel band quantification in four RA and five OA patients. Low constitutive expression of H19 RNA by RA-SFBs cultured in medium/10% FCS (A, B) was strongly (3.5-fold) induced after starvation for 96 hours in medium/1% FCS (B). This induction was significantly lower in OA-SFBs (approximately twofold). In neither RA- nor OA-SFBs, starvation-induced H19 expression was significantly influenced by the addition of IL-1β, PDGF, or TNF-α. M, molecular size standard given in bp. §, significant change.
In both RA-SFBs (Figure 6, A and B) ▶ and OA-SFBs (Figure 6B) ▶ , the MAP-kinases ERK-1/2 and the phosphatidylinositol-3 kinase significantly influenced H19 RNA expression after IL-1β or PDGF-BB stimulation, as shown by inhibitor studies with U0126 and Wortmannin, respectively.
Figure 6.
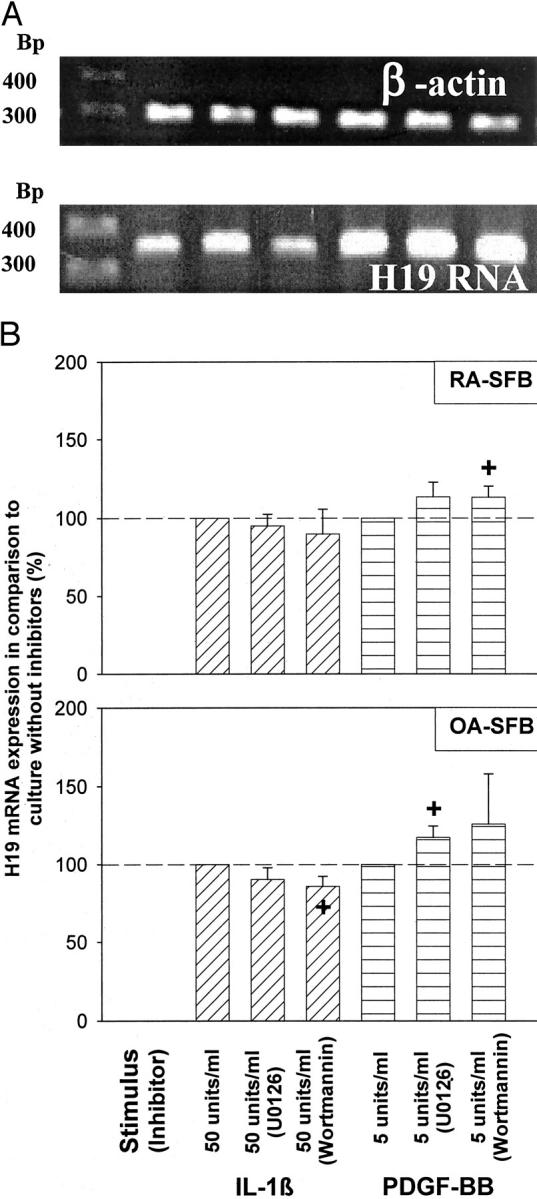
Influence of the blockade of MAP-kinase or PI3K signal transduction on the H19 RNA expression in RA-SFBs/OA-SFBs after stimulation with IL-1β or PDGF. A: Gel analysis of the H19 and β-actin PCR products from one RA patient. B: Means and standard errors of the mean (SEM) of the gel band quantification in three RA and three OA patients. In both RA- and OA-SFBs, the MAP-kinase ERK-1/2 and the phosphatidylinositol-3 kinase significantly influenced H19 RNA expression after IL-1β or PDGF-BB stimulation, as shown by inhibitor studies with U0126 and Wortmannin, respectively. M, molecular size standard given in bp.
Discussion
H19 RNA is variably, but strongly, expressed in RA-ST, with significantly higher levels than in N/JT-ST (P = 0.000), and numerically higher levels than in OA-ST and ReA-ST. In addition, the percentage of strongly H19-positive cells in the lining layer of RA-ST is significantly higher than that in OA- and N/JT-ST. This shows that H19 RNA is expressed in chronically inflamed ST, with some specificity for RA, providing one of the first reports on H19 expression in chronic, nontumoral disease. Specificity appears to be quantitative and not qualitative because H19 RNA is also expressed in inflammatory controls (OA, ReA) and even in normal ST. Although the physiological/pathophysiological role of H19 RNA remains elusive, 11 high expression in RA raises several considerations.
H19 RNA is abundantly expressed in embryonal tissue 38 and is therefore considered an indicator of embryonal dedifferentiation of adult tissue. Such dedifferentiation may also occur in RA-ST, a hypothesis supported by other fetal features, for example expression of embryonic growth factors of the wingless and frizzled families 39 and presence of mesenchymal stem cells. 40
H19 RNA is considered to be a tumor marker, because it is repressed after birth in all tissues except skeletal muscle, 15 and only re-expressed in a number of different tumors. 11 Although the hyperplastic RA-ST displays some features of a semitransformed tissue, 1-3 the expression of H19 RNA in OA and ReA (two unquestionably nonmalignant diseases) excludes that H19 RNA expression can be taken as a marker of malignancy in RA. By extension, the expression of H19 RNA not only in tumors but also in chronic inflammatory diseases questions the validity of this RNA species as a pure tumor marker.
Further support for this view derives from the finding that H19 expression is also increased in atherosclerotic plaque cells. 41 Because atherosclerosis is increasingly regarded as a chronic inflammatory disease based on the correlation of high CRP values with the grade of the disease and the local overproduction of chemoattractants, chemokines, and nitric oxide, 42 H19 RNA overexpression in atherosclerotic plaques may be partially driven by local proinflammatory mediators. In the case of RA, a contribution of the inflammatory status to the expression of H19 was indeed indicated by a correlation between H19 expression in the SM and rheumatoid factor positivity in patients with RA, OA, and ReA.
H19 RNA possibly exerts tumor-suppressor activity, because it is capable of inhibiting proliferation, clonogenicity, and tumorigenicity on transfection of cell lines 13,25 and because it is down-regulated in several tumors. 19,21-24 Up-regulation of H19 RNA expression in RA-ST may therefore reflect increased tumor-suppressor activity. Indeed, other tumor-suppressor factors are expressed in RA-ST, for example PTEN, 43 and p53. 44 However, PTEN is expressed at very low levels in RA-ST and p53 contains dominant-negative mutations, 4,45-47 rather suggesting a lack of tumor suppression. Because the H19 TATA-less promoter is efficiently repressed by wild-type p53, 48 such dominant-negative mutations of P53 may even directly contribute to increased H19 RNA expression in RA-ST. Clarification of the role of H19 RNA in RA awaits functional studies in transfected synovial cells and mutation analysis in ST.
H19 RNA expression is increased on stress induced by serum starvation (Figure 5) ▶ ; 26 its up-regulation in RA-ST may therefore mimic inflammatory/oxidative stress. Indeed, major pathological stress factors, including heat stress, shear stress, cytokine stress, and/or oxidative stress likely contribute to joint inflammation in RA. 7 In addition, an elevated H19 RNA expression is frequent in drug-resistant tumor cells after toxic influence 49 and in U937 cells exposed to concentrations of methotrexate (MTX) used for tumor therapy (data not shown). In the present study, however, there was no statistical indication for an influence of MTX treatment on the expression of H19 RNA in RA-ST; also, the potential contribution of other forms of stress to H19 RNA expression remains to be assessed.
H19 RNA expression was demonstrated in synovial Mφ and SFBs (Figures 2 and 4) ▶ , both strongly activated in RA, 1-3,50,51 raising the possibility that H19 RNA expression is a sign of cell activation. In RA- and OA-SFBs, this possibility was addressed by in vitro serum starvation (1% FCS) and subsequent stimulation with IL-1β, PDGF-BB, or TNF-α. Serum starvation strongly induced H19 RNA expression, in agreement with previous studies. 29 Interestingly, RA-SFBs showed a significantly (or borderline significantly) higher induction of H19 expression than OA-SFBs after serum starvation with or without the addition of IL-1β and PDGF-BB (Figure 5) ▶ , indicating that RA-SFBs may be intrinsically more susceptible to inflammatory/oxidative stress. On the other hand, no significant differences were observed in either RA- or OA-SFBs among groups subjected to serum starvation alone or including the addition of the above cytokines/growth factors. Therefore, an influence of the inflammatory micromilieu in RA-ST on the H19 RNA expression in SFBs is unlikely, although recent studies have demonstrated the regulation of H19 expression by hepatocyte growth factor in epithelial cells 52 or by interferon-α in melanoma cells. 53
In both RA- and OA-SFBs, the H19 expression in serum-starved and IL-1β- or PDGF-stimulated cells was significantly altered by inhibitors of the MAP-kinases ERK-1/2 and the phosphatidylinositol-3 kinase (Figure 6) ▶ . This is in agreement with the inhibition of hepatocyte growth factor-induced H19 expression by MAP-kinase inhibitors in epithelial cells 52 and underlines the importance of these two signal transduction pathways for H19 promoter transactivation.
Regarding possible functional consequences of H19 overexpression in RA-ST, recent studies have shown up-regulation of thioredoxin, MAP-kinase kinase 1 (MKK1), nuclear factor-κB, c-jun N-terminal kinase 2 (JNK2), TNF-α, and IL-6 in H19-transfected cells. 26,28 Also in the present study, the expression of H19 RNA in RA- and OA-SFBs showed a highly significant correlation with the mRNA expression for TIMP-2 in the same cells (r = 0.893; P = 0.007; n = 7), suggesting that the level of H19 expression in SFBs may influence the balance between tissue-degrading matrix metalloproteinases and their inhibitors.
Methylation of CpG islands in promoter regions is a common mechanism for the inactivation of imprinted genes (such as H19 and IGF-2), oncogenes, and proto-oncogenes. 54,55 Recent studies indicate that hypomethylation of such regions can lead to enhanced expression of c-myc and c-myb proteins. 56,57 Because DNA hypomethylation has been reported in peripheral blood and ST from patients with RA or SLE, 58,59 this mechanism could also contribute to the observed increase in H19 RNA expression. 60,61
In summary, significant overexpression of H19 RNA and its significantly increased sensitivity to starvation/cytokine regulation suggests a pathogenetic role of this oncofetal gene in RA, possibly reflecting embryonal dedifferentiation of the adult ST and/or ongoing inflammatory/oxidative stress.
Acknowledgments
We thank Dr. Ernesta Palombo-Kinne for critical revision of the manuscript and Dr. Dirk Pohlers for helpful suggestions and assistance with the preparation of the manuscript.
Footnotes
Address reprint requests to Bruno Stuhlmüller, Ph.D., Department of Internal Medicine, Rheumatology and Clinical Immunology, Charité University Hospital, Tucholskystrasse 2, D-10117-Berlin, Germany. E-mail: bruno.stuhlmueller@charite.de.
Supported by grants from the Deutsche Forschungsgemeinschaft (STU-224/1-1 and ER-73/6-1), the Bundesministerium für Bildung und Forschung (01-VM-9705/6 to B. S. and 01-ZZ-9602 to R. W. K.), and funds from the Chemische Industrie e. V. (to V. A. E.).
References
- 1.Gay S: Rheumatoid arthritis. Curr Opin Rheumatol 2001, 13:191-192 [DOI] [PubMed] [Google Scholar]
- 2.Kinne RW, Palombo-Kinne E, Emmrich F: Activation of synovial fibroblasts in rheumatoid arthritis. Ann Rheum Dis 1995, 54:501-504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamanishi Y, Firestein GS: Pathogenesis of rheumatoid arthritis: the role of synoviocytes. Rheum Dis Clin North Am 2001, 27:355-371 [DOI] [PubMed] [Google Scholar]
- 4.Aupperle KR, Boyle DL, Hendrix M, Seftor EA, Zvaifler NJ, Barbosa M, Firestein GS: Regulation of synoviocyte proliferation, apoptosis, and invasion by the p53 tumor suppressor gene. Am J Pathol 1998, 152:1091-1098 [PMC free article] [PubMed] [Google Scholar]
- 5.Tak PP, Zvaifler NJ, Green DR, Firestein GS: Rheumatoid arthritis and p53: how oxidative stress might alter the course of inflammatory diseases. Immunol Today 2000, 21:78-82 [DOI] [PubMed] [Google Scholar]
- 6.Dunger S, Neumann S, Zell R, Birch-Hischfeld E, Stelzner A, Paschke R, Kinne RW, Sickinger S: Mutation detection in mosaic situations: RNA mismatch assay and denaturing gradient gel electrophoresis are more sensitive than conventional cycle sequencing. Anal Biochem 2001, 294:89-93 [DOI] [PubMed] [Google Scholar]
- 7.Schett G, Tohidast-Akrad M, Steiner G, Smolen J: The stressed synovium. Arthritis Res 2001, 3:80-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burmester GR, Stuhlmuller B, Keyszer G, Kinne RW: Mononuclear phagocytes and rheumatoid synovitis. Mastermind or workhorse in arthritis? Arthritis Rheum 1997, 40:5-18 [DOI] [PubMed] [Google Scholar]
- 9.Erdmann VA, Szymanski M, Hochberg A, Groot N, Barciszewski J: Non-coding, mRNA-like RNAs database Y2K. Nucleic Acids Res 2000, 28:197-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erdmann VA, Barciszewska MZ, Hochberg A, de Groot N, Barciszewski J: Regulatory RNAs. Cell Mol Life Sci 2001, 58:960-977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ariel I, de Groot N, Hochberg A: Imprinted H19 gene expression in embryogenesis and human cancer: the oncofetal connection. Am J Med Genet 2000, 91:46-50 [DOI] [PubMed] [Google Scholar]
- 12.Hurst LD, Smith NG: Molecular evolutionary evidence that H19 mRNA is functional. Trends Genet 1999, 15:134-135 [DOI] [PubMed] [Google Scholar]
- 13.Juan V, Crain C, Wilson C: Evidence for evolutionarily conserved secondary structure in the H19 tumor suppressor RNA. Nucl Acids Res 2000, 28:1221-1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goshen R, Rachmilewitz J, Schneider T, de Groot N, Ariel I, Palti Z, Hochberg AA: The expression of the H-19 and IGF-2 genes during human embryogenesis and placental development. Mol Reprod Dev 1993, 34:374-379 [DOI] [PubMed] [Google Scholar]
- 15.Lustig O, Ariel I, Ilan J, Lev-Lehman E, de Groot N, Hochberg A: Expression of the imprinted gene H19 in the human fetus. Mol Reprod Dev 1994, 38:239-246 [DOI] [PubMed] [Google Scholar]
- 16.Ariel I, Sughayer M, Fellig Y, Pizov G, Ayesh S, Podeh D, Libdeh BA, Levy C, Birman T, Tykocinski ML, de Groot N, Hochberg A: The imprinted H19 gene is a marker of early recurrence in human bladder carcinoma. Mol Pathol 2000, 53:320-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ariel I, Ayesh S, Perlman EJ, Pizov G, Tanos V, Schneider T, Erdmann VA, Podeh D, Komitowski D, Quasem AS, de Groot N, Hochberg A: The product of the imprinted H19 gene is an oncofetal RNA. Mol Pathol 1997, 50:34-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biran H, Ariel I, de Groot N, Shani A, Hochberg A: Human imprinted genes as oncodevelopmental markers. Tumour Biol 1994, 15:123-134 [DOI] [PubMed] [Google Scholar]
- 19.Bertherat J, Logie A, Gicquel C, Mourrieras F, Luton JP, Le Bouc Y: Alterations of the 11p15 imprinted region and the IGFs system in a case of recurrent non-islet-cell tumour hypoglycaemia (NICTH). Clin Endocrinol 2000, 53:213-220 [DOI] [PubMed] [Google Scholar]
- 20.Casola S, Pedone PV, Cavazzana AO, Basso G, Luksch R, d’Amore ES, Carli M, Bruni CB, Riccio A: Expression and parental imprinting of the H19 gene in human rhabdomyosarcoma. Oncogene 1997, 14:1503-1510 [DOI] [PubMed] [Google Scholar]
- 21.Moulton T, Crenshaw T, Hao Y, Moosikasuwan J, Lin N, Dembitzer F, Hensle T, Weiss L, McMorrow L, Loew T, et al: Epigenetic lesions at the H19 locus in Wilms’ tumour patients. Nat Genet 1994, 7:440-447 [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Kahri AI, Heikkila P, Ilvesmaki V, Voutilainen R: H19 and insulin-like growth factor-II gene expression in adrenal tumors and cultured adrenal cells. J Clin Endocrinol Metab 1995, 80:492-496 [DOI] [PubMed] [Google Scholar]
- 23.Yballe CM, Vu TH, Hoffman AR: Imprinting and expression of insulin-like growth factor-II and H19 in normal breast tissue and breast tumor. J Clin Endocrinol Metab 1996, 81:1607-1612 [DOI] [PubMed] [Google Scholar]
- 24.Adriaenssens E, Lottin S, Dugimont T, Fauquette W, Coll J, Dupouy JP, Boilly B, Curgy JJ: Steroid hormones modulate H19 gene expression in both mammary gland and uterus. Oncogene 1999, 18:4460-4473 [DOI] [PubMed] [Google Scholar]
- 25.Hao Y, Crenshaw T, Moulton T, Newcomb E, Tycko B: Tumour-suppressor activity of H19 RNA. Nature 1993, 365:764-767 [DOI] [PubMed] [Google Scholar]
- 26.Ayesh S, Matouk I, Schneider T, Ohana P, Laster M, Al-Sharef W, De-Groot N, Hochberg A: Possible physiological role of H19 RNA. Mol Carcinog 2002, 35:63-74 [DOI] [PubMed] [Google Scholar]
- 27.Lottin S, Adriaenssens E, Dupressoir T, Berteaux N, Montpellier C, Coll J, Dugimont T, Curgy JJ: Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis 2002, 23:1885-1895 [DOI] [PubMed] [Google Scholar]
- 28.Adriaenssens E, Lemoine J, El Yazidi-Belkoura I, Hondermarck H: Growth signaling in breast cancer cells: outcomes and promises of proteomics. Biochem Pharmacol 2002, 64:797-803 [DOI] [PubMed] [Google Scholar]
- 29.Hayashida T, Eversole-Cire P, Jones PA, Sasaki H: Imprinted genes are up-regulated by growth arrest in embryonic fibroblasts. J Biochem 1997, 122:901-903 [DOI] [PubMed] [Google Scholar]
- 30.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, Medsger TA, Jr, Mitchell DM, Neustadt DH, Pinals RS, Schaller JG, Sharp JT, Wilder RL, Hunder GG: The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 1988, 31:315-324 [DOI] [PubMed] [Google Scholar]
- 31.Altman R, Asch E, Bloch D, Bole G, Borenstein D, Brandt K, Christy W, Cooke TD, Greenwald R, Hochberg M, Howell D, Kaplan D, Koopman W, Longley S, III, Mankin H, McShane DJ, Medsger T, Jr, Meenan R, Mikkelsen W, Moskowitz R, Murphy W, Rothschild B, Segal M, Sokoloff L, Wolfe F: Development of criteria for the classification and reporting of osteoarthritis. Classification of osteoarthritis of the knee. Diagnostic and Therapeutic Criteria Committee of the American Rheumatism Association. Arthritis Rheum 1986, 29:1039-1049 [DOI] [PubMed] [Google Scholar]
- 32.Zimmermann T, Kunisch E, Pfeiffer R, Hirth A, Stahl H-D, Sack U, Laube A, Liesaus E, Roth A, Palambo-Kinne E, Emmrich F, Kinne RW: Isolation and characterization of rheumatoid arthritis synovial fibroblasts from primary culture—primary culture cells markedly differ from fourth-passage cells. Arthritis Res 2001, 3:72-76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, Copeland RA, Magolda RL, Scherle PA, Trzaskos JM: Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem 1998, 273:18623-18632 [DOI] [PubMed] [Google Scholar]
- 34.Wymann MP, Bulgarelli-Leva G, Zvelebil MJ, Pirola L, Vanhaesebroeck B, Waterfield MD, Panayotou G: Wortmannin inactivates phosphoinositide 3-kinase by covalent modification of Lys-802, a residue involved in the phosphate transfer reaction. Mol Cell Biol 1996, 16:1722-1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chirgwin JM, Pryzbyla AE, MacDonald J, Rutter WJ: Isolation of biologically active ribonucleic acid from sources enriched in ribonucleases. Biochemistry 1979, 18:5294-5299 [DOI] [PubMed] [Google Scholar]
- 36.Kriegsmann J, Keyszer G, Geiler T, Gay RE, Gay S: A new double labeling technique for combined in situ hybridization and immunohistochemical analysis. Lab Invest 1994, 71:911-917 [PubMed] [Google Scholar]
- 37.Franz JK, Pap T, Hummel KM, Nawrath M, Aicher WK, Shigeyama Y, Müller-Ladner U, Gay RE, Gay S: Expression of sentrin, a novel antiapoptotic molecule, at sites of synovial invasion in rheumatoid arthritis. Arthritis Rheum 2000, 43:599-607 [DOI] [PubMed] [Google Scholar]
- 38.Goshen R, Rachmilewitz J, Schneider T, de-Groot N, Ariel I, Palti Z, Hochberg AA: The expression of the H-19 and IGF-2 genes during human embryogenesis and placental development. Mol Reprod Dev 1993, 34:374-379 [DOI] [PubMed] [Google Scholar]
- 39.Sen M, Lauterbach K, El-Gabalawy H, Firestein GS, Corr M, Carson DA: Expression and function of wingless and frizzled homologs in rheumatoid arthritis. Proc Natl Acad Sci USA 2000, 97:2791-2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Bari C, Dell’Accio F, Tylzanowski P, Luyten FP: Multipotent mesenchymal stem cells from adult human synovial membrane. Arthritis Rheum 2001, 44:1928-1942 [DOI] [PubMed] [Google Scholar]
- 41.Han DK, Khaing ZZ, Pollock RA, Haudenschild CC, Liau G: H19, a marker of developmental transition, is reexpressed in human atherosclerotic plaques and is regulated by the insulin family of growth factors in cultured rabbit smooth muscle cells. J Clin Invest 1996, 97:1276-1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shoenfeld Y, Sherer Y, Harats D: Artherosclerosis as an infectious, inflammatory and autoimmune disease. Trends Immunol 2001, 22:293-295 [DOI] [PubMed] [Google Scholar]
- 43.Pap T, Franz JK, Hummel KM, Jeisy E, Gay R, Gay S: Activation of synovial fibroblasts in rheumatoid arthritis: lack of expression of the tumour suppressor PTEN at sites of invasive growth and destruction. Arthritis Res 2000, 2:59-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Firestein GS, Nguyen K, Aupperle KR, Yeo M, Boyle DL, Zvaifler NJ: Apoptosis in rheumatoid arthritis: p53 overexpression in rheumatoid arthritis synovium. Am J Pathol 1996, 149:2143-2151 [PMC free article] [PubMed] [Google Scholar]
- 45.Firestein GS, Echeverri F, Yeo M, Zwaifler NJ, Green DR: Somatic mutations in the p53 tumor suppressor gene in rheumatoid arthritis synovium. Proc Natl Acad Sci USA 1997, 94:10985-10900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han Z, Boyle DL, Shi Y, Green DR, Firestein GS: Dominant-negative p53 mutations in rheumatoid arthritis. Arthritis Rheum 1999, 42:1088-1092 [DOI] [PubMed] [Google Scholar]
- 47.Yamanashi Y, Boyle DL, Pinkosi MJ, Mahboubi A, Lin T, Han Z, Zvaifler NJ, Green DR, Firestein GS: Regulation of joint destruction and inflammation by p53 in collagen-induced arthritis. Am J Pathol 2002, 160:123-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dugimont T, Montpellier C, Adriaenssens E, Lottin S, Dumont L, Iotsova V, Lagrou C, Stehelin D, Coll J, Curgy JJ: The H19 TATA-less promoter is efficiently repressed by wild-type tumor suppressor gene product p53. Oncogene 1998, 16:2395-2401 [DOI] [PubMed] [Google Scholar]
- 49.Doyle LA, Yang W, Rishi AK, Gao Y, Ross DD: H19 gene overexpression in atypical multidrug-resistant cells associated with expression of a 95-kilodalton membrane glycoprotein. Cancer Res 1996, 56:2904-2907 [PubMed] [Google Scholar]
- 50.Kinne RW, Bräuer R, Stuhlmüller B, Palombo-Kinne E, Burmester GR: Macrophages in rheumatoid arthritis. Arthritis Res 2000, 2:189-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kinne RW, Liehr T, Beensen V, Kunisch E, Zimmermann T, Holland H, Pfeiffer R, Stahl H-D, Lungershausen W, Hein G, Roth A, Emmrich F, Claussen U, Froster UG: Mosaic chromosomal aberrations in synovial fibroblasts of patients with rheumatoid arthritis, osteoarthritis, and other inflammatory joint diseases. Arthritis Res 2001, 3:319-330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adriaenssens E, Lottin S, Berteaux N, Hornez L, Fauquette W, Fafeur V, Peyrat JP, LeBourhis X, Hondermarck H, Coll J, Dugimont T, Curgy JJ: Cross-talk between mesenchyme and epithelium increases H19 gene expression during scattering and morphogenesis of epithelial cells. Exp Cell Res 2002, 275:215-229 [DOI] [PubMed] [Google Scholar]
- 53.Certa U, Seiler M, Padovan E, Spagnoli GC: Interferon-α sensitivity in melanoma cells: detection of potential response marker genes. Recent Results Cancer Res 2002, 160:85-91 [DOI] [PubMed] [Google Scholar]
- 54.Veilleux C, Bernardino J, Gibaud A, Niveleau A, Malfoy B, Dutrillaux B, Bourgeois CA: Changes in methylation of tumor cells: a new in situ quantitative approach on interphase nuclei and chromosomes. Bull Cancer 1995, 82:939-945 [PubMed] [Google Scholar]
- 55.Nyce J: Drug-induced DNA hypermethylation and drug resistance in human tumors. Cancer Res 1989, 49:5829-5836 [PubMed] [Google Scholar]
- 56.Stephenson J, Akdag R, Ozbek N, Mufti GJ: Methylation status within exon 3 of the c-myb gene as a prognostic marker in myeloma and leukemia. Leuk Res 1993, 17:291-293 [DOI] [PubMed] [Google Scholar]
- 57.Evans JL, Boyle WJ, Ting JP: Molecular basis of elevated c-myb expression in the abnormal L3T4-, Lyt-2- T lymphocytes of autoimmune mice. J Immunol 1987, 139:3497-3505 [PubMed] [Google Scholar]
- 58.Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M: Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum 1990, 33:1665-1673 [DOI] [PubMed] [Google Scholar]
- 59.Corvetta A, Della Bitta R, Luchetti MM, Pomponio G: 5-Methylcytosine content of DNA in blood, synovial mononuclear cells and synovial tissue from patients affected by autoimmune rheumatic diseases. J Chromatogr 1991, 566:481-491 [DOI] [PubMed] [Google Scholar]
- 60.Kim YI, Logan JW, Mason JB, Roubenoff R: DNA hypomethylation in inflammatory arthritis: reversal with methotrexate. J Lab Clin Med 1996, 128:165-172 [DOI] [PubMed] [Google Scholar]
- 61.Neidhart M, Rethage J, Kuchen S, Kunzler P, Crowl RM, Billingham ME, Gay RE, Gay S: Retrotransposable L1 elements expressed in rheumatoid arthritis synovial tissue: association with genomic DNA hypomethylation and influence on gene expression. Arthritis Rheum 2000, 43:2634-2647 [DOI] [PubMed] [Google Scholar]