Abstract
Type II restriction endonucleases (REases) are deoxyribonucleases that cleave DNA sequences with remarkable specificity. Type II REases are highly divergent in sequence as well as in topology, i.e. the connectivity of secondary structure elements. A widely held assumption is that a structural core of five β-strands flanked by two α-helices is common to these enzymes. We introduce a systematic procedure to enumerate secondary structure elements in an unambiguous and reproducible way, and use it to analyze the currently available X-ray structures of Type II REases. Based on this analysis, we propose an alternative definition of the core, which we term the αβα-core. The αβα-core includes the most frequently observed secondary structure elements and is not a sandwich, as it consists of a five-strand β-sheet and two α-helices on the same face of the β-sheet. We use the αβα-core connectivity as a basis for grouping the Type II REases into distinct structural classes. In these new structural classes, the connectivity correlates with the angles between the secondary structure elements and with the cleavage patterns of the REases. We show that there exists a substructure of the αβα-core, namely a common conserved core, ccc, defined here as one α-helix and four β-strands common to all Type II REase of known structure.
INTRODUCTION
Restriction endonucleases (REases) are components of restriction modification systems that protect bacteria and archaea against invading foreign DNA. Bacteria initially resist infections by new viruses because REases within the cells destroy foreign DNA molecules by hydrolyzing the ester bonds of the sugar-phosphate backbone at a particular recognition sequence. Bacterial DNA is protected by the methylation modification produced by the corresponding bacterial methylase specific towards the same recognition sequence. The restriction–modification (R–M) systems are named Types I to IV depending on the number and organization of their functional (restriction, modification, specificity) subunits (1). The Type II REases are the most common among biochemically characterized REases. Type II REases recognize specific unmethylated DNA sequences and cleave at constant positions, at or close to the recognition sequence to produce 5′-phosphates and 3′-hydroxyls (1–3). The specificity of Type II REases has made them indispensable tools in recombinant DNA technologies (3,4), and recent reviews have focused on their structure and function, the role of metal ions and mechanisms of catalysis (2,5,6). After initial identification of a PD-(D/E)XK motif in Type II REases, this motif was identified in many enzymes involved in DNA recombination and repair (7–9). Typically, the sequence similarity between these proteins is so low that most of the relationships between known members of the PD-(D/E)XK superfamily were identified only after the corresponding structures were determined experimentally (7,9).
Type II REases also vary dramatically; they range in size from 157 amino acids (PvuII) to 1250 amino acids (CjeI) and even beyond (3); typically they have very low sequence identity, in the so-called ‘twilight’ or ‘midnight’ zone of sequence similarity. In this zone, the sequences of homologous proteins are identical to the extent of <10–15%, and genuine similarities disappear in the random noise (10–12), making structure-based tools (such as structure-based alignment and threading) crucially important for the investigation of REases (7,13–17). Crystallographic structures of 22 Type II REases have been solved to date. In the hierarchical classification of all protein domain structures, which clusters proteins at four major levels [Class (C), Architecture (A), Topology (T) and Homologous superfamily (H) (CATH) (18)], the Type II REases are assigned to the 3.40 (three-layer αβα sandwich) architecture (18). However, the connectivity of the secondary structural elements (the ‘topology’) of these proteins varies, and this increases even further the difficulty in comparing them caused by their highly dissimilar sequences.
The Structural Classification of Proteins (SCOP) database (19,20), that offers a comprehensive ordering of all proteins of known structure according to their evolutionary and structural relationships, assigns all REases to one family (restriction-endonuclease-like, 52980) with a common core. The core is defined in terms of the components of an αβα sandwich structure: a mixed β-sheet consisting of five strands in the order 12345 (where strand 2 and, in some families, strand 5 is antiparallel to the rest) flanked by helices on both sides (10,21,22). Additional versions of the common core have been suggested (5,11,21–23). Here, we analyze the structures of 22 different Type II REases, introduce a nomenclature for the secondary structure elements, and show that two helices that occur most frequently in the existing structures do not flank the β-sheet, but are positioned on the same face of that sheet. Thus, we propose an αβα-core that consists of two helices and five β-strands that do not constitute an αβα structural sandwich. We also show here that the sequential order and connectivity of the secondary structural elements in the αβα-core are not strictly preserved in the Type II REases, and suggest a new classification of Type II REase structures based on the connectivity of the αβα-core secondary structural elements.
REases have been divided into an EcoRI-like (α-class) and an EcoRV-like (β-class), in which the fifth β-strand is parallel or antiparallel to the first β-strand, respectively. The α-class REases were found to cleave DNA to produce 5′ ‘sticky’ ends (5′ overhang) and the β-class REases were found to produce 3′ overhangs and blunt ends (11,21,23–25). We reevaluate this classification in the context of the new crystal structures and the newly defined αβα-core-based structural classes.
Finally, we show that a substructure of the αβα-core, namely the one helix/four-strand structure, is conserved in all 22 structures. This substructure is, therefore, designated here as the conserved common core (ccc). We illustrate the usefulness of the cores for structural comparison and for the prediction of function for proteins obtained from structural genomics approaches.
MATERIALS AND METHODS
Numbering of secondary structure elements
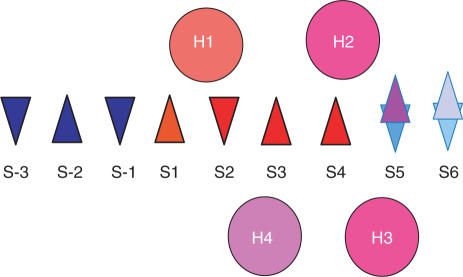
We use the following convention for the numbering of the secondary structure elements of the αβα-core: using the structure of EcoRV as a reference, the β-strand containing the catalytic residue D74 (the first aspartate in the PD-(D/E)XK motif) was defined as S2. One of the neighboring β-strands of S2, containing the catalytic residues D90 and K92, was named S3, while the second neighboring β-strand was named S1. By matching these three β-strands to the schematic diagram shown in Figure 1, β-strands to the left of S1 were assigned consecutive negative numbers (i.e. S-1, S-2 and S-3) and the remaining strands to the right were assigned consecutive positive numbers (i.e. S4 to S6). By using the β-strands S1 to S4 for alignments, all the REase structures were mapped to the αβα-core. Type II REases were structurally aligned using the Automated Pairwise Structural Superposition tool provided with the ICM program (Molsoft, Inc.) (26) and the combinatorial extension method (27). The sequence alignments and the PDB files of the secondary structure elements of the core in aligned orientation are available in the Supplementary Data (accessible via the journal website and at http://physiology.med.cornell.edu/faculty/niv/3D_alignments.zip).
Figure 1.
Schematic diagram of shared secondary structure elements observed after a structural alignment of Type II REases. Triangles represent β-strands and circles represent the α-helices. Up and down triangles are used to indicate the direction of one β-strand with respect to another. The letters S and H followed by a digit label the β-strands and α-helices, respectively, according to their position. Elements are colored using a scale in which those that appear with higher frequency in the structures of the 22 proteins are shown in longer wavelength colors. The red triangles designate the strands that are common to all Type II REase structures. The frequency of appearance of each secondary structure element is the following: H1 = 22/22; H2 = 20/22, H3 = 20/22, H4 = 16/22, S-3 = 1/22, S-2 = 1/22, S-1 = 2/22, S1 = 22/22, S2 = 22/22, S3 = 22/22, S4 = 22/22, S5 = 22/22 (with S5 parallel to S1 or S5up = 14/22, and S5 anti parallel to S1 or S5down = 8/22) and S6 = 13/22 (with S6 parallel to S1 or S6up = 9/13, and S6 anti parallel to S1 or S6down = 4/13).
The convention for numbering the α-helices was based on the frequency with which these secondary structure elements were found in similar regions of the 3D structure with respect to the conserved set of strands described above, under the condition that those α-helices must involve at least six consecutive amino acid residues, i.e. a minimal α-helix as defined by Kabsch and Sander (28). The most frequently occurring helices were named in the sequence H1, H2, H3 and H4.
Characterization of the endonuclease structures
Analysis of the α/β packing arrangement of the RE family of proteins considered in this study was carried out with the methodology developed by Chou et al. (29). The relative orientation of the axis of an α-helix with respect to the central axis of the plane formed by the β-sheet can be specified in terms of two parameters, namely, the horizontal projected angle Ωαβ and the distance between the midpoints, DM, i.e. representing the distance from the origin of the β-sheet coordinate system to the midpoint of the α-helix. The parameters Ωαβ and DM were computed from the Cα coordinates of the REases.
Analysis of the packing arrangement of the α-helices of the REases follows the methodology of Chou et al. (30), with the relative positions of two α-helix axes specified by two parameters: the distance D connecting the midpoint of the two α-helices, and the angle Ωo describing the relative orientation of the two axes. The parameters D and Ωo were computed by using the Cα coordinates of the proteins.
Enzyme classification
The Enzyme Classification (EC) (31) codes serve for discussion of functionally related enzymes. The EC assigns a specific numerical identifier, the EC number, which identifies the enzyme in terms of the reaction catalyzed. The first digit represents the type of reaction catalyzed. The second digit of the EC number refers to the subclass, which generally contains information about the type of compound or group involved. The third digit, the sub-subclass, further specifies the nature of the reaction and the fourth digit is a serial number that is used to identify the individual enzyme within a sub-subclass. For example, Type II REases have EC code 3.1.21.4, and the hierarchy of EC classification is
3.-.-.-Hydrolases.
3.1.-.-Hydrolases acting on ester bonds.
3.1.21.-Endodeoxyribonucleases producing 5′-phosphomonoesters.
3.1.21.4 Type II site-specific deoxyribonuclease.
The EC of proteins of known structures can be accessed via http://www.ebi.ac.uk/thornton-srv/databases/enzymes/
RESULTS
Definition of structural classes
Visual inspection of the representative entries for each of the 22 PD-(D/E)XK superfamily Type II REases for which structures have been solved (listed in Table 1) reveals a set of secondary structure elements shared by these proteins, as summarized in Figure 1.
Table 1.
Classification of REase structures
| REase | PDB code boundaries | Order of secondary-structure elements | Class notation | Class | CATH topology | FSSP/DALI fold | α/β | Recognition sequence | Blunt/sticky | Subtype |
|---|---|---|---|---|---|---|---|---|---|---|
| MunI | 1d02B | H1S1S2S3H4S4H3S5(↑)H2 | H1-S-H2-up | I | 3.40.580 | 263 | α | C⁁AATTG | 5′ end | IIP |
| K3-D202 | (ECORI) | |||||||||
| Cfr10I | 1cfr | H1S1S2S3H4S4H3S5(↑)H2 | H1-S-H2-up | I | 3.40.91 | 263 | α | R⁁CCGGY | 5′ end | IIF |
| M1-L283 | (Restr. Endonuc) | |||||||||
| Bse634I | 1knvA | H1S1S2S3H4S4H3S5(↑)H2 | H1-S-H2-up | I | 3.40.91 | 263 | α | R⁁CCGGY | 5′ end | IIF |
| N4-K293 | (Restr. Endonuc) | |||||||||
| EcoRII | 1na6B | H1S1S2S3H4S4H3S5(↑)H2 | H1-S-H2-up | I | not in CATH | 404 | α | ⁁CCWGG | 5′ end | IIE |
| Y183-D402 | ||||||||||
| EcoO109I | 1wtdB | H1S1S2S3H4S4H3S5(↑)S6(↑)H2 | H1-S-H2-up | I | not in CATH | 2501 | α | RG⁁GNCCY | 5′ end | IIP |
| M1-E272 | ||||||||||
| BsoBI | 1dc1A | H1S-2(↑)S-3(↓)S-1(↓)S1S2S3H4S4H3S5(↑)H2 | H1-S-H2-up | I | 3.40.91 | 263 | α | C⁁YCGRG | 5′ end | IIP |
| K5-I323 | (Restr. Endonuc) | |||||||||
| FokI | 2fokA | H1S1S2S3H4S4H3S5(↑)H2S6(↑) | H1-S-H2-up | I | 3.40.91 | 263 | α | GGATGNNNNNNNNN⁁NNNN | 5′ end | IIS |
| P405-F579 | (Restr. Endonuc) | |||||||||
| NgoMIV | 1fiuA | H1S1S2S3H4S4H3S5(↑)S6(↑)H2 | H1-S-H2-up | I | 3.40.50 | 263 | α | G⁁CCGGC | 5′ end | IIF |
| M1-V286 | (Rossmann fold) | |||||||||
| EcoRI | 1qc9A | H1S1S2S3H4S4H3S5(↑)H2 | H1-S-H2-up | I | 3.40.580 | 263 | α | G⁁AATTC | 5′ end | IIP |
| S17-K277 | (ECORI) | |||||||||
| Ecl18kI | 2fqzA | H1S1S2S3H4S4H3S5(↑)S6(↑) H2 | H1-S-H2-up | I | not in CATH | not in FSSP | α | ⁁CCNGG | 5′ end | IIP |
| L4-A302 | ||||||||||
| BstYI | 1sdoA | S6(↓)H2H1S1S2S3H4S4H3S5(↑) | H2-H1-S-up | II | not in CATH | 264 | α | R⁁GATCY | 5′ end | IIP |
| M1-P203 | ||||||||||
| BamHI | 2bamA | S6(↓)H2H1S1S2S3H4S4H3S5(↑) | H2-H1-S-up | II | 3.40.91 | 264 | α | G⁁GATCC | 5′ end | IIP |
| M1-K207 | (Restr. Endonuc) | |||||||||
| BglII | 1dfmA | S6(↓)H2H1S1S2S3H4S4H3S5(↑) | H2-H1-S-up | II | 3.40.91 | 264 | α | A⁁GATCT | 5′ end | IIP |
| M1-M30;D49-E172 | (Restr. Endonuc) | |||||||||
| MspI | 1sa3A | H1H2S1S2S3H4S4S5(↓)H3 | H1-H2-S-down | III | not in CATH | 275 | β | C⁁CGG | 5′ end | IIP |
| M1-G262 | ||||||||||
| HinP1I | 1ynmA | S6(↑)H1H2S1S2S3H4S4S5(↓)H3 | H1-H2-S-down | III | not in CATH | not in FSSP | β | G⁁CGC | 5′ end | IIP |
| G7-I247 | ||||||||||
| HincII | 1xhvA | H2S6(↑)H1S1S2S3H4S4S5(↓) | H2-H1-S-down | IV | 3.40.600 | not in FSSP | β | GTY⁁RAC | blunt | IIP |
| S2-L258 | (ECORV) | |||||||||
| EcoRV | 1b94A | H2H1S1S2S3S4S5(↓)H3 | H2-H1-S-down | IV | 3.40.600 | 270 | β | GAT⁁ATC | blunt | IIP |
| S2-K245 | (ECORV) | |||||||||
| NaeI | 1iawA | H2H1S1S2S3S4S5(↓)H3S6(↑) | H2-H1-S-down | IV | 3.40.600 | 268 | β | GCC⁁GGC | blunt | IIE |
| E10-E171 | (ECORV) | |||||||||
| BgII | 1dmuA | H2H1S1S2S3S4S5(↓)S6(↑) | H2-H1-S-down | IV | 3.40.600 | 268 | β | GCCNNNN⁁NGGC | 3′ end | IIP |
| M1-K299 | (ECORV) | |||||||||
| SfiI | 2ezvA | H2H1S1S2S3S4S5(↓)H3S6(↑) | H2-H1-S-down | IV | not in CATH | not in FSSP | β | GGCCNNNN⁁NGGCC | 3′ end | IIF |
| M1-R269 | ||||||||||
| PvuII | 3pviA | H1S1S2S3H4S4S5(↓)H3S6(↑) | H1-S-down | V | 3.40.210 | 268 | β | CAG⁁CTG | blunt | IIP |
| S2-Y157 | (PvuII) | |||||||||
| SdaI | 2ixsA | H1S1S-1(↓)S2S3H4S4H3S5(↑)S6(↓) | H1-S-up | VI | not in CATH | not in FSSP | α | CCTGCA⁁GG | 3′ end | IIP |
| P175-R323 |
The columns are ordered as follows: REase column lists the names of the REases; PDB code column lists the pdb files and the residues used in the analysis; Order of secondary structure elements is given in terms of elements that are defined in the ‘material send methods’ section, (↑) and (↓) designate S5 parallel and antiparallel to S1, respectively. Class notation describes the sequential order of the elements and the direction of the S5 strand with respect to the S1 strand. For example, H1-S-H2-up means that H1 is followed by five β-strands (S), followed by helix H2. S5 is parallel to S1 (up); Class indicates the Class number. These are followed (up); Class indicates the Class number. These are followed by CATH topology classification (18), 3.40 is three-layer αβα sandwich; DALI/FSSP fold classification (32); α/β notation, where α-class stands for S5 strand parallel to S1 strand and β-class stands for S5 antiparallel to S1 (11,21,23–25); Recognition sequences (and cleavage sites, designated by ⁁), obtained from REBASE http://rebase.neb.com/rebase/rebase.html; Blunt/sticky classification, where ‘sticky’ leave 5′ or 3′ DNA end overhang and blunt leave no overhangs upon cleavage; Subtype lists the functional subtypes as defined in the REase nomenclature (1).
Alignment and numbering of secondary structure elements were performed as described in the Materials and methods section. The number of strands forming the β-sheet varies from 5 to 9, of which the strands labeled S1 to S4 are common to all of the structures. Strands S5 and S6 may run parallel or antiparallel to S1. The orientation of S5 with respect to the central β-sheet has been used to classify REase into classes α (parallel to S1, EcoRI-like) and β (antiparallel to S1, EcoRV-like) (11,21,23–25). Helix H1 is conserved in all 22 structures. Helices H2 and H3 occur in 20 out of 22 structures. H4, which is positioned on the other face of the β-sheet and thus completes a structural sandwich, is absent in the structures of NaeI (1iawA), EcoRV (1b94A), SfiI (2ezvA), and BglI (1dmuA). The assignment in MspI (1sa3A) and HinplI (1ynmA) is ambiguous because there are four α-helices between strands S3 and S4.
Interestingly, even secondary structure elements which have structurally conserved positions and occur in most of the Type II REases, vary in their sequential order. Table 1 summarizes the connectivity of the secondary structure elements.
We assigned each protein to a specific structural class based on the connectivity (18,32) of the most conserved secondary elements and the direction of S5. Specifically, the conserved elements H1, H2 and S1 to S5 were included in the analysis and called the αβα-core. We found that the H3 sequential position correlates with the orientation of S5: for all the cases in which H3 precedes S5, S5 is parallel to S1. For all the cases in which H3 follows S5 or is absent, S5 is antiparallel to S1 (see column 4 of Table 1). The H3 connectivity was, therefore, not included in the structural class definition (columns 5 and 6 of Table 1). The connectivity of H4 was also not included in the structural class definition, because the H4 connectivity is conserved: H4, when present, appears between strands S3 and S4. H4 is absent in REases in which H2 is followed by H1, followed by β-sheet with S5 antiparallel to S1, but is present in HincII.
Class I is defined by the elements of the αβα-core having the following sequential order: H1 is followed by β-strands S1 to S5, and then by H2, with the fifth β-strand always in the ‘up’ orientation (i.e. parallel to strands S1, S3 and S4). For example, endonuclease Cfr10I belongs to this class and its secondary structure, as defined by the Kabsch and Sander algorithm (28), is as follows:
Helices: H1 (residues 59–84), H2 (residues 272–281), H3 (residues 238–244) and H4 (residues 195–218);
Strands: S1 (residues 89–93), S2 (residues 135–139), S3 (residues 183–190), S4 (residues 228–233) and S5 (residues 265–268). Regarding the numbering of the secondary structure elements, it should be noted, for example, that residues 272–281 are C-terminal to residues 238–244, but are designated H2 and H3, respectively, based on the frequency with which those elements were found at a particular 3D position relative to the β-sheet in the structural template (details described in the Materials and methods section). Consequently, the overall sequential order of the secondary structure elements in Cfr10I is H1-S1-S2-S3-H4-S4-H3-S5-H2, where the αβα-core elements are underlined. In our notation, this ordering is H1-S-H2-up: H1 is followed by five β-strands (S), followed by helix H2. S5 is parallel to S1 (up) and thus structural class I belongs to the broader α-class (24,25) (EcoRI-like (2)).
