Abstract
Cloned embryos produced by somatic cell nuclear transfer (SCNT) display a plethora of phenotypic characteristics that make them different from fertilized embryos, indicating defects in the process of nuclear reprogramming by the recipient ooplasm. To elucidate the extent and timing of nuclear reprogramming, we used microarrays to analyze the transcriptome of mouse SCNT embryos during the first two cell cycles. We identified a large number of genes mis-expressed in SCNT embryos. We found that genes involved in transcription and regulation of transcription are prominent among affected genes, and thus may be particularly difficult to reprogram, and these likely cause a ripple effect that alters the transcriptome of many other functions, including oxidative phosphorylation, transport across membrane, and mRNA transport and processing. Interestingly, we also uncovered widespread alterations in the maternal (i.e. non transcribed) mRNA population of SCNT embryos. We conclude that gene expression in early SCNT embryos is grossly abnormal, and that this is at least in part the result of incomplete reprogramming of transcription factor genes.
Keywords: embryo, somatic cell nuclear transfer, reprogramming, microarray, transcription, transcriptome
Introduction
Cloning by somatic cell nuclear transfer (SCNT) is a remarkable process that relies on the oocyte’s ability to act upon the somatic nucleus and to transform it into a nucleus compatible with long-term embryonic development. This process of nuclear “reprogramming” is particularly remarkable considering the dramatic differences between somatic and early embryonic cells. These include fundamentally different cell cycles and cell cycle regulation (e.g., cleavage without growth), strikingly different gene expression profiles (Latham et al, 1991) revealed by two-dimensional gel electrophoresis, diverging modes of carbohydrate metabolism and energy production, a different array of amino acid transporters, glucose transporters, and ion transporters, (Aghayan et al, 1992; Baltz et al, 1991a, 1991b, 1993; Carayannopoulos et al, 2000, 2004; Chi et al, 2000; Hogan et al, 1991; Leppens-Luisier et al, 2001; Moley et al, 1998; Morita et al, 1994; Pantaleon and Kaye, 1998; Pantaleon et al, 2001; Van Winkle, 2001), different mechanisms of osmoregulation and pH regulation (Baltz et al, 1991a, 1991b, 1993; Edwards et al, 1998a,b; Zhao and Baltz, 1996; Zhao et al, 2005), and dramatic differences in mitochondrial ultrastructure and activity (Hillman and Tasca, 1969; Matsumoto et al, 1998; Shepard et al, 1998, 2000; Sathananthan and Trouson, 2000).
Over the course of the 50 years during which SCNT studies have been performed, first in amphibians (King and Briggs, 1955) and more recently in mammals (for review see Latham KE, 2004 and references therein; Campbell et al, 2005), it has become clear that the rate of success (i.e., development to term) is quite low (1–5%). Although incomplete nuclear reprogramming is often put forth as an explanation for this poor success, the nature of such a deficiency has never been defined.
The cell type-specific expression of transcription factors (both activators and repressors) likely results in a distinct global pattern of gene expression that provides a molecular signature that defines the differentiated state of a somatic cell. The expression of these transcription regulators, a priori, must be stable in order to maintain a stable state of differentiation, and indeed such seems to be the case (e.g., Hox genes in Drosophila). Thus, genes encoding transcription factors may be among the most difficult genes for the oocyte to reprogram during cloning. Failure to reprogram even a small number of key transcription factor genes could readily lead to a “ripple effect” resulting in aberrant expression of entire networks of downstream target genes.
To determine the degree to which inefficient reprogramming of transcription factor genes may underlie poor cloning success, to examine clones for disruption in the expression of other genes, and to identify specific biological processes that are likely disrupted as a consequence, we analyzed the transcriptome of clones immediately following SCNT using microarrays. In contrast to previous studies that focused on surviving clones of advanced development (Humpherys et al, 2002; Smith et al, 2005), we focused on the first two cell cycles, because these stages encompass the earliest interactions between ooplasm and donor nuclei, and because aberrant gene regulation at these early stages can have profound consequences for long-term development. Our goal was therefore to determine to what degree SCNT embryos at these early stages resemble normal embryos of high developmental potential, and to what degree the somatic cell program might remain expressed.
We find that, although the transcript profiles of SCNT and fertilized embryos are quite similar at the one-cell stage, aberrant gene transcription is nevertheless evident even at this early stage, along with apparent disruptions in the regulation of maternally encoded (i.e., oocyte-accumulated) mRNAs. During the two-cell stage, as transcriptional activation ensues, the number of aberrantly transcribed genes in SCNT embryos increases by nearly two orders of magnitude to nearly 1,000 genes, indicating a substantial continued expression of the somatic cell program. As predicted, the aberrantly expressed mRNAs include many involved in transcription, and also many involved in mRNA processing, oxidative phosphorylation, metabolism, protein biosynthesis, protein degradation, protein modification, and transmembrane solute transport.
Materials and Methods
Preparation and collection of mouse embryos
Ovulated eggs were obtained from adult (B6D2)F1 females 8–12 weeks of age by superovulation as described (Chung et al., 2002; Gao et al., 2003, 2004b). Adherent cumulus cells were removed by hyaluronidase treatment and the eggs were cultured in CZB medium supplemented with glucose (Chung et al., 2002). SCNT was performed as described (Chung et al., 2002; Gao et al., 2003, 2004b). At the end of the procedure, cloned constructs were activated by 5.5 h of culture in Ca2+-free CZB medium supplemented with 10 mM Sr2+ and 5 μg/ml cytochalasin B (Chung et al., 2002). Cloned constructs were cultured in minimal essential medium alpha formulation (MEMα) medium as described (Gao et al., 2004b) with or without α-amanitin (24 μg/ml). For SCNT, adherent adult cumulus cells (presumably G1 phase) from ovulated oocytes were employed as nuclear donors. Diploid parthenogenetically activated embryos were obtained using the same activation protocol of clones. The parthenotes were obtained from the same pools of oocytes used to make cloned embryos and were activated at the same time. Parthenogenetic embryos resemble normal fertilized embryos with respect to culture requirements, but have the added advantage that they are activated and develop in close temporal synchrony with the activated cloned embryos. Embryos fertilized in vivo (henceforth referred to as fertilized) were obtained by mating (B6D2)F1 mice after injection of females 8–12 weeks of age with Pregnant Mare Serum Gonadotropin (PMSG) and human Chorionic Gonadotrophin (hCG), as described (Chung et al., 2002; Gao et al., 2003, 2004b). Cloned, parthenogenetic, and fertilized embryos were cultured at 37°C in an atmosphere of 5% CO2 in air.
RNA extraction, labeling, and hybridization
For each experimental/treatment group, four pools of 20 embryos were collected and transferred to 20 μl of extraction buffer (Picopure, Arcturus). The tube was incubated at 42° C for 30 min and then stored at −70 °C. RNA extraction was performed with the Picopure RNA extraction kit according to manufacturer instructions for small sample preparation. For each sample, the mRNA population was reverse transcribed. The cDNA was employed for a first round of in vitro transcription, followed by random priming and a second round of reverse transcription and in vitro transcription to achieve a linear amplification (Affymetrix Small Sample Technical Bulletin, www.affymetrix.com) with the following minor modifications: the initial volume for mRNA annealing was raised to 5 μl, and the conditions for reverse transcription were 30 min at 42° C followed by 30 min at 45° C to increase the reaction efficiency in GC rich regions of mRNA. The final yield of biotinylated cRNA was 28.5 to 83.4 μg for one-cell stage embryos and 26 to 88.5 μg for two-cell stage embryos; 20 μg of cRNA per replicate were fragmented and 10 μg hybridized to Affymetrix MOE 430 2.0 Gene Chips in the Penn Microarray Facility, then washed and stained on fluidic stations, and scanned according to the manufacturer’s instructions.
Microarray data analysis
Microarray Analysis Suite 5.0 (MAS, Affymetrix) was used to quantify microarray signals with default analysis parameters and global scaling to target a mean equal to 150 signal units. Quality control parameters for all samples were within ranges shown in Table 1. Tabular data for all samples are available at the Gene Expression Omnibus (GEO) repository (www.ncbi.nlm.gih.gov/geo). The MAS metric output was loaded into GeneSpring v7 (Silicon genetics) with per chip normalization to the 50th percentile and per gene normalization to the median. To minimize false positive signals, only genes called “Present” in at least three out of four replicates in one embryo kind/condition were used for further analysis with all statistical packages. The K-means hierarchical clustering (HCL) of GeneSpring v7 was used among samples at the same developmental stage to divide them into groups based on their expression patterns and to produce groups with a high degree of similarity within groups and low degree of similarity between groups. The outputs for these analyses are shown in Figs. 1 and 2.
Table 1.
Quality control parameter for array hybridization in different kind of embryos and treatment.
| one-cell | two-cell | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | Fert. | Fert.+a | SCNT | SCNT+a | Parth. | Fert. | Fert.+a | SCNT | SCNT+a | Parth. |
| Scale factor | 1.4–2.9 | 0.6–1 | 1.3–1.9 | 1.7–2.3 | 0.9–1.1 | 0.8–1 | 1.3–1.76 | 0.7–1 | 1.6–2.2 | 1.4–1.7 |
| Background | 35.8–50.5 | 47.6–63.1 | 35.8–40.7 | 37.5–47.4 | 55.1–63.4 | 46.1–64.5 | 51.8–62.5 | 53.2–63.9 | 41–49.8 | 49.5–60.3 |
| % P call | 33.5–36.1 | 38–39 | 35.5–37.6 | 33.2–35.2 | 37.2–38.2 | 36.2–38.8 | 31.4–32.3 | 39.5–41.1 | 29.4–31.4 | 34.5–36 |
| Actin 3/5 | 3.3–12.4 | 3.7–4.4 | 4.8–15 | 4.2–13.9 | 3.9–5.5 | 4.4–7.2 | 4.2–5.9 | 4.9–7.4 | 4.2–4.9 | 4.7–5.2 |
| GAPDH 3/5 | 1.5–6.2 | 5.6–6.2 | 1.7–6.2 | 1.9–7.7 | 5.1–6.2 | 5.6–7.2 | 4.2–6.1 | 4.2–6.8 | 4–6.2 | 4.7–6.6 |
a = α-amanitin in culture medium; Fert. = fertilized embryos; SCNT = somatic cell nuclear transfer embryos; Parth. = parthenogenetic embryos.
Figure 1.

K means hierarchical cluster (HCL) of one-cell samples compiled after filtering for presence call in at least 3 of 4 replicates of at least one of the conditions. C cloned, C+a cloned + α-amanitin, P parthenotes, F in vivo fertilized, F+a in vivo fertilized + α-amanitin.
Figure 2.
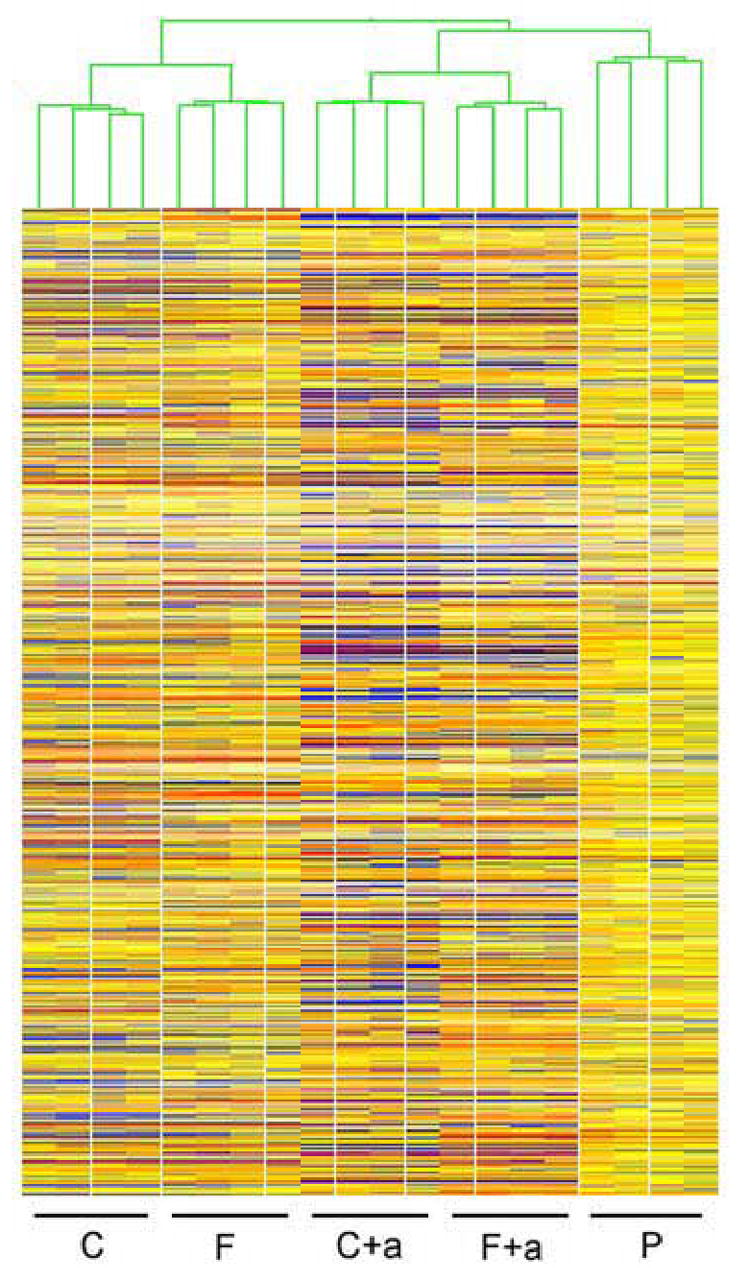
K means hierarchical cluster (HCL) of two-cell samples compiled after filtering for presence call in at least 3 of 4 replicates of at least one of the conditions. C cloned, C+a cloned + α-amanitin, P parthenotes, F in vivo fertilized, F+a in vivo fertilized + α-amanitin.
It is important to note that, although the Affymetrix MOE430 2.0 array interrogates one gene with every probe set, 14.7% of the genes present on the array are represented by more than one probe set. All analyses described were performed using the Affymetrix probe set lists, except when noted where gene numbers were used to avoid redundancy.
The filtered MAS metrics output was loaded into TIGR-MEV v3.0.3 (Saeed et al, 2003). The Statistical Analysis of Microarray (SAM; Tusher et al., 2001) algorithm was applied to identify genes with significant differences among samples at the 1% false discovery rate (FDR).
Fold-changes of expression differences between stages and conditions were calculated following SAM analysis. The resulting lists of differentially expressed (≥ two-fold) genes (Supplemental Material, Tables S1–S6) were imported into Expression Analysis Systematic Explorer (EASE, version 2.0) to analyze gene ontology for over-representation (Hosack et al., 2003). EASE is an algorithm designed to analyze a list of candidate genes against a set population (in our case the list of genes detected on the GeneChip) and to report a score that is the expression of the likelihood of over-representation in the Gene Ontology (GO) annotation categories for biological process, molecular function, or cellular component. The EASE score was calculated for likelihood of over-representation of annotation classes, and only GO biological processes with an EASE score less than 5% are shown. It is important to note that a significant EASE score does not relate to an increased fold-change or overall expression significance, but merely a higher than expected number of transcripts falling into a GO annotation category. The filtered list of transcripts over-expressed in clones versus fertilized and parthenogenetic embryos, and also with α-amanitin sensitive (i.e., reduced by α-amanitin treatment) expression, and different in expression from parthenogenetic embryos at the two-cell stage was further imported into Ingenuity Pathway Analysis (IPA, www.ingenuity.com) in order to detect networks detailing physical association or functional interaction among transcripts falling into different GO annotation categories.
Quantitative RT-PCR analysis
Groups of 25–50 embryos were collected, and total RNA was isolated as described above. Thirteen genes were selected for analysis at the one-cell and two-cell stage, and their mRNAs quantified by reverse transcription followed by real time PCR (qRT-PCR). The corresponding ABI TaqMan gene expression IDs were: Zar1 (Mm-00558078), Yy1 (Mm-00456392_m1), Fos (Mm00487425_m1), Cpa1 (Mm_00465942_m1), H1foo (Mm00506768_m1), Zfp352 (Mm-02528443_s1), Por (Mm00435876_m1), Eif3s12 (Mm-00503812_m1), Maf1 (Mm-00593524_g1), Klf4 (Mm-00516104_m1), Sra1 (Mm-00491755_m1), Uqcrb (Mm-00835346_gH), Psmc3 (Mm-00477177_m1). Three replicates were used for each qRT-PCR reaction, and each mRNA was analyzed 2–3 times per replicate. Minus RT and minus primers/probe reactions served as controls. Quantification was normalized to the endogenous histone H2A [Mm-00501974_s1, (Hisst2ah2aa10)] within the log linear phase of the amplification curve using the comparative Ct method (ABI PRISM 7700 Sequence detection System, user bulletin 32). These mRNAs were selected to be examined by qRT-PCR because of their apparent abundances as judged by the micorarray hybridization signals and as representatives of specific functional categories (see Results).
Experimental design
The objectives of this study were to determine the timing and extent of nuclear reprogramming during the first two cell cycles of SCNT embryo development, and to identify specific genes or categories of genes that could account for the observed differences in phenotype between SCNT and fertilized embryos. To meet these objectives, we adopted a microarray-based approach for transcript profiling that has been used successfully for mouse oocytes and preimplantation embryos (Zeng et al, 2004; Zeng and Schultz, 2005; Pan et al, 2005).
Although simple in concept, such studies are complicated by technical aspects of SCNT embryo production and culture. First, it is difficult to obtain in vivo fertilized embryos that are developing in close synchrony with SCNT embryos, so that effects of asynchrony on relative mRNA abundances could arise. To control for possible effects of asynchrony, we employed parthenogenetic controls, which are activated at the same time as SCNT embryos using the same method, and from the same pools of eggs as those employed to prepare the SCNT embryos. The use of parthenogenetic controls also accounts for possible differences that might be related to absence of a fertilizing sperm and activation in response to chemical treatment rather than sperm factors. For this reason, parthenogenetic controls provided a significant advantage over, for example, in vitro fertilized embryos, as a control for possible asynchrony, because they addressed additional aspects of the procedures used to produce SCNT embryos.
Second, SCNT embryos display radically altered culture medium preferences when compared to normal embryos (Chung et al, 2002). No single culture medium has yet been identified that is optimized for both SCNT and normal embryos. In fact, many SCNT embryos arrest in media optimized for embryo culture, and many fertilized embryos arrest in the somatic cell culture media favored by SCNT embryos (Chung et al, 2002; Gao et al., 2004b). Because our objective was to explore the limits and timing of reprogramming, it was essential that the analyses be performed on embryos of the highest developmental potential and cultured in the best media available for each type of embryo. This would avoid comparisons between embryos that are developmentally viable and embryos that are already developmentally arrested, or between two kinds of embryos both of which are known a priori to be compromised. Such comparisons would yield artifactual results that would be unrelated to basic questions related to nuclear reprogramming and how well clones resemble normal embryos. We therefore adopted the strategy of employing the best available culture media for each kind of embryo, namely MEMα for SCNT embryos and KSOM for parthenotes and fertilized embryos. SCNT embryos develop very poorly in KSOM even to the four-cell stage, making an analysis of SCNT embryo in this medium uninformative (Chung et al, 2002; Gao et al., 2004b). Fertilized embryos and parthenogenetic embryos have been cultured in MEMα. Although this medium has been found to be superior to a number of grossly sub-optimum media, KSOM remains superior to MEMα for such embryos (Chung et al, 2002). Our strategy therefore allowed us to compare embryos of all three classes under those culture conditions that support the highest in vitro efficiency achievable beyond the first two cell cycles and, more importantly, to display the greatest rates of development to the blastocyst stage, the highest quality of blastocysts, and the most consistent rates of development to term achievable. This permitted our microarray analysis to reveal specific effects of SCNT and nuclear function without concern that such differences were being contributed by less specific deficiencies related to simple developmental arrest.
This strategy, however, creates a secondary need to account for possible effect of the different culture media. To resolve this issue, we applied two sets of controls. In one control study, we undertook an independent microarray comparison between fertilized two-cell embryos cultured in either KSOM or MEMα, using the same developmental time point and data analysis parameters described above (Fig. 3). This comparison between fertilized embryos cultured in the two media yielded a set of 145 genes, the expression of which could potentially be altered by the choice of culture medium (Supplemental material, Table S7). This set of media-sensitive genes was later compared to the lists of genes differentially expressed between two-cell stage SCNT and normal embryos in order to reveal potential effects of culture medium. We observed only 12 genes in common between the media-sensitive list (Table S7) and the lists of genes altered in two-cell SCNT embryos (see Supplemental material, Tables S3–S6), indicating that the potential effect of the culture systems on the overall microarray results is highly limited. As a second test for possible effects of culture medium, we employed qRT-PCR analysis to compare gene expression between SCNT, fertilized, and parthenogenetic control embryos cultured either in KSOM or MEMα (Fig. 4). These analyses revealed little if any variation between samples of fertilized control embryos cultured in different media (compare FK and FM in Fig. 4). Although for some of the genes assayed slightly greater differences were observed between parthenotes cultured in the two media, qualitatively identical directional differences in gene expression were seen even between SCNT and parthenotes, regardless of the media employed. Collectively, these data indicate that the culture media employed for maintaining the highest developmental potential amongst SCNT and control embryos while in culture did not adversely affect the discovery of differences in gene expression. This result confirms the robustness of the statistical analysis.
Figure 3.

K means hierarchical cluster (HCL) of two-cell samples compiled after filtering for presence call in at least 3 of 4 replicates of at least one of the conditions. F KSOM, two-cell stage embryos cultured in KSOM medium, F MEMa, two-cell stage embryos cultured in MEMα.
Figure 4.
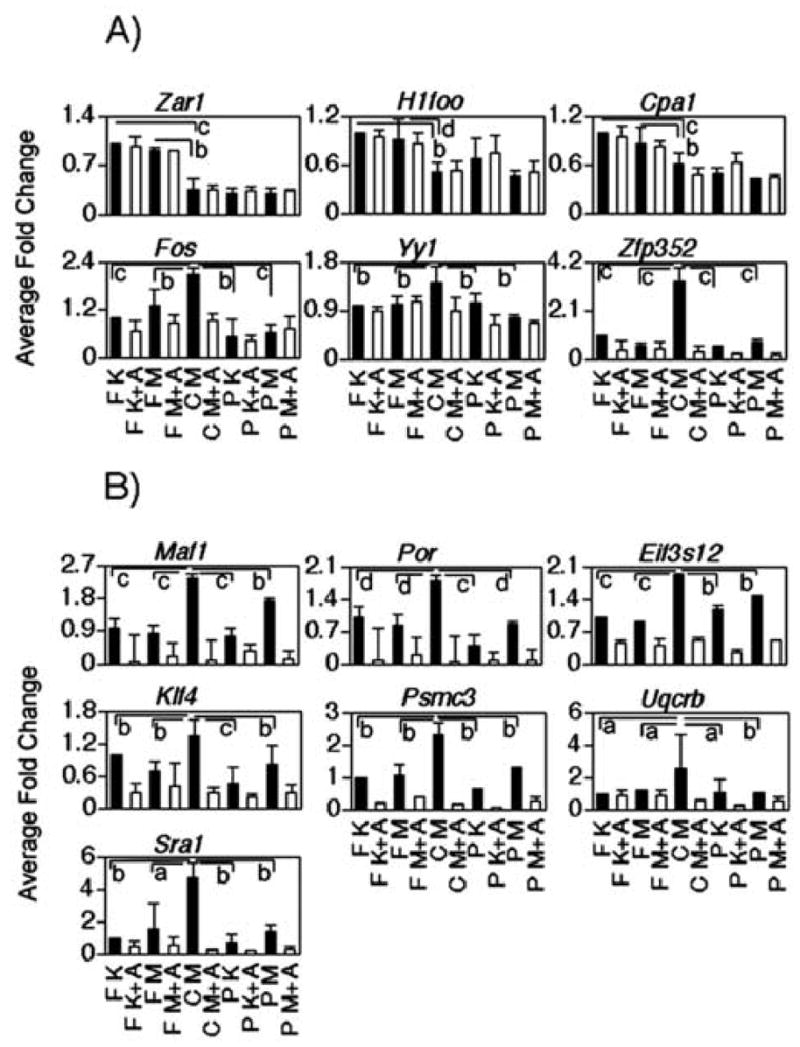
Real time PCR derived expression patterns of selected genes at the one-cell (Panel A) and two-cell (Panel B) stage cultured in different media with or without α-amanitin. Y axes indicate the relative fold change to fertilized embryos cultured in KSOM (reference treatment, expression adjusted to = 1.0). F = fertilized embryos; C = SCNT embryos, P = parthenotes; A= amanitin treatment, K = KSOM culture medium, M = MEMα culture medium. Significant difference among kind of embryos and culture media are indicated as follows: a: p<0.1; b: p<0.05; c: p<0.01; d: p<0.001.
The final requirement for our array analysis was to be able to distinguish between effects on maternal transcript populations and effects on transcribed genes. To address this requirement, we included in our experimental design for both microarray and qRT-PCR experiments SCNT and fertilized embryos that were cultured in the presence of α-amanitin, a potent RNA polymerase II inhibitor. The treated embryos would thus display α-amanitin-dependent reductions in mRNA abundance for transcribed genes.
Last, it is worth noting that our approach to identify sets of differentially expressed genes incorporated stringent parameters for false discovery rate, statistical significance of difference, and fold cutoff, combined with sequential filtering of gene sets based on differential expression first between SCNT and fertilized embryos, then between SCNT and parthenogenetic controls, and finally distinctions based on α-amanitin sensitivity. The gene sets obtained are therefore highly reliable, and thus capable of providing significant new insight into how genes are differentially regulated between SCNT and control embryos, and hence the extent and timing of nuclear reprogramming.
Results
Overview of microarray results
The microarray data sets obtained in this study are available in tabular form from the Gene Expression Omnibus Repository (www.ncbi.nlm.nih.gov/geo). Among the entire series of samples (Figs 1–2, one-cell, two-cell, α-amanitin-treated and untreated) expression of between 13,230 and 18,500 mRNAs was detected (Table 1). This range reflects differences in the complexity of the mRNA populations of different stages/treatments of embryos. The quality control parameter for all the samples were within the following ranges: scale factor 0.6 to 1.9 (accepted range: 0.5 to 5.0), and background 35.8 to 64.5 (accepted range: 20 to 100); percent IDs detected 29.4 to 41.1; actin 3′/5′ signal ratio 3.3 to 12.4; GADPH 3′/5′ signal ratio 1.5 to 7.7 (Table 1). The quality control data are in agreement with that reported in two other studies using the same array platform (Zeng et al, 2004; Pan et al, 2005) as well as within the ranges recommended by Affymetrix. All the quality control parameters, as well as the internal and spiked controls in place to ensure correct mRNA processing and preparation, confirmed that the datasets obtained were of high quality.
It is often assumed that reprogramming must occur within hours of nuclear transfer. Published studies, however, indicate that clones manifest unusual characteristics during these early stages indicative of slow or incomplete reprogramming (Gao et al., 2003, 2004b; review, Latham, 2004, 2005). No study to date has attempted to measure the degree of similarity or difference between SCNT and fertilized embryos. We used K-means hierarchical clustering (HCL) to ascertain the overall similarities/differences of embryos derived from the different treatments (Figs. 1 and 2). At both developmental stages, replicate samples of the same kind/condition clustered together and apart from other embryo kinds/conditions, which indicates that SCNT are indeed significantly different from control embryos with respect to transcriptome composition. Additionally, this clustering pattern indicates a high degree of reproducibility and small biological variability among samples of a given kind of embryo. It is noteworthy that the HCL output of one-cell stage embryos grouped embryos by kind and treatment, indicating that SCNT embryos at this stage of development are already different from both normal and parthenogenetic embryos (Fig. 1). Moreover, the clustering of the α-amanitin treated samples apart from non-treated ones indicates that the α-amanitin effect is already sizeable at this early stage.
Three other aspects of the data argue for an early effect of the donor nucleus on the SCNT embryo phenotype. First, we see that the two-cell stage samples treated with α-amanitin (both fertilized and SCNT embryos) are distinct from the three non α-amanitin-treated groups, but that the α-amanitin treated samples retain their cluster grouping by kind of embryo (i.e., SCNT embryos remain separate from fertilized embryos). This indicates that the maternal (i.e., not diminished by α-amanitin treatment) mRNA population is regulated differently between SCNT and fertilized embryos due to the difference in nuclear origin, a point that will be addressed further below. Second, one-cell parthenogenetic embryos cluster apart from both SCNT and fertilized embryos, at a position intermediate between the latter two groups. This indicates that even before the first cleavage division, the cloned embryo transcriptome has diverged even from that of parthenogenetic controls, which are activated simultaneously from the same pool of eggs and developing in close synchrony with SCNT embryos. Third, we observe that the degree of difference between SCNT and fertilized embryos increases between the one-cell and two-cell stages. If nuclear reprogramming occurred rapidly after SCNT, then we would not expect a large increase in the degree of difference between SCNT, parthenogenetic, and fertilized embryos as development proceeds. The two-cell HCL plot instead reveals an increasing divergence between the three classes of embryos, indicating that the donor cell nuclei exert a strong effect on phenotype as the embryo proceeds through embryonic genome activation (Fig. 2).
Global changes in mRNA population during the first embryonic cell cycle
A most interesting question that arises from SCNT is how well the donor cell genome is silenced after transfer into recipient eggs. Two scenarios could be envisioned. In the first one, as the one-cell embryo acquires the capacity to undertake gene transcription (Latham et al, 1992), an array of donor cell genes could be transcribed before the first cell division. Indeed, the overall rate of transcription in clones might be increased due to the original chromatin state of the donor genome. Alternatively, because the ooplasm establishes a transcriptionally repressive state within the early embryo (Latham et al, 1992), the donor cell genome may become highly transcriptionally repressed. Our microarray data distinguish between these alternatives, and also provide an opportunity for identifying aberrantly expressed genes. Moreover, they provide new information about the fate of maternal transcripts in clones.
We found 259 mRNAs that were differentially expressed between SCNT and fertilized embryos at the one-cell stage using the cut-off filter of 2.0-fold or greater difference (Fig. 5, 1A+1B). This corresponds to only ~1.6 % of the detected transcripts, indicating that the transcriptome of cloned one-cell embryos is very close to that of controls. Of the 259 differentially expressed mRNAs, 137 were higher in SCNT than in fertilized embryos (Fig. 5, 1A), whereas 122 were lower (Fig. 5, 1B). When considering the transcripts that are different and also sensitive to the α-amanitin treatment, however, the numbers decreased to 45 and 8, respectively. Three mRNAs (Fos, Yy1, Zfp352) were tested by qRT-PCR and all confirmed to be elevated and α-amanitin-sensitive in SCNT embryos, indicating aberrant transcription and mRNA accumulation even at this early stage. As many as 80% of the differentially expressed mRNAs (206 out of 259) were indeed not diminished by α-amanitin treatment, and thus were likely of maternal origin. Three well-known maternal transcripts (Zar1, H1foo and Cpa1) were confirmed by qRT-PCR to be present at a reduced abundance in SCNT embryos when compared to normal embryos (Fig. 4), providing further evidence that these maternal mRNAs are indeed affected. It should be noted that the real time RT-PCR data did not reveal any effect of culture media in this experiment for H1foo or any of these three maternal mRNAs (Fig. 4). These observations indicate that the donor cell genome is markedly silenced by the ooplasm at this point in development, and that regulation of maternal mRNA stability, and possibly translation, is altered in clones, with some maternal mRNAs being stabilized and others being precociously degraded.
Figure 5.
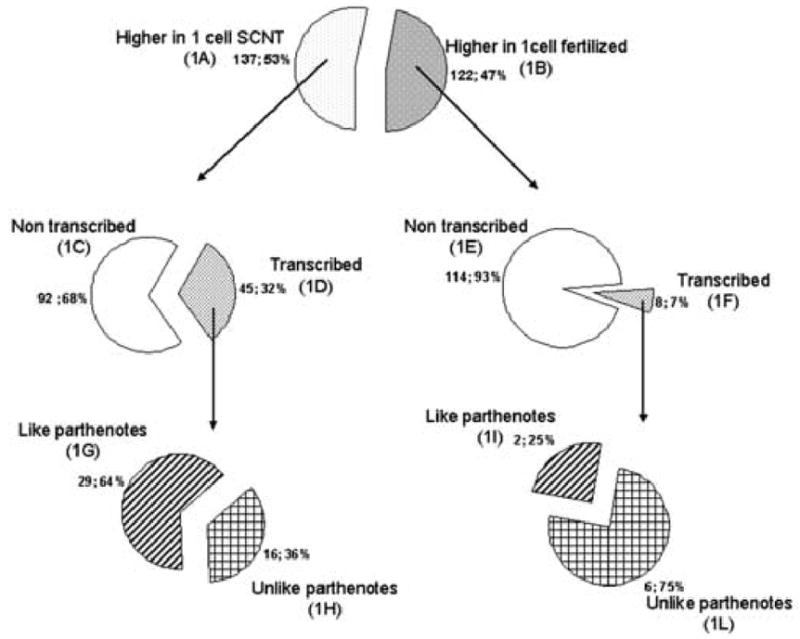
Number of genes differentially expressed in SCNT, in vivo fertilized and parthenogenetic embryos at the late one-cell stage. Those genes that displayed α-amanitin dependent reductions in mRNA abundance were judged to be transcribed, while those that did not were judged to be non-transcribed.
Relationship between genes affected at the one-cell stage and specific biological processes
We next sought to determine whether any specific biological processes were likely affected by the differential effects on the maternal mRNA population. We attempted to divide the list of differentially expressed maternal mRNAs into functional categories. Of the 114 maternal mRNAs that were of lower abundance in clones (Fig. 5, 1E), 59 had some annotation information attached to them. We did not, however, find any specific gene ontology (GO) category that included more than four transcripts in the list.
Out of the 16 mRNAs (Fig. 5, 1H; Supplemental Table S1) that were expressed more highly in SCNT embryos as compared to both fertilized and parthenogenetic embryos in an α-amanitin-sensitive manner, ten were annotated. In sharp contrast to the maternal mRNAs, these ten mRNAs displayed a clear bias in functional category, four encoding transcription factors (9030612M13Rik, Dbp, Fos, Gadd45g), and one additional mRNA (Zfp352) encoding a putative transcription factor (Liu et al., 2003a). We tested and confirmed the differential expression of two of these transcripts by qRT-PCR (Fos and Zfp352; Fig. 4). Among the six mRNAs that were more highly expressed in fertilized embryos as compared to either SCNT or parthenogenetic embryos in an α-amanitin-sensitive manner (Fig. 5, 1L; Supplemental Table S2), none encoded transcription factors.
To determine whether the 16 genes examined in Fig. 5 and Supplemental Table S1, and over-expressed in SCNT embryos, reflected gene activity of the donor nuclei, we examined a microarray data set for cumulus cells generously shared with us by Dr. John Eppig (The Jackson Laboratory). These cumulus cells were isolated from cumulus-oocyte complexes (COCs) obtained from PMSG-primed 22-day-old females. Additional samples corresponded to cells isolated from the COCs of 12 d old females and cultured for 10 d in vitro as described (O’Brien et al., 2003). Of the 16 genes overexpressed in SCNT embryos, 13 were among those detected as being expressed in samples of cells isolated directly from 22 d COCs, and one additional gene was expressed in the in vitro cultured cells. One additional gene (Zfp352) was confirmed by qRT-PCR (Fig. 4) to be expressed in cumulus cells from ovulated cumulus-oocyte complexes (donors employed for SCNT). The remaining transcript (C130047D21Rik) was not detected in the Eppig array data, and is not included among available ABI TaqMan gene expression IDs, and so was not tested by qRT-PCR. Thus, of the 16 genes that were transcribed and over-expressed in one-cell SCNT embryos, at least 15 are expressed in cumulus cells. This indicates that the array of genes overexpressed in one-cell SCNT embryos correlates highly with the gene activity of the donor nuclei.
Global changes in gene expression during the second embryonic cell cycle
The overall array of different transcripts in both SCNT and fertilized embryos increased at the two-cell stage compared to the one-cell stage. In fertilized embryos, for example, the percent P-call increased from an average of 34.9 to an average of 37.5. Similarly, for SCNT embryos this value increased from 36.9 to 40.1 (Table 1, “% P call”). By contrast, for α-amanitin treated samples, no such increases were seen, and in fact the overall transcriptome complexity diminished during this period. We also observed a much larger difference between the average number of transcripts detected in untreated and α-amanitin treated SCNT embryos than between untreated and α-amanitin treated fertilized embryos (9.9% and 5.7 %, respectively), and SCNT embryos exhibited a larger array of transcripts than fertilized embryos (p < 0.01). These results reflect activation of the embryonic genome, leading to a net increase in the complexity of the transcript population, and indicate that SCNT embryos transcribe an expanded array of genes at the two-cell stage as compared to fertilized or parthenogenetic controls.
We indeed observed substantial differences between the transcriptomes of SCNT embryos and fertilized embryos (Fig. 6, Tables S3–S6), and this was about an order of magnitude greater than the difference observed at the one-cell stage. We found 2427 mRNAs differentially expressed between SCNT and normal embryos (Fig. 6, 2A+2B). Of these, ~67% (1633) were over-expressed in SCNT embryos (Fig. 6, 2A), and 33% (794) were under-expressed relative to fertilized embryos (Fig. 6, 2B). Of the 1633 over-expressed mRNAs in SCNT embryos, 1087 (67%) were α-amanitin-sensitive (Fig. 6, 2D), and hence actively transcribed, whereas 546 (33%) were not diminished by α-amanitin treatment (Fig. 6, 2C). Of the 794 mRNAs that were expressed at reduced abundances in SCNT embryos (Fig. 6, 2E+2F), 452 (57%) were transcribed (Fig. 6, 2F) and 342 (43%) were not diminished by α-amanitin treatment (Fig. 6, 2E).
Figure 6.
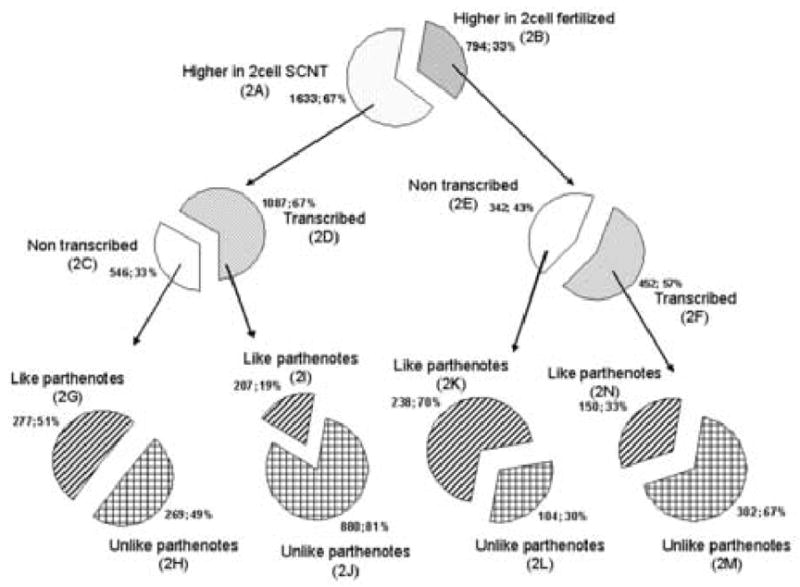
Number of genes differentially expressed in SCNT, in vivo fertilized and parthenogenetic embryos at the two-cell stage. Those genes that displayed α-amanitin dependent reductions in mRNA abundance were judged to be transcribed, while those that did not were judged to be non-transcribed.
To determine the degree to which the large differences between SCNT and fertilized embryos was the result of unique properties of SCNT embryos, or instead might be due to differences related to egg activation, absence of a fertilizing sperm, or simple effects of developmental timing, we examined in parthenogenetic embryos expression of mRNAs that were differentially transcribed between SCNT and fertilized embryos. Parthenogenetic embryos were prepared from the same pools of oocytes as SCNT embryos, activated in synchrony, and cultured in parallel, and also lack any fertilizing sperm contribution. The expression of 880 (81%) of 1087 mRNAs that were transcriptionally elevated in SCNT embryos relative to fertilized embryos was also elevated relative to parthenogenetic controls (Fig. 6, 2J and Supplemental Table S4). None of these was media-sensitive. Of the 452 transcribed mRNAs that were reduced in expression in SCNT embryos relative to fertilized embryos, a majority (302, 67%) was likewise reduced in SCNT embryos relative to parthenogenetic embryos (Fig. 6, 2M and supplemental Table S6). Seven of these were among the media-sensitive list of genes (supplemental Table S7). These results indicate that the defects in gene expression detected in SCNT embryos are due to unique features of cloned embryos, and not due to absence of a sperm, or an effect of the egg activation protocol or developmental timing.
In addition to the above effects on transcribed genes, we observed significant differences between clones and both fertilized and parthenogenetic controls in the population of non- transcribed, maternal mRNAs (Fig. 6, 2H and 2L, supplemental Tables S3 and S5). The vast majority of these differences were insensitive to culture media (i.e., only 5 out of 373 appear in Table S7, < 1%).
Relationship between genes differentially transcribed at the two-cell stage and specific biological processes
The large number of genes differentially expressed between SCNT and control two-cell embryos raises the question as to whether the aberrant regulation of these affected genes alters specific biological processes in SCNT embryos, and hence can account for some of the unusual characteristics observed for SCNT embryos. We analyzed the lists of differentially expressed genes using three different computational approaches. The first approach applied the Expression Analysis Systematic Explorer (EASE) software (Table 2). Among the transcripts over-represented in SCNT embryos, EASE analysis identified 13 Gene Ontology (GO) categories with an EASE score <0.05 (Table 2). Oxidoreductase activity was the category identified with the most significant level of over-representation, and the transporter activity category presented the largest number (68) of affected genes within a category. According to the EASE analysis of the 302 mRNAs that were reduced in expression in SCNT embryos relative to control embryos, there was only one GO category (nucleic acid binding) significantly over-represented (EASE score, 0.00295, n=155 genes).
Table 2.
EASE analysis output for genes upregulated at the two-cell stage in SCNT embryos and sensitive to α-amanitin treatment (Fig. 6, Set 2J). Characteristic molecular functions are listed for annotated genes with an EASE score < 0.05.
| GO Molecular Function | EASE score | N. genes |
|---|---|---|
| oxidoreductase activity | 1.20E-05 | 50 |
| electron transporter activity | 1.28E-04 | 18 |
| NADH dehydrogenase activity | 6.96E-05 | 9 |
| oxidoreductase activity\, acting on NADH or NADPH | 1.57E-04 | 11 |
| transporter activity | 2.53E-03 | 68 |
| primary active transporter activity | 2.05E-03 | 19 |
| carrier activity | 5.37E-04 | 32 |
| ion transporter activity | 2.49E-03 | 33 |
| cation transporter activity | 2.52E-03 | 30 |
| monovalent inorganic cation transporter activity | 5.75E-05 | 20 |
| sodium ion transporter activity | 2.28E-04 | 8 |
| hydrogen ion transporter activity | 1.34E-04 | 19 |
EASE analysis is limited by the degree and accuracy of annotations within category. Moreover it relies solely on numerical relationships between genes lists, it does not account for magnitudes of changes of individual genes, and cannot account for differences in arrays of genes within categories. Hence, although a positive result with EASE analysis provides clear evidence that a specific process is affected, a negative result does not exclude other biologically relevant differences. Accordingly, we evaluated the lists of differentially expressed mRNAs using a second approach to understand what processes may be operating during early embryogenesis and altered by SCNT. The transcripts in each list of differentially expressed mRNAs were assigned to functional categories and then the categories with the higher number of entries analyzed, regardless of their relative overrepresentation (EASE) value (Fig. 7). We also took into account the array of genes within each category.
Figure 7.
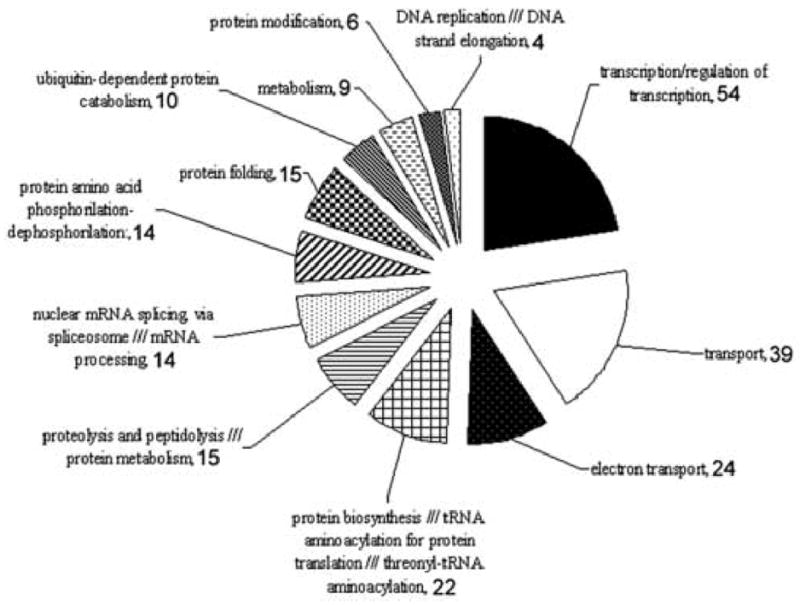
GO functional annotation of α-amanitin sensitive transcripts upregulated in SCNT compared to fertilized and parthenogenetic embryos at the two-cell stage. Numbers beside each category indicate the number of mRNA in that category.
Of the 466 transcripts that have a GO annotation assigned to them, the most abundant category represented was that of transcription factors (TF) and transcriptional regulators (54 transcripts). The 54 TF mRNAs over-expressed in SCNT embryos were elevated by ratios ranging from 2 to 12.7 fold (Supplementary Table S8). We tested and confirmed by qRT-PCR analysis the increased expression of Klf4, Maf1 and Sra1 mRNAs (Figs. 4, 8). The next largest categories encompassed transcripts involved in transport across membranes (39 transcripts) and by transcripts involved in the oxidative phosphorylation pathway (24 transcripts), thus confirming the results of the EASE analysis for these two categories. Our qRT-PCR analysis confirmed increased expression of Uqcrb and Por (electron transport), Psmc3 (transport) and Eif3e12 (protein biosynthesis; Figs. 4, 8). It is noteworthy that the 24 transcribed and overexpressed members of the oxidative phosphorylation category are all encoded by nuclear genes and are distributed among all of the OXPHOS protein complexes. Additional categories up-regulated in SCNT embryos were those of proteolysis, peptidolysis, protein phosphorylation and dephosphorylation, and protein folding.
Figure 8.
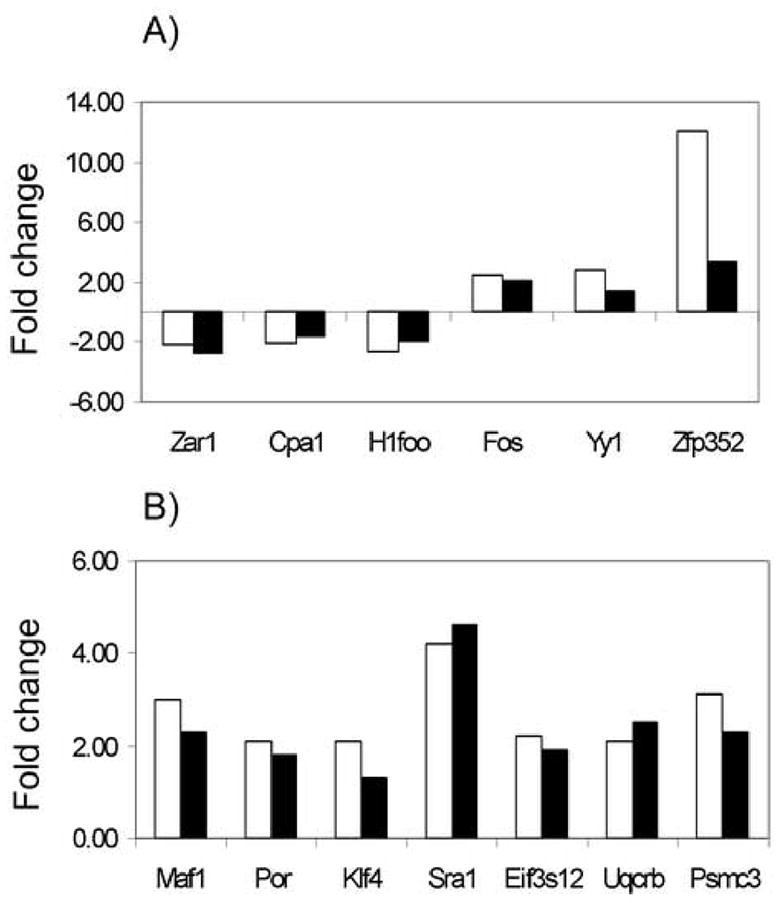
Comparison between expected fold change based on microarray analysis (black bars) and observed fold change by real time PCR (white bars) for selected genes in SCNT embryos at the one-cell (Panel A) and two-cell (Panel B) stage.
Among the 302 α-amanitin-sensitive mRNAs that were reduced in expression in SCNT embryos (Fig. 6, 2M) relative to control embryos, 169 were annotated. Interestingly, the transcription factor category was once again the largest category (n=35), indicating further deficiencies in transcription regulation in SCNT embryos. This category was followed by transport across membrane (n=18), and by proteolysis (n=8) and protein biosynthesis (n=7).
As described above, the TF category was the largest category of affected genes identified by our manual assignment of genes to functional categories. The combinatorial nature of interactions among transcription factors raises the potential that perturbations in TF expression could have a far-reaching effect on the overall process of nuclear reprogramming. We therefore used Ingenuity Pathway Analysis (IPA) to determine networks of genes that may interact with the transcription factors whose expression was perturbed in SCNT embryos. IPA identified 15 networks linking the affected TFs either directly or indirectly to other affected target genes, or indicating direct interaction between different TFs within the affected list. In the list of 54 TFs (Supplementary Table S8), 42 had scientific literature and annotation available, while 12 lacked information on interaction with other transcripts. Thirty-three of the 42 annotated TFs (79%) were identified by IPA as interacting with other TFs (31, 74%) and/or other genes in the list of upregulated transcripts (16, 38%). A representative example of such networks is presented in Fig. 9.
Figure 9.
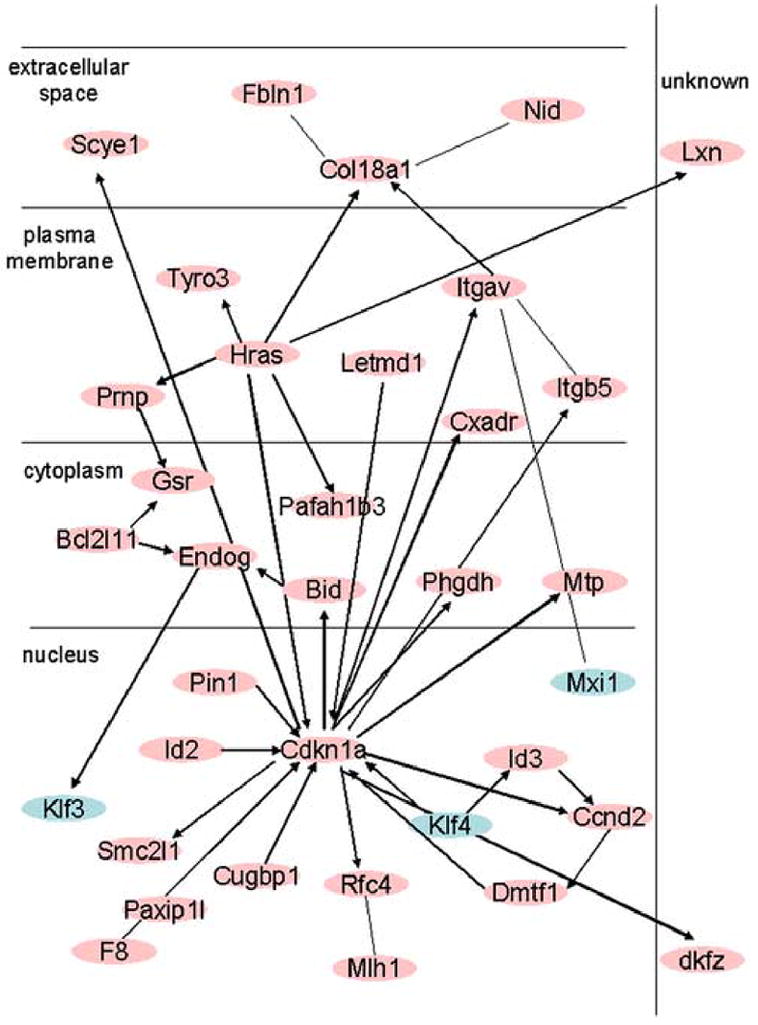
Ingenuity Pathway Analysis output example of an interaction network between transcription factors (light blue) and other upregulated genes in SCNT two-cell embryos.
Discussion
The data presented here provide for the first time in any species a detailed insight into the extent and timing of nuclear reprogramming during the first two cell cycles of development, reveal substantial disregulation of both transcription and maternal mRNA handling, and identify specific cellular processes that are affected by these defects. With respect to the extent of nuclear reprogramming, our data reveal that, although transcription in the donor nucleus appears to be greatly extinguished by the late one-cell, the donor cell genome nevertheless manifests itself via transcription and accumulation of a small array of transcripts. During the second cell cycle, when the rate of embryonic gene transcription normally increases, the donor cell genome directs the aberrant expression of over 1000 different transcripts (880 also elevated relative to parthenotes), and deficient transcription of many other genes. These results are consistent with the previously reported dramatic differences in SCNT embryo phenotype as compared to fertilized or parthenogenetic control embryos (Gao et al., 2003, 2004b; Ng and Gurdon, 2005). Indeed, we previously reported that clones display altered phenotypes even before the first cell division (Chung et al., 2002), and this early effect of the donor cell genome is evident in the microarray data as well.
Superimposed on this deficiency in transcriptional reprogramming is a substantial disruption in the maternal mRNA population, with a large number of maternal mRNAs being either precociously degraded or failing to undergo degradation. Among the transcripts differentially expressed between SCNT and fertilized embryos at the two-cell stage, 888 (37% of the total) were not diminished by α-amanitin treatment, and therefore were likely of maternal origin (Fig. 6, 2C+2E); over 40% (373) of these are also affected relative to parthenotes (Fig. 6, 2H+2L). This effect on the maternal mRNA population appears to be an intrinsic feature of clones, and not an effect of the culture system, because only 5 of these mRNAs was affected at the two-cell stage by choice of culture medium, and one of these (H1foo) was also reduced in one-cell SNCT embryos, but was not media-sensitive at that stage. Of the 373 affected maternal mRNAs, 104 were reduced in SCNT embryos and thus appeared to be precociously degraded. This accelerated degradation at the two-cell stage may be of comparatively little consequence to the embryo, because it may have little effect on expression of proteins that are being eliminated at that stage. For example, the H1foo mRNA encodes a protein that becomes undetectable in embryonic nuclei at the two-cell stage in both controls and SCNT embryos (Gao et al., 2004a). Of much greater potential significance, we observed a large number of maternal mRNAs that were elevated in SCNT embryos (269 mRNAs elevated in clones relative to both normal and parthenogenetic embryos; Fig. 6, 2H). These mRNAs most likely represent maternal transcripts that are inappropriately stabilized in the SCNT embryo. Although it is possible that some of these mRNAs correspond to abundant mRNAs in the donor cell cytoplasm and are transferred along with the nucleus, this is unlikely for several reasons. First, the donor cell is quite small in comparison to the oocyte and much of its cytoplasm is removed before injection. Thus, it is unlikely that mRNAs in the cumulus donor can make a substantial contribution to the array result. Second, we observe that many mRNAs that are expressed in somatic cells (even at high levels) but present at very low abundances in eggs (e.g., actin, Hprt, Pdha1, Pgk1, Prps1, Xist) are not elevated in clones. Third, it is most unlikely that such a large number of affected mRNAs would be abundant enough in cumulus cells to raise the observed abundance in clones. Fourth, we observe that only 92 mRNAs are elevated and α-amanitin-insensitive at the one-cell stage, but 269 are affected at the two-cell stage (Fig. 5, 1C and Fig. 6, 2H), an unlikely pattern if the source was solely the donor cell. Last, in favor of the explanation that these mRNAs are stabilized in clones, we find that 159 (59%) of the 269 α-amanitin-insensitive, affected mRNAs increase in relative abundance between the one-cell and two-cell stage, indicating a greater stability relative to the rest of the maternal mRNA population. Of the remaining mRNAs, 87 (32%) do not change significantly in abundance from the one-cell to the two-cell stage, also indicating long-term stability. Only 23 (9%) decrease in abundance during this period. These observations indicate that the majority of elevated, α-amanitin-insensitive mRNAs in SCNT embryos are very likely maternal in origin rather than imported with the somatic nucleus. Thus, cloned embryos do not undergo the normal elimination of a large number of maternal mRNAs that occurs in fertilized and parthenogenetic control embryos.
The precocious loss or stabilization of a large number of maternal mRNAs in clones was totally unanticipated. Although the molecular basis for this phenomenon is unknown, it is possible that the embryonic genome coordinates maternal mRNA degradation. Consistent with this proposal is that α-amanitin treatment apparently stabilizes some maternal mRNAs (Worrad and Schultz, 1997; Rambhatla et al, 1995). Replacing an embryonic genome with a somatic cell genome, with attendant aberrant gene regulation, could therefore lead to such defects. This explanation seems less likely for the one-cell stage, because only a small number of genes are aberrantly transcribed at this stage.
Depletion of factors associated with the spindle-chromosome complex (SCC), which is removed during the first step of cloning, could be a contributing factor. Tetraploid embryos, prepared identically to clones but without SCC removal, display ameliorated effects of the somatic cell genome (Gao et al, 2003), including a lack of aberrantly expressed somatic cell type DNMT1 (Chung et al., 2003), reduced glucose uptake, reduced requirement for glucose in the culture medium, reduced expression of GLUT4, correct regulation of GLUT1 localization to the plasma membrane, and a much greater tolerance for embryo culture medium (Gao et al, 2003). In addition, the presence or absence of the SCC affects the pace at which the oocyte loses the ability to direct changes in histone H1 composition (Gao et al, 2004a). Thus, absence of the regulatory functions of the SCC could contribute to the observed disruption in maternal mRNA stability, particularly at the one-cell stage.
The combined effects of aberrant transcription and mRNA handling disrupt the array of mRNAs that direct a range of specific cellular processes. The largest group of affected transcripts encodes transcription and mRNA processing factors—we observe this at both the one- and two-cell stages—such that some transcription factor genes normally transcribed in fertilized embryos are under-expressed in SCNT embryos. The relative abundances of mRNAs that regulate mRNA localization and transport were also reduced in SCNT embryos. Thus, SCNT embryos exhibit profound deficiencies in transcriptional reprogramming. This, coupled with a deficiency in post-transcriptional processes, could readily result in the observed aberrant phenotype of SCNT embryos.
As briefly discussed in the Introduction, reprogramming of transcription factors may be a difficult step in cloning because these proteins are responsible for establishing and maintaining a stable differentiated state of the donor somatic cell, and thus must themselves be programmed for stable expression. Genes that define a cell state are often among the most stable with respect to expression programming. In Drosophila for example, genes involved in egg polarity, and gap, pair rule, and segmentation genes act in a sequential manner to establish a combinatorial program of expression of target transcription regulatory genes (e.g., Hox genes), which become programmed for expression in a stable spatial pattern even after the patterning genes cease to be expressed (Gilbert, 2000). This involves the actions of chromatin regulatory genes (e.g., Polycomb) that establish a stable chromatin structure. Thus, cloned embryos may be predisposed to over-express genes encoding transcription factors. This would lead to aberrant expression of numerous other downstream target genes, thus affecting cloned embryo phenotype. Conversely, clones should also exhibit deficiencies in expression of TF genes associated with the embryonic state.
The results presented here support this proposal. For example, we observe an entire network of transcriptional regulators and their affected downstream genes to be upregulated in clones. Moreover, several of the aberrantly transcribed transcription factor genes, either in this network or otherwise, fit the profile of genes that establish cell state by regulating a wide array of target genes. Excellent examples of these are Sra1, Klf4, and Cbx4. Sra1 is expressed in all human tissues examined and encodes an RNA component of ribonucleoprotein complexes that contain steroid receptor coactivator-1 and may confer specificity on these transcriptional complexes (Lanz et al, 1999). KLF4 (GKLF) is likewise widely expressed, participates in epithelial cell differentiation (Jaubert et al, 2003; Segre et al, 1999), exerts anti-proliferative, pro-differentiative effects in many cell types (Higaki et al, 2002; Hinnebusch et al, 2004; Chen et al, 2002a,b, 2003; Foster et al, 2005; Katz et al, 2005; Li et al, 2005; Liu et al, 2003b, 2005; Siddique et al, 2003; Wu et al, 2004; Yoon et al, 2005), and regulates a wide variety of genes (Ai et al, 2004;). Basu et al, 2004; Blanchon et al, 2001; Chen et al, 2002a, 2003; Chiambaretta et al, 2004; Higaki et al, 2002; Hinnebusch et al, 2004; Jaubert et al, 2003; Liu et al, 2003b, 2005; Miller et al, 2001; Mao et al, 2003;Piccinni et al, 2004; Reidling et al, 2003; Siddique et al, 2003; Yasuda et al, 2002; Zhang et al, 2005). KLF proteins also interact with multiple other transcription factors, such as FLH3 and CtBP2 (Crossley et al, 1996; Gallagher et al, 2000; Sabath et al, 1996; Scohy et al, 2000; Turner et al, 1998, 2003; van Vliet et al, 2001; Yang et al, 2005). We also observed increased expression of the Cbx4 mRNA in our microarray data. The CBX4 protein, like KLF4, affects the expression of a myriad of genes, through its role in the formation of Polycomb bodies, effects on chromatin structure, recruitment of various factors to these complexes, and a combination of either activating or repressive effects (e.g., Kagey et al, 2003, 2005; Long et al, 2005; Satijn et al, 1997). The ability of both KLF4 and CBX4 to recruit CtBP to regulatory complexes suggests possible cooperative interactions between these proteins.
Another striking category of aberrantly expressed genes included those involved in oxidative phosphorylation. Genes encoding components of all of the OXPHOS protein complexes are up-regulated in clones, with some mRNAs overexpressed as a result of transcription and some elevated as a result of maternal mRNA stabilization. This may exert an effect on carbohydrate metabolism and energy production in clones. Indeed, we have reported previously that clones display increased glucose uptake and a strong preference for glucose-containing media. In this regard, it is interesting to note that one of the genes known to affect mitochondria transcription, Tfam, is present in the list of elevated genes in two-cell SCNT embryos (Supplemental Table S4), further supporting the concept of a “ripple effect” of altered reprogramming of transcription factor on downstream genes and embryonic phenotype.
Another prominent affected category encodes proteins related to solute transport and homeostasis. We observe a large number of over-expressed mRNAs at the two-cell stage in this category, and also a large number of maternal mRNAs that are aberrantly stabilized at the two-cell stage. This indicates that the cellular mechanisms regulating ion transport, amino acid transport, intracellular pH, and osmolarity are likely altered. This would likely contribute to the previously reported preference of clones for somatic cell culture media (Chung et al, 2002), which differ a great deal from embryo culture media with respect to ionic and amino acid composition.
With such a large number of aberrantly transcribed genes, the question arises whether so many genes are mis-expressed under the control of a large number or a limited number of transcription regulatory mechanisms. In addition to the possible “ripple effect” that may arise downstream of mis-regulated transcription factor encoding genes, there exists the possibility that factors expressed in the oocyte may contribute to aberrant gene regulation. The two-cell stage constitutes a period of transcriptional promiscuity during which very little histone H1 linker of any type exists, and during which the ability to regulate gene transcription is evolving (Wiekowsky et al., 1997). Given the different chromatin structure of somatic cell nuclei as compared to gamete genomes, these conditions establish the possibility that ooplasmic factors may initially activate a range of genes in the somatic nucleus that might not otherwise be activated in the normal embryo. Such activation could have broad-reaching effects, particularly when combined with the downstream consequences of aberrant transcription factor gene expression.
The observations presented here provide vital new information for evaluating the mechanisms and limitations of nuclear reprogramming during somatic cell nuclear transfer. These data also provide a rich foundation for understanding the basic biology of ooplasmic-nuclear interactions, the biology of cloning, and specific factors that must be considered if the process is to be improved.
Supplementary Material
Acknowledgments
This research was supported by grants from the NIH (HD 43092 to KEL and HD22681 to RMS). We sincerely thank Dr. Hua Pan for help with GeneSpring v7 use and Prof. John Eppig for sharing his microarray data.
Abbreviations
- SCNT
Somatic Cell Nuclear Transfer
- EGA
Embryonic Genome Activation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aghayan M, Rao LV, Smith RM, Jarett L, Charron MJ, Thorens B, Heyner S. Developmental expression and cellular localization of glucose transporter molecules during mouse preimplantation development. Development. 1992;115:305–312. doi: 10.1242/dev.115.1.305. [DOI] [PubMed] [Google Scholar]
- Ai W, Liu Y, Langlois M, Wang TC. Kruppel-like factor 4 (KLF4) represses histidine decarboxylase gene expression through an upstream Sp1 site and downstream gastrin responsive elements. J Biol Chem. 2004;279:8684–8693. doi: 10.1074/jbc.M308278200. [DOI] [PubMed] [Google Scholar]
- Baltz JM, Biggers JD, Lechene C. Relief from alkaline load in two-cell stage mouse embryos by bicarbonate/chloride exchange. J Biol Chem. 1991a;266:17212–17217. [PubMed] [Google Scholar]
- Baltz JM, Biggers JD, Lechene C. Two-cell stage mouse embryos appear to lack mechanisms for alleviating intracellular acid loads. J Biol Chem. 1991b;266:6052–6057. [PubMed] [Google Scholar]
- Baltz JM, Biggers JD, Lechene C. A novel H+ permeability dominating intracellular pH in the early mouse embryo. Development. 1993;118:1353–1361. doi: 10.1242/dev.118.4.1353. [DOI] [PubMed] [Google Scholar]
- Basu P, Sargent TG, Redmond LC, Aisenberg JC, Kransdorf EP, Wang SZ, Ginder GD, Lloyd JA. Evolutionary conservation of KLF transcription factors and functional conservation of human gamma-globin gene regulation in chicken. Genomics. 2004;84:311–319. doi: 10.1016/j.ygeno.2004.02.013. [DOI] [PubMed] [Google Scholar]
- Blanchon L, Bocco JL, Gallot D, Gachon AM, Lemery D, Dechelotte P, Dastugue B, Sapin V. Co-localization of KLF6 and KLF4 with pregnancy-specific glycoproteins during human placenta development. Mech Dev. 2001;105:185–189. doi: 10.1016/s0925-4773(01)00391-4. [DOI] [PubMed] [Google Scholar]
- Campbell KH, Alberio R, Choi I, Fisher P, Kelly RD, Lee JH, Maalouf W. Cloning: eight years after Dolly. Reprod Dometic Anim. 2005;40:256–268. doi: 10.1111/j.1439-0531.2005.00591.x. [DOI] [PubMed] [Google Scholar]
- Carayannopoulos MO, Schlein A, Wyman A, Chi M, Keembiyehetty C, Moley KH. GLUT9 is differentially expressed and targeted in the preimplantation embryo. Endocrinology. 2004;145:1435–1443. doi: 10.1210/en.2003-1264. [DOI] [PubMed] [Google Scholar]
- Carayannopoulos MO, Chi MM, Cui Y, Pingsterhaus JM, McKnight RA, Mueckler M, Devaskar SU, Moley KH. GLUT8 is a glucose transporter responsible for insulin-stimulated glucose uptake in the blastocyst. PNAS. 2000;97:7313–7318. doi: 10.1073/pnas.97.13.7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Shie JL, Tseng CC. STAT1 is required for IFN-gamma-mediated gut-enriched Kruppel-like factor expression. Exp Cell Res. 2002a;281:19–27. doi: 10.1006/excr.2002.5633. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Shie JL, Tseng CC. Gut-enriched Kruppel-like factor represses ornithine decarboxylase gene expression and functions as checkpoint regulator in colonic cancer cells. J Biol Chem. 2002b;277:46831–46839. doi: 10.1074/jbc.M204816200. [DOI] [PubMed] [Google Scholar]
- Chen X, Whitney EM, Gao SY, Yang VW. Transcriptional profiling of Kruppel-like factor 4 reveals a function in cell cycle regulation and epithelial differentiation. J Mol Biol. 2003;326:665–677. doi: 10.1016/S0022-2836(02)01449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi MM, Pingsterhaus J, Carayannopoulos M, Moley KH. Decreased glucose transporter expression triggers BAX-dependent apoptosis in the murine blastocyst. J Biol Chem. 2000;275:40252–7. doi: 10.1074/jbc.M005508200. [DOI] [PubMed] [Google Scholar]
- Chiambaretta F, De Graeve F, Turet G, Marceau G, Gain P, Dastugue B, Rigal D, Sapin V. Cell and tissue specific expression of human Kruppel-like transcription factors in human ocular surface. Mol Vis. 2004;10:901–909. [PubMed] [Google Scholar]
- Chung YG, Mann MRW, Bartolomei MS, Latham KE. Nuclear-cytoplasmic ‘tug-of war’ during cloning: effects of somatic cell nuclei on culture medium preferences of preimplantation cloned mouse embryos. Biol Repro. 2002;66:1178–1184. doi: 10.1095/biolreprod66.4.1178. [DOI] [PubMed] [Google Scholar]
- Chung YG, Ratnam S, Chaillet JR, Latham KE. Abnormal regulation of DNA methyltransferase expression in cloned mouse embryos. Biol Reprod. 2003;69:146–153. doi: 10.1095/biolreprod.102.014076. [DOI] [PubMed] [Google Scholar]
- Crossley M, Whitelaw E, Perkins A, Williams G, Fujiwara Y, Orkin SH. Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol Cell Biol. 1996;16:1695–1705. doi: 10.1128/mcb.16.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards LJ, Williams DA, Gardner DK. Intracellular pH of the mouse preimplantation embryo: amino acids act as buffers of intracellular pH. Hum Reprod. 1998a;13:3441–3448. doi: 10.1093/humrep/13.12.3441. [DOI] [PubMed] [Google Scholar]
- Edwards LJ, Williams DA, Gardner DK. Intracellular pH of the preimplantation mouse embryo: effects of extracellular pH and weak acids. Mol Reprod Dev. 1998b;50:434–442. doi: 10.1002/(SICI)1098-2795(199808)50:4<434::AID-MRD7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Foster KW, Liu Z, Nail CD, Li X, Fitzgerald TJ, Bailey SK, Frost AR, Louro ID, Townes TM, Paterson AJ, Kudlow JE, Lobo-Ruppert SM, Ruppert JM. Induction of KLF4 in basal keratinocytes blocks the proliferation-differentiation switch and initiates squamous epithelial dysplasia. Oncogene. 2005;24:1491–1500. doi: 10.1038/sj.onc.1208307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher PG, Romana M, Tse WT, Lux SE, Forget BG. The human ankyrin-1 gene is selectively transcribed in erythroid cell lines despite the presence of a housekeeping-like promoter. Blood. 2000;96:1136–1143. [PubMed] [Google Scholar]
- Gao S, Chung YG, Parseghian MH, King GJ, Adashi EY, Latham KE. Rapid H1 linker histone transitions following fertilization or somatic cell nuclear transfer: evidence for a uniform developmental program in mice. Dev Biol. 2004a;266:62–75. doi: 10.1016/j.ydbio.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Gao S, Chung G, Williams JW, Riley J, Moley K, Latham KE. Somatic cell-like features of cloned mouse embryos prepared with cultured myoblast nuclei. Biol Reprod. 2003;69:48–56. doi: 10.1095/biolreprod.102.014522. [DOI] [PubMed] [Google Scholar]
- Gao S, Czirr E, Chung YG, Han Z, Latham KE. Genetic variation in oocytes phenotype revealed through parthenogenesis and cloning: correlation with differences in pronuclear epigenetic modification. Biol Reprod. 2004b;70:1162–1170. doi: 10.1095/biolreprod.103.024216. [DOI] [PubMed] [Google Scholar]
- Gilbert SF. Developmental Biology Sinauer Associates Publishers. Sunderland; Massachusetts: 2000. pp. 287–289. [Google Scholar]
- Higaki Y, Schullery D, Kawata Y, Shnyreva M, Abrass C, Bomsztyk K. Synergistic activation of the rat laminin gamma1 chain promoter by the gut-enriched Kruppel-like factor (GKLF/KLF4) and Sp1. Nucleic Acids Res. 2002;30:2270–2279. doi: 10.1093/nar/30.11.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman N, Tasca RJ. Ultrastructural and autoradiographic studies of mouse cleavage stages. Am J Anat. 1969;126:151–173. doi: 10.1002/aja.1001260203. [DOI] [PubMed] [Google Scholar]
- Hinnebusch BF, Siddique A, Henderson JW, Malo MS, Zhang W, Athaide CP, Abedrapo MA, Chen X, Yang VW, Hodin RA. Enterocyte differentiation marker intestinal alkaline phosphatase is a target gene of the gut-enriched Kruppel-like factor. Am J Physiol Gastrointest Liver Physiol. 2004;286:G23–30. doi: 10.1152/ajpgi.00203.2003. [DOI] [PubMed] [Google Scholar]
- Hogan A, Heyner S, Charron MJ, Copeland NG, Gilbert DJ, Jenkins NA, Thorens B, Schultz GA. Glucose transporter gene expression in early mouse embryos. Development. 1991;113:363–372. doi: 10.1242/dev.113.1.363. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humpherys D, Eggan K, Akutsu H, Friedman A, Hochedlinger K, Yanagimachi R, Lander ES, Golub TR, Jaenish R. Abnormal gene expression in cloned mice derived from embryonic stem and cumulus cell nuclei. PNAS. 2002;99:12889–12894. doi: 10.1073/pnas.192433399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaubert J, Cheng J, Segre JA. Ectopic expression of kruppel like factor 4 (Klf4) accelerates formation of the epidermal permeability barrier. Development. 2003;130:2767–2777. doi: 10.1242/dev.00477. [DOI] [PubMed] [Google Scholar]
- Kagey MH, Melhuish TA, Powers SE, Wotton D. Multiple activities contribute to Pc2 E3 function. EMBO J. 2005;24:108–119. doi: 10.1038/sj.emboj.7600506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- Katz JP, Perreault N, Goldstein BG, Actman L, McNally SR, Silberg DG, Furth EE, Kaestner KH. Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology. 2005;128:9359–45. doi: 10.1053/j.gastro.2005.02.022. [DOI] [PubMed] [Google Scholar]
- King TJ, Briggs R. Changes in the nuclei of differentiating gastrula cells, as demonstrated by nuclear transplantation. PNAS. 1955;41:321–325. doi: 10.1073/pnas.41.5.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O’Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- Latham KE. Cloning: question answered and unsolved. Differentiation. 2004;72:11–22. doi: 10.1111/j.1432-0436.2004.07201003.x. [DOI] [PubMed] [Google Scholar]
- Latham KE. Early and delayed aspects of nuclear reprogramming during cloning. Biol Cell. 2005;97:119–132. doi: 10.1042/BC20040068. [DOI] [PubMed] [Google Scholar]
- Latham KE, Garrels JI, Chang C, Solter D. Quantitative analysis of protein synthesis in mouse embryos I. Extensive re-programming at the one- and two-cell stages. Development. 1991;112:921–932. doi: 10.1242/dev.112.4.921. [DOI] [PubMed] [Google Scholar]
- Latham KE, Solter D, Schultz RM. Acquisition of a transcriptionally permissive state during the one-cell stage of mouse embryogenesis. Dev Biol. 1992;149:457–562. doi: 10.1016/0012-1606(92)90300-6. [DOI] [PubMed] [Google Scholar]
- Leppens-Luisier G, Urner F, Sakkas D. Facilitated glucose transporters play a crucial role throughout mouse preimplantation embryo development. Hum Reprod. 2001;16:1229–1236. doi: 10.1093/humrep/16.6.1229. [DOI] [PubMed] [Google Scholar]
- Li Y, McClintick J, Zhong L, Edenberg HJ, Yoder MC, Chan RJ. Murine embryonic stem cell differentiation is promoted by SOCS-3 and inhibited by the zinc finger transcription factor Klf4. Blood. 2005;105:635–637. doi: 10.1182/blood-2004-07-2681. [DOI] [PubMed] [Google Scholar]
- Liu TY, Chen HH, Lee KH, Choo KB. Display of different modes of transcription by the promoters of an early embryonic gene, zfp352, in preimplantation embryos and in somatic cells. Mol Reprod Dev. 2003a;64:52–60. doi: 10.1002/mrd.10218. [DOI] [PubMed] [Google Scholar]
- Liu Y, Sinha S, Owens G. A transforming growth factor-beta control element required for SM alpha-actin expression in vivo also partially mediates GKLF-dependent transcriptional repression. J Biol Chem. 2003b;278:48004–48011. doi: 10.1074/jbc.M301902200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Sinha S, McDonald OG, Shang Y, Hoofnagle MH, Owens GK. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J Biol Chem. 2005;280:9719–9727. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- Long J, Zuo D, Park M. Pc2-mediated sumoylation of Smad-interacting protein 1 attenuates transcriptional repression of E-cadherin. J Biol Chem. 2005;280:35477–35489. doi: 10.1074/jbc.M504477200. [DOI] [PubMed] [Google Scholar]
- Mao Z, Song S, Zhu Y, Yi X, Zhang H, Shang Y, Tong T. Transcriptional regulation of A33 antigen expression by gut-enriched Kruppel-like factor. Oncogene. 2003;22:4434–4443. doi: 10.1038/sj.onc.1206508. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Shoji N, Sugawara S, Umezu M, Sato E. Microscopic analysis of enzyme activity, mitochondrial distribution and hydrogen peroxide in two-cell rat embryos. J Reprod Fertil. 1998;113:231–238. doi: 10.1530/jrf.0.1130231. [DOI] [PubMed] [Google Scholar]
- Miller KA, Eklund EA, Peddinghaus ML, Cao Z, Fernandes N, Turk PW, Thimmapaya B, Weitzman SA. Kruppel-like factor 4 regulates laminin alpha 3A expression in mammary epithelial cells. J Biol Chem. 2001;276:42863–42868. doi: 10.1074/jbc.M108130200. [DOI] [PubMed] [Google Scholar]
- Moley KH, Chi MM, Mueckler MM. Maternal hyperglycemia alters glucose transport and utilization in mouse preimplantation embryos. Am J Physiol. 1998;275:E38–47. doi: 10.1152/ajpendo.1998.275.1.E38. [DOI] [PubMed] [Google Scholar]
- Morita Y, Tsutsumi O, Oka Y, Taketani Y. Glucose transporter GLUT1 mRNA expression in the ontogeny of glucose incorporation in mouse preimplantation embryos. Biochem Biophys Res Commun. 1994;199:1525–1531. doi: 10.1006/bbrc.1994.1404. [DOI] [PubMed] [Google Scholar]
- Ng RK, Gurdon JB. Epigenetic memory of active gene transcription is inherited through somatic cell nuclear transfer. PNAS. 2005;102:1957–1962. doi: 10.1073/pnas.0409813102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien MJ, Pendola JK, Eppig JJ. A revised protocol for in vitro development of mouse oocytes from primordial follicles dramatically improves their developmental competence. Biol Reprod. 2003;68:1682–1686. doi: 10.1095/biolreprod.102.013029. [DOI] [PubMed] [Google Scholar]
- Pan H, O’brien MJ, Wigglesworth K, Eppig JJ, Schultz RM. Transcript profiling during mouse oocytes development and the effect of gonadotropin priming and development in vitro. Dev Biol. 2005;286:493–506. doi: 10.1016/j.ydbio.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Pantaleon M, Kaye PL. Glucose transporters in preimplantation development. Rev Reprod. 1998;3:77–81. doi: 10.1530/ror.0.0030077. [DOI] [PubMed] [Google Scholar]
- Pantaleon M, Ryan JP, Gil M, Kaye PL. An unusual subcellular localization of GLUT1 and link with metabolism in oocytes and preimplantation mouse embryos. Biol Reprod. 2001;64:1247–1254. doi: 10.1095/biolreprod64.4.1247. [DOI] [PubMed] [Google Scholar]
- Piccinni SA, Bolcato-Bellemin AL, Klein A, Yang VW, Kedinger M, Simon-Assmann P, Lefebvre O. Kruppel-like factors regulate the Lama1 gene encoding the laminin alpha1 chain. J Biol Chem. 2004;279:9103–9114. doi: 10.1074/jbc.M305804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambhatla L, Patel B, Dhanasekeran N, Latham KE. Analysis of G protein alpha subunit mRNA abundance in preimplantation mouse embryos using a rapid, quantitative RT- PCR approach. Mol Reprod Dev. 1995;41:314–324. doi: 10.1002/mrd.1080410306. [DOI] [PubMed] [Google Scholar]
- Reidling JC, Said HM. In vitro and in vivo characterization of the minimal promoter region of the human thiamin transporter SLC19A2. Am J Physiol Cell Physiol. 2003;285:633–641. doi: 10.1152/ajpcell.00076.2003. [DOI] [PubMed] [Google Scholar]
- Sabath DE, Koehler KM, Yang WQ. Structure and function of the zeta-globin upstream regulatory element. Nucleic Acids Res. 1996;24:4978–4986. doi: 10.1093/nar/24.24.4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Satijn DP, Olson DJ, van der Vlag J, Hamer KM, Lambrechts C, Masselink H, Gunster MJ, Sewalt RG, van Driel R, Otte AP. Interference with theexpression of a novel human polycomb protein, hPc2, results in cellular transformation and apoptosis. Mol Cell Biol. 1997;17:6076–6086. doi: 10.1128/mcb.17.10.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scohy S, Gabant P, Szpirer C, Szpirer J. Identification of an enhancer and an alternative promoter in the first intron of the alpha-fetoprotein gene. Nucleic Acids Res. 2000;28:3743–3751. doi: 10.1093/nar/28.19.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- Sathananthan AH, Trounson AO. Mitochondrial morphology during preimplantational human embryogenesis. Hum Reprod. 2000;15:148–59. doi: 10.1093/humrep/15.suppl_2.148. [DOI] [PubMed] [Google Scholar]
- Shepard TH, Muffley LA, Smith LT. Ultrastructural study of mitochondria and their cristae in embryonic rats and primate (N. nemistrina) Anat Rec. 1998;252:383–392. doi: 10.1002/(SICI)1097-0185(199811)252:3<383::AID-AR6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Shepard TH, Muffley LA, Smith LT. Mitochondrial ultrastructure in embryos after implantation. Hum Reprod. 2000;15:218–228. doi: 10.1093/humrep/15.suppl_2.218. [DOI] [PubMed] [Google Scholar]
- Siddique A, Malo MS, Ocuin LM, Hinnebusch BF, Abedrapo MA, Henderson JW, Zhang W, Mozumder M, Yang VW, Hodin RA. Convergence of the thyroid hormone and gut-enriched Kruppel-like factor pathways in the context of enterocyte differentiation. J Gastrointest Surg. 2003;7:1053–1061. doi: 10.1016/j.gassur.2003.09.006. [DOI] [PubMed] [Google Scholar]
- Smith SL, Everts RE, Tian XC, Du F, Sung LY, Rodriguez-Zas SL, Jeong BS, Renard JP, Lewin HA, Yang X. Global gene expression profiles reveal significant nuclear reprogramming by the blastocyst stage after cloning. PNAS. 2005;102:17582–17587. doi: 10.1073/pnas.0508952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J, Crossley M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J, Nicholas H, Bishop D, Matthews JM, Crossley M. The LIM protein FHL3 binds basic Kruppel-like factor/Kruppel-like factor 3 and its co-repressor C-terminal-binding protein 2. J Biol Chem. 2003;278:12786–12795. doi: 10.1074/jbc.M300587200. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. PNAS. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Winkle LJ. Amino acid transport regulation and early embryo development. Biol Reprod. 2001;64:1–12. doi: 10.1095/biolreprod64.1.1. [DOI] [PubMed] [Google Scholar]
- van Vliet J, Turner J, Crossley M. Human Kruppel-like factor 8: a CACCC-box binding protein that associates with CtBP and represses transcription. Nucleic Acids Res. 2000;28:1955–1962. doi: 10.1093/nar/28.9.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, Nothias JY, DePamphilis ML. Changes in histone synthesis and modification at the beginning of mouse development correlate with the establishment of chromatin mediated repression of transcription. JCell Science. 1997;110:1147–1158. doi: 10.1242/jcs.110.10.1147. [DOI] [PubMed] [Google Scholar]
- Worrad DM, Schultz RM. Regulation of gene expression in the premplantation mouse embryo: temporal and spatial pattern of expression of the transcription factor Sp1. Mol Reprod Dev. 1997;46:268–277. doi: 10.1002/(SICI)1098-2795(199703)46:3<268::AID-MRD5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Wu J, Lingrel JB. KLF2 inhibits Jurkat T leukemia cell growth via upregulation of cyclin-dependent kinase inhibitor p21WAF1/CIP1. Oncogene. 2004;23:8088–8096. doi: 10.1038/sj.onc.1207996. [DOI] [PubMed] [Google Scholar]
- Yang XO, Doty RT, Hicks JS, Willerford DM. Regulation of T-cell receptor D beta 1 promoter by KLF5 through reiterated GC-rich motifs. Blood. 2003;101:4492–4499. doi: 10.1182/blood-2002-08-2579. [DOI] [PubMed] [Google Scholar]
- Yasuda K, Hirayoshi K, Hirata H, Kubota H, Hosokawa N, Nagata K. The Kruppel-like factor Zf9 and proteins in the Sp1 family regulate the expression of HSP47, a collagen-specific molecular chaperone. J Biol Chem. 2002;277:44613–44622. doi: 10.1074/jbc.M208558200. [DOI] [PubMed] [Google Scholar]
- Yoon HS, Ghaleb AM, Nandan MO, Hisamuddin IM, Dalton WB, Yang VW. Kruppel-like factor 4 prevents centrosome amplification following gamma-irradiation-induced DNA damage. Oncogene. 2005;24:4017–4025. doi: 10.1038/sj.onc.1208576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Zeng F, Schultz RM. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev Biol. 2005;283:464–457. doi: 10.1016/j.ydbio.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Zhang P, Basu P, Redmond LC, Morris PE, Rupon JW, Ginder GD, Lloyd JA. A functional screen for Kruppel-like factors that regulate the human gamma-globin gene through the CACCC promoter element. Blood Cells Mol Dis. 2005;35:227–235. doi: 10.1016/j.bcmd.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Baltz JM. Bicarbonate/chloride exchange and intracellular pH throughout preimplantation mouse embryo development. Am J Physiol. 1996;271:C1512–1520. doi: 10.1152/ajpcell.1996.271.5.C1512. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chauvet PJ, Alper SL, Baltz JM. Expression and function of bicarbonate/chloride exchangers in the preimplantation mouse embryo. J Biol Chem. 1995;270:24428–24434. doi: 10.1074/jbc.270.41.24428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