Class II: helix H1 follows helix H2 in sequence, and S5 is parallel to S1. This connectivity is denoted H2-H1-S-up and belongs to the broader α-class.
Class III: helix H1 is followed by helix H2 in sequence, and then by the β-strands. In this class (which is not a sandwich, neither in structure nor in sequence), the fifth β-strand is directed antiparallel to strand S1. This connectivity is denoted as H1-H2-S-down and belongs to the broader β-class.
Class IV: helix H2 is followed by helix H1 and then by the strands. S5 is antiparallel to S1, and the connectivity is denoted H2-H1-S-down and belongs to the broader β-class.
Class V contains only PvuII (33,34), the smallest (157 residues) among the Type II REases with known structures. It contains only four β strands and one helix (H1), and the connectivity is denoted H1-S-down and belongs to the broader β-class.
Class VI contains only SdaI. The connectivity is denoted H1-S-up and belongs to the broader α-class.
Comparison of structural classifications
The results of our structural classification are summarized in Table 1, along with the corresponding CATH and FSSP/DALI classifications, αβ classification and functional data.
Manual structure classification schemes of SCOP (19) and CATH (18) databases present a hierarchical view in which ‘structure space’ is divided into isolated, non-overlapping ‘islands’ that are denoted by categories such as folds. SCOP unites all Type II REases in a restriction endonuclease-like fold (52980), while CATH assigns 14 REases to the same architecture (3.40, three-layer αβα sandwich), but to five different topologies within this architecture. Notably, our class IV includes all REases with CATH topology 3.40.600 (named after EcoRV endonuclease) and only these. On the other hand, class I combines members from three different CATH topologies, and CATH topology 3.40.91 members are divided between classes I and III. Our classification differs from the CATH topology classification because we base it on the topology of αβα-core elements only, and these are more conserved than the highly variable overall structures of REases. DALI/FSSP classification of folds (32,35) is based on tight clusters of domains in fold space. We note that DALI/FSSP fold 264 (ENDONUCLEASE BAMHI) maps to our class III, and DALI/FSSP fold 263 (TYPE II RESTRICTION ENZYME NGOMIV) maps to our class I with two exceptions.
In summary, Type II REases are grouped differently using different classification methods. This is not surprising, and agrees with the current view that there is no unambiguous way to cluster proteins into discrete groups (36,37). Our classification has the advantage of being based on the most conserved, and thus potentially functionally important, structural elements in the family.
Quantitative geometrical characterization of classes I–VI
Figure 2 shows the structural alignments of the αβα-core elements of REases for each of the six structural classes. The names of the REases and their structures in each class are listed in Table 1. Great variability of angles between secondary structure elements is evident (see Figures 2 and 3) even for the αβα-core elements of the same structural class.
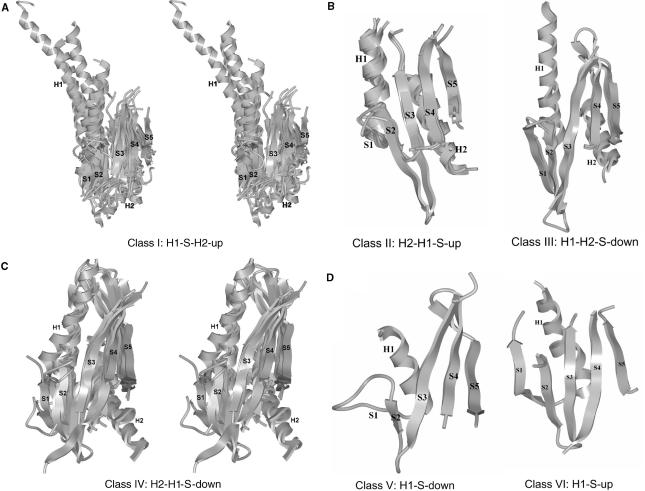
Figure 2.
Ribbon representation of the six αβα-core connectivity-based structural classes. The α-helices H1 and H2, and β-strands S1 to S5, constituting the αβα-core, are shown for superposed structures belonging to each one of the new structural classes: (A) Structures 1cfr, 1knvA, 1na6B, 1wtdB, 1dc1A, 2fokA, 1fiuA and 1qc9A for class I in stereo view; (B) 1sdoA, 2bamA and 1dfmA for class II; 1sa3A and 1ynmA for class III; (C) 1xhvA, 1b94A, 1iawA and 1dmuA for class IV in stereo view; (D) 3pviA for class V and 2ixs for class VI.
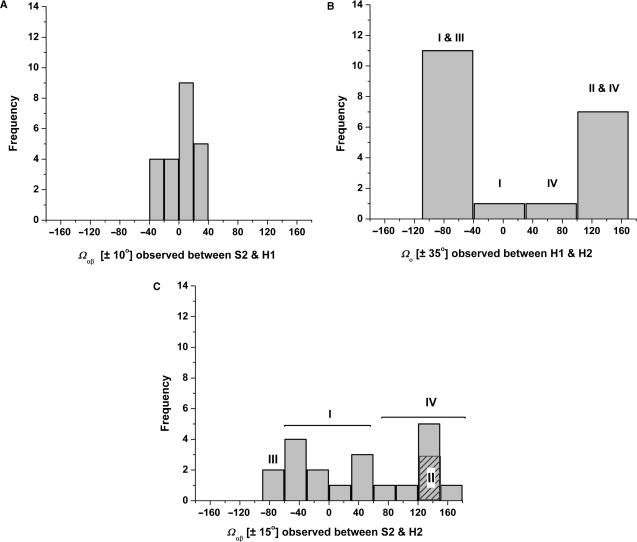
Figure 3.
Distribution of angles between secondary structure elements. (A) S2–H1 angle distribution (based on 22 structures). (B) H1–H2 angle distribution (based on 20 structures). (C) S2–H2 angle distribution (based on 20 structures).
To characterize the arrangement of the pairs of secondary structure elements of the αβα-core, the structures of Type II REases were analyzed, following the procedure of Chou et al. (29,30), in terms of a set of parameters, including distances and angles (as defined in the Materials and methods section). The distribution of distances, D, between the centers of H1 and H2 ranges between 9.5 and 29.4 Å, with an average value of 15.0 Å. The distribution of distances, DM, between the centers of S2 and H1 ranges between 10.5 and 22.3 Å, with an average value of 14.4 Å. Similarly, the distribution of distances, DM, between the centers of S2 and H2 ranges between 10.8 and 18.0 Å with an average value of 13.7 Å. The distributions of the horizontal projected angles Ωo and Ωαβ formed by pairs of secondary structure elements are plotted in Figure 3 (the same scale is used in panels A–C to facilitate visual comparison).
The following patterns emerge from the angular distributions: (a) The frequency of occurrence of Ωαβ for S2–H1 (computed by using a step size of 20°) (Figure 3A) is narrowly concentrated between +40 and −40° for all the structural classes; (b) In contrast, the frequency of occurrence of values of the angle Ωo between H1 and H2 (computed by using a step size of 70°) (Figure 3B) has a bimodal distribution around −60° and around 120° that contains 90% of the structures; the Ωo angle of the structures belonging to classes I and III is in the range [−90, 30] while, for classes II and IV, the Ωo values are within the range [30, 150]. Classes V and VI have only H1 or only H2, respectively. (c) The frequency of occurrence of Ωαβ for S2–H2 (computed by using a step size of 30°; Figure 3C) is widely spread over the ±180° range. This large variation in the angles between core secondary structure elements is rather unique: compare these values, for example, with the 30° range of secondary structure orientations in the superfamilies of globins and immunoglobulins (38,39). Interestingly, the distribution of S2-H2 angles shows a distinctive preferred arrangement for each structural class. This is indicated by the roman numerals used in Figure 3C to show which class is contributing to each bin.
In summary, the orientation of H1 with respect to strand S2 lies within ±40° for all structural classes (Figure 3A), while the orientations of helix H1 with respect to helix H2 (Figure 3B) and of H2 with respect to strand S2 (Figure 3C) correlate with the individual structural classes that we have defined. The orientation of H3 with respect to H1 and to S2 is widespread (−130 to 160 and −120 to 160°, respectively). The orientation of H4 with respect to H1 is narrower and varies between −80 and −20°, whereas the orientation of H4 with respect to S2 is between −50 and −10° (except in PvuII). It is interesting to note that although H4 occurs less frequently in the structures than helices H1, H2 and H3, its connectivity is fully conserved, namely between strands S3 and S4, and its angular distribution with respect to S2 is narrower than that of the other helices. The angular distributions are depicted in Supplementary Figure 1.
Structure–function relations in Type II REases
An intriguing question is whether a correlation exists between structure (as defined by the structural classes and the angles of the secondary elements) and modes of function of Type II REases (as defined by the recognition length, the cleavage site, type and number of recognition sites and the arrangement of the REase monomeric units). The α-class REases (S5 parallel to S1) include our classes I and II, while β-class REases (S5 antiparallel to S1) include our classes III, IV and V. Pingoud and Jeltsch (24) and others (11,21,23–25) have noted that EcoRI-like α-class REases are typically 5′ end cutters (that leave 5′ overhangs of the DNA strand), while EcoRV-like β-class REases, are typically blunt cutters, cutting exactly in the middle and thus leave no ‘sticky’ ends. The structural data set available today is somewhat enriched as compared to the one used by Bujnicki in 2004 (11). In this larger set of REases with known structures, the correlation between α-class and 5′ end cutters is still apparent, with the exception of SdaI, which belongs to α-class but is a 3′ end cutter. The β-class no longer consists mostly of blunt cutters. There are now two examples of 5′ end cutters (MspI and HinP1I; class III) and two examples of 3′ end cutters (BglI, SfiI; class IV), in addition to the three blunt cutters (EcoRV, NaeI and HincII; class IV).
Overall, the newly defined classes correlate rather well with the cleavage patterns, where classes I, II and III are 5′ end cutters, and class IV consists of both 3′ end and blunt cutters. We checked whether the connectivity of helices H3 and H4 would correlate with cleavage patterns and found that this is not the case: Both HincII (H4 between S3 and S4, H3 not present) and EcoRV (H3 after S5, H4 not present) are blunt cutters, but SfiI (H3 after S5, H4 not present as in EcoRV) is a 3′ end cutter (see Table 1). Notably, blunt cutters (except NaeI) have larger H1/H2 and S2/H2 angles than the 3′ end cutters (Table 2).
Table 2.
Class IV angles and cleavage patterns
| REase | PDB code | Class notation | Blunt/sticky | Subtype | Ωo H1/H2 | Ωαβ S2/H2 | Ωαβ S2/H1 |
|---|---|---|---|---|---|---|---|
| HincII | 1xhvA | H2-H1-S-down | blunt | IIP | 138.1 | 154.5 | 18.1 |
| EcoRV | 1b94A | H2-H1-S-down | blunt | IIP | 131.4 | 143.6 | 12.7 |
| NaeI | 1iawA | H2-H1-S-down | blunt | IIE | 73.5 | 85.8 | 37.6 |
| BgII | 1dmuA | H2-H1-S-down | 3′ end | IIP | 104.3 | 109.3 | 12.3 |
| SfiI | 2ezvA | H2-H1-S-down | 3′ end | IIF | 108.1 | 122.4 | 16.6 |
The columns are ordered as follows: REase column lists the names of the REases; PDB code column lists the pdb files used in the analysis; Class notation describes the sequential order of the elements and the direction of the S5 strand with respect to the S1 strand; Blunt/sticky classification, where ‘sticky’ leave 5′ or 3′ DNA end overhang and blunt leave no overhangs upon cleavage; Subtype lists the functional subtypes as defined in the REase nomenclature (1); Ω columns list the angles, calculated as described in the Material and methods section.
In two structural classes, we find a correlation between structural class and the length of the recognition site as well as the particular cleavage position within that sequence: Class II REases recognize a six base-pair site that is cleaved after the first base pair; MspI (C⁁CGG) and HinP1I (G⁁CGC) are the only enzymes that belong to class III, the only ones that recognize sequences of four bases (cleaving after the first base pair) and interact with the recognition sequences as monomers. However, we note that these correlations are derived based on the currently available small data set and should be reevaluated as more REase structures become available.
Roberts and colleagues have introduced a nomenclature that categorizes the different REases according to the type and number of recognition sites and the arrangement of the REase monomeric units (1). Several REase subtypes are represented in our data set (listed in Table 1), namely IIP (homodimers recognizing palindromic sequences), IIE (REases that require two recognition sites but only one is cleaved and the other serves as an allosteric activator), IIF (homotetrameric enzyme concertedly cleaving two recognition sites) and IIS (REases with asymmetric recognition sites that cleave at a defined distance). Interestingly, there is no apparent correlation of these functional subtypes with any of the previous structural classifications of REases, or the one introduced here. For example, the tetrameric Type IIF enzyme SfiI is structurally more similar to the dimeric Type IIP REase BglI (both are class IV in our classification) than to the other Type IIF enzymes, NgoMIV, Cfr10I, Bse634I and Ecl18kI (class I). Class I includes functional subtypes IIP, IIF, IIE and IIS.
The common conserved core (ccc)
Though the angular distribution between secondary structure elements correlates with the structural classes (Figure 3), the variability of the angle between H1 and H2 and of the angle between S2 and H2 is high even within individual structural classes. Therefore, we defined a substructure of the αβα-core that consists of the helix H1 and the strands S1 to S4 as the conserved common core (the ccc). The ccc is conserved in all structures and does not include S5 (which can have an up or down orientation, Figure 3C), or the orientationally variable H2 (Figure 3B).
Various definitions of conserved substructures in Type II REases have been used: Pingoud and Jeltsch noted the absolutely conserved four β-strands (5). Huai and coworkers proposed three conserved β-strands as the common core (23), and Bujnicki discussed the most conserved αβββ core (11). Venclovas and coworkers (21) and Pingoud and Jeltsch (5) have previously highlighted the α-helix and four β-strands structurally conserved between EcoRV and EcoRI, the same substructure that here we call the ccc.
We checked whether other proteins with overall structural similarity to Type II REases share the ccc. We obtained structurally related proteins by submitting each of the Type II REase structures as a query in the DALI (40) server at the European Bioinformatics Institute http://www.ebi.ac.uk/msd-srv/dali/cgi-bin/dali_align.cgi?mode=DBsearch. In the resulting set of 72 proteins, only 11 share the ccc of the Type II REase family, and those are listed in Table 3. Notably, 7 of these proteins are classified into the 263 (restriction endonuclease-like) DALI/FSSP fold and 8 are ester bond hydrolases. Not all of them have been classified in the EC (31) database (which we accessed via http://www.ebi.ac.uk/thornton-srv/databases/enzymes/), but they can be assigned the EC (31) code 3.1.-.- (‘3’ for hydrolases, ‘3.1.’ for hydrolases acting on ester bond) based on their names. Since these hydrolases act on the nucleotide backbone, the ccc emerges as an important structural component common to a range of enzymes that hydrolyze a nucleotide backbone. Many DNA nucleases do not have the ccc, and some are all-α proteins (41).
Table 3.
Structurally related proteins that harbor the ccc substructure
| Protein | PDB code | Class notation | CATH topology | DALI/FSSP fold | EC code |
|---|---|---|---|---|---|
| TnsA endonuclease | 1f1z | H1-S* | 3.40.1350 | 263 | N/A |
| lambda exonuclease | 1avq | H1-S-down | 3.90.320 | 263 | 3.1.11.3 |
| VSR endonuclease | 1vsr | H1-S-H2-up | 3.40.960 | 263 | 3.1.-.- |
| T7 endonuclease I | 1fzr | H1′-S1′- S2-S3-S4-S5-H2-up | 3.40.91 | 263 | 3.1.21.2 |
| Archaeal HJC hydrolase | 1gef | H1-S-H2-down | 3.40.1350 | 263 | N/A |
| XPF/RAD1/MUS81 nuclease | 1j23 | S1-H1-S2-S3-S4-S5-H2-up | 3.40.50 | 263 | N/A |
| HJC hydrolase | 1hh1 | H1-S-H2-down | 3.40.1350 | 263 | N/A |
| RecB endonuclease | 1w36 | H1-H2-S-up | 1.10.3170 | N/A | 3.1.11.5 |
| structural genomics | 1xmx | H1-S-H2-up | N/A | N/A | N/A |
| structural genomics | 1y88 | H1-S-H2-up | 3.40.91 | N/A | N/A |
| structural genomics | 1wdj | H1-S-down | 3.90.1570 | N/A | N/A |
The columns are ordered as follows: Protein column lists the names of the proteins; PDB code column lists the pdb files of structurally related proteins that share the ccc substructure; Class notation describes the sequential order of the elements and the direction of the S5 strand with respect to the S1 strand. The prime is used to indicate that the secondary structure element belongs to a different monomer. Only four strands from the β-sheet in TnsA endonuclease; CATH topology classification (18) DALI/FSSP fold classification (32); EC code, Enzyme Classification code obtained from http://www.ebi.ac.uk/thornton-srv/databases/enzymes/. Some enzymes are not available (N/A) in the database, but all of them can be assigned to EC code 3.1.-.-, hydrolases acting on ester bonds.
A closer inspection of the proteins that share the ccc, reveals that some of them present a novel connectivity of helices and strands, and do not fall into classes I to VI defined above. In bacteriophage T7 endonuclease I (PDB code: 1fzr), for example, the αβα-core is formed by secondary structure elements contributed by two different monomers, with the connectivity H1′ S1′ S2 S3 S4 S5 H2, where the prime is used to indicate that the secondary structure element belongs to a different monomer. We denote this configuration as class VII. Another interesting case is the archaeal nuclease (PDB code: 1j23), whose structure contains the seven secondary structure elements H1, H2 and S1 to S5, but these are connected differently from the classes defined here for Type II REases. In this class, which we denote VIII, H1 is inserted in the sequence between S1 and S2. In the rest of the proteins shown in Table 3, the angles have similar distributions to those found in the Type II REases (not shown).
Three of the proteins identified by these criteria (1y88, the C-terminal domain of 1xmx, and 1wdj) were obtained from structural genomics approaches and have unknown function. To estimate whether the proteins identified by structural genomics are likely to act as nuclease or related hydrolases, we checked the residues that structurally align with the Type II REase catalytic residues. All Type II REases used in this study, as well as the vast majority of the characterized Type II REases and many additional nucleases, harbor the catalytic motif PD-(D/E)XK or variations thereof (7–9,42). Residues D38, E51, K53 in 1y88; D293, E306 and K308 in 1xmx and D86, E116 and R118 in 1wdj are in structural alignment with the catalytic resides D, (D/E) and K. Proteins 1y88 and 1xmx comply with the PD-(D/E)XK motif, and the unorthodox D86–E116XR118 motif found in 1wdj was previously noted by Kosinski et al. (9), who have included the respective protein in the PD-(D/E)XK superfamily (9).
A database of predicted enzymatic functions for unannotated protein structures from structural genomics, termed PDB-UF, was introduced recently (43). The predictions in this web-accessible database are generated by a 3D-Hit approach (44), in which the function is inferred from that of the most similar structure to the unannotated structure, and the 3D-Fun approach, in which the function is determined by analogy to clusters of all proteins structurally similar to the unannotated structure (43). In PDB-UF, 1wdj is classified as ‘hydrolase acting on ester bonds’, EC code 3.1.-.-; 1xmx is not classified; 1y88 is classified as ‘exodeoxyribonucleases producing 5′-phosphomonoesters’ (EC code 3.1.11.-, exonucleases). This is in some disagreement with the sequence-based Conserved Domain Search (45), which identified predicted endonuclease domains (Mrr_cat and RecB) in the 1y88 sequence, suggesting that the 1y88 annotation should be 3.1.21.- (endonuclease, cutting within the DNA strand) rather than 3.1.11.- (exonuclease, chopping a single nucleotide at the end of the strand). Notably, the PDB-UF database flags the assignments of 1wdj and of 1y88 as ‘insignificant or inconsistent’. Our results provide support for the functional annotation of 1y88, 1wdj and the C-terminal part of 1xmx as ‘hydrolases acting on ester bonds’ (EC 3.1.-.-), based on the similarity with the Type II REase ccc, and compatibility with the catalytic hallmark PD-(D/E)XK motif. Futhermore, we assigned the structural genomics hits to the newly defined structural classes based on αβα-core secondary structure elements: 1xmx and 1y88 belong to class I (H1-S-H2-up) and we, therefore, predict that, if these proteins are REases, they are likely to cleave their DNA targets leaving 5′ end overhangs. 1wdj belongs to class V (H1-S-down), and by analogy to PvuII may cleave leaving blunt ends.
DISCUSSION
From a survey of the available X-ray structures of Type II REases, and quantitative analysis of the mutual orientation of their secondary structure elements, we propose the ccc as the largest common unit of all REase structures. Notably, among 72 proteins that share structural similarity with Type II REases, the proteins that have the ccc are nucleases, stressing the functional importance of this substructure for DNA cleavage events, and highlighting a common substructure in such different nucleases as endo- and exonucleases, Holliday Junction resolvases and other ester bond hydrolases. The other three similar proteins sharing the ccc are structural genomics proteins, and the presence of the catalytic residues suggests that these also function as ester-bond hydrolases.
Type II REases have an α/β sandwich architecture, for which the structural space was previously suggested to be described better as a continuum than as distinct folds (46–49). The three-layer sandwich architecture αβα (3.40) is among the most tolerant towards structural change, in terms of ranges of secondary structure orientations, and insertions and deletions of secondary structure elements (50). Thornton and coworkers (46) have introduced the measure of fold similarity, termed gregariousness. Gregarious folds have significant structural overlap with many other folds. Indeed, the average gregariousness for the α/β sandwich architecture is among the highest (46). Interestingly, circular permutation of the overall topology has been demonstrated for some DNA methylases (51,52) (EC code 2.1.1.37) which are functionally coupled to REases and also have α/β sandwich architecture. There is an increasing number of alternative approaches to view folds and describe their plasticity: a gallery of contiguous substructures (53) and a dictionary of ‘protein parts’ (superstructures of secondary elements ignoring sequence order, to describe full-sized proteins) (54) were recently compiled. Grishin and coworkers described a pathway for fold change through ‘structural drift’, in which a protein is a hybrid of two overlapping subdomains, and the old core of any of the subdomains is changed with respect to the parent cores (47–49). The modular and repetitive nature of protein structure, in which the original core is preserved, has been described as the Russian Doll effect, in which a protein at every level is formed by the addition of secondary structural elements to the core of the previous level (55). In this terminology, the smallest Russian Doll of Type II REases is the ccc, which is inserted into the αβα-core in different ways, resulting in the six structural classes that we defined, I to VI.
In analyzing the packing of the αβα-core secondary structure elements, we found that the distribution of Ωαβ for S2/H1 is narrow and similar for all classes, while the distributions of Ωαβ for S2/H2 and Ωo for H1/H2 are widely spread, with particular regions correlating with individual structural classes. Thus, structural classes that were defined based on connectivity of secondary structure elements of αβα-core seem to correlate with mutual orientation of these elements and with cleavage types: Classes I, II and III are 5′ end cutters, while class IV REases are 3′ end or blunt-end cutters. With the exception of NaeI, class IV blunt-end cutters have larger H1/H2 and H1/S2 angles than class IV 3′ end cutters. In addition, class III REases are the only ones that recognize sequences of four base pairs. We suggest that additional structural information for the blunt and 3′ end cutters, for the four base recognizers and for REases of subtypes other than IIP, will be required to further explore the complex structure–function relations of these fascinating proteins.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online and at http://physiology.med.cornell.edu/faculty/niv/3D_alignments.zip.
ACKNOWLEDGEMENTS
We thank Dr Richard J. Roberts for helpful discussions. This research was supported in part by NIH-GM-14312 and NIH-TW-6335 grants to H.A.S., NIH-GM-44006 grant to A.K.A., a Cornell/Weill grant to H.A.S. and H.W. and with the resources of the Computational Biology Service Unit from Cornell University that is partially funded by Microsoft Corporation, and of the Institute for Computational Biomedicine at Weill Cornell Medical College. Funding to pay the Open Access publication charge was provided by a Cornell/Weill grant to H.A.S and H.W.
Conflict of interest statement. None declared.
REFERENCES
- 1.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PubMed] [Google Scholar]
- 3.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE – restriction enzymes and DNA methyltransferases. Nucleic Acids Res. 2005;33:D230–D232. doi: 10.1093/nar/gki029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts RJ. How restriction enzymes became the workhorses of molecular biology. Proc. Natl. Acad. Sci. U.S.A. 2005;102:5905–5908. doi: 10.1073/pnas.0500923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vanamee ES, Aggarwal AK. In: Handbook of Metalloproteins. Messerschmidt A, Cygler M, Bode W, editors. Vol. 3. John Wiley and Sons Ltd; 2004. pp. 742–756. [Google Scholar]
- 7.Bujnicki JM, Rychlewski L. Grouping together highly diverged PD-(D/E)XK nucleases and identification of novel superfamily members using structure-guided alignment of sequence profiles. J. Mol. Microbiol. Biotechnol. 2001;3:69–72. [PubMed] [Google Scholar]
- 8.Bujnicki JM, Rychlewski L. Identification of a PD-(D/E)XK-like domain with a novel configuration of the endonuclease active site in the methyl-directed restriction enzyme Mrr and its homologs. Gene. 2001;267:183–191. doi: 10.1016/s0378-1119(01)00405-x. [DOI] [PubMed] [Google Scholar]
- 9.Kosinski J, Feder M, Bujnicki JM. The PD-(D/E)XK superfamily revisited: identification of new members among proteins involved in DNA metabolism and functional predictions for domains of (hitherto) unknown function. BMC Bioinformatics. 2005;6:172. doi: 10.1186/1471-2105-6-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bujnicki JM. Crystallographic and bioinformatic studies on restriction endonucleases: inference of evolutionary relationships in the ‘midnight zone’ of homology. Curr. Protein Pept. Sci. 2003;4:327–337. doi: 10.2174/1389203033487072. [DOI] [PubMed] [Google Scholar]
- 11.Bujnicki JM. In: Restriction Endonucleases. Pingoud A, editor. Springer, Berlin-Heidelberg; 2004. pp. 63–93. [Google Scholar]
- 12.Pawlak SD, Radlinska M, Chmiel AA, Bujnicki JM, Skowronek KJ. Inference of relationships in the ‘twilight zone’ of homology using a combination of bioinformatics and site-directed mutagenesis: a case study of restriction endonucleases Bsp6I and PvuII. Nucleic Acids Res. 2005;33:661–671. doi: 10.1093/nar/gki213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bujnicki JM. Phylogeny of the restriction endonuclease-like superfamily inferred from comparison of protein structures. J. Mol. Evol. 2000;50:39–44. doi: 10.1007/s002399910005. [DOI] [PubMed] [Google Scholar]
- 14.Deva T, Krishnaswamy S. Structure-based sequence alignment of type-II restriction endonucleases. Biochim. Biophys. Acta. 2001;1544:217–228. doi: 10.1016/s0167-4838(00)00223-5. [DOI] [PubMed] [Google Scholar]
- 15.Bujnicki JM. Understanding the evolution of restriction-modification systems: clues from sequence and structure comparisons. Acta Biochim. Pol. 2001;48:935–967. [PubMed] [Google Scholar]
- 16.Chmiel AA, Bujnicki JM, Skowronek KJ. A homology model of restriction endonuclease SfiI in complex with DNA. BMC Struct. Biol. 2005;5:2. doi: 10.1186/1472-6807-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinch LN, Ginalski K, Rychlewski L, Grishin NV. Identification of novel restriction endonuclease-like fold families among hypothetical proteins. Nucleic Acids Res. 2005;33:3598–3605. doi: 10.1093/nar/gki676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH – a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 19.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 20.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venclovas C, Timinskas A, Siksnys V. Five-stranded beta-sheet sandwiched with two alpha-helices: a structural link between restriction endonucleases EcoRI and EcoRV. Proteins. 1994;20:279–282. doi: 10.1002/prot.340200308. [DOI] [PubMed] [Google Scholar]
- 22.Aggarwal AK. Structure and function of restriction endonucleases. Curr. Opin. Struct. Biol. 1995;5:11–19. doi: 10.1016/0959-440x(95)80004-k. [DOI] [PubMed] [Google Scholar]
- 23.Huai Q, Colandene JD, Chen Y, Luo F, Zhao Y, Topal MD, Ke H. Crystal structure of NaeI – an evolutionary bridge between DNA endonuclease and topoisomerase. EMBO J. 2000;19:3110–3118. doi: 10.1093/emboj/19.12.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pingoud A, Jeltsch A. Recognition and cleavage of DNA by type-II restriction endonucleases. Eur. J. Biochem. 1997;246:1–22. doi: 10.1111/j.1432-1033.1997.t01-6-00001.x. [DOI] [PubMed] [Google Scholar]
- 25.Kovall RA, Matthews BW. Type II restriction endonucleases: structural, functional and evolutionary relationships. Curr. Opin. Chem. Biol. 1999;3:578–583. doi: 10.1016/s1367-5931(99)00012-5. [DOI] [PubMed] [Google Scholar]
- 26.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 27.Abagyan RA, Totrov MM, Kuznetsov DA. ICM: a new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J. Comp. Chem. 1994;15:488–506. [Google Scholar]
- 28.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 29.Chou KC, Nemethy G, Rumsey S, Tuttle RW, Scheraga HA. Interactions between an alpha-helix and a beta-sheet. Energetics of alpha/beta packing in proteins. J. Mol. Biol. 1985;186:591–609. doi: 10.1016/0022-2836(85)90133-0. [DOI] [PubMed] [Google Scholar]
- 30.Chou KC, Nemethy G, Scheraga HA. Energetic approach to the packing of alpha-helices. 1. Equivalent helices. J. Phys. Chem. 1983;87:2869–2881. [Google Scholar]
- 31.Fleischmann A, Darsow M, Degtyarenko K, Fleischmann W, Boyce S, Axelsen KB, Bairoch A, Schomburg D, Tipton KF, et al. IntEnz, the integrated relational enzyme database. Nucleic Acids Res. 2004;32:D434–D437. doi: 10.1093/nar/gkh119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 33.Cheng X, Balendiran K, Schildkraut I, Anderson JE. Crystal structure of the PvuII restriction endonuclease. Gene. 1995;157:139–140. doi: 10.1016/0378-1119(95)00670-2. [DOI] [PubMed] [Google Scholar]
- 34.Athanasiadis A, Vlassi M, Kotsifaki D, Tucker PA, Wilson KS, Kokkinidis M. Crystal structure of PvuII endonuclease reveals extensive structural homologies to EcoRV. Nat. Struct. Biol. 1994;1:469–475. doi: 10.1038/nsb0794-469. [DOI] [PubMed] [Google Scholar]
- 35.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang AS, Honig B. An integrated approach to the analysis and modeling of protein sequences and structures. I. Protein structural alignment and a quantitative measure for protein structural distance. J. Mol. Biol. 2000;301:665–678. doi: 10.1006/jmbi.2000.3973. [DOI] [PubMed] [Google Scholar]
- 37.Kolodny R, Petrey D, Honig B. Protein structure comparison: implications for the nature of ‘fold space’, and structure and function prediction. Curr. Opin. Struct. Biol. 2006;16:393–398. doi: 10.1016/j.sbi.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 38.Lesk AM, Chothia C. How different amino acid sequences determine similar protein structures: the structure and evolutionary dynamics of the globins. J. Mol. Biol. 1980;136:225–270. doi: 10.1016/0022-2836(80)90373-3. [DOI] [PubMed] [Google Scholar]
- 39.Lesk AM, Chothia C. Evolution of proteins formed by beta-sheets. II. The core of the immunoglobulin domains. J. Mol. Biol. 1982;160:325–342. doi: 10.1016/0022-2836(82)90179-6. [DOI] [PubMed] [Google Scholar]
- 40.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 41.Romier C, Dominguez R, Lahm A, Dahl O, Suck D. Recognition of single-stranded DNA by nuclease P1: high resolution crystal structures of complexes with substrate analogs. Proteins. 1998;32:414–424. [PubMed] [Google Scholar]
- 42.Dupureur CM, Dominguez MA., Jr The PD …(D/E)XK motif in restriction enzymes: a link between function and conformation. Biochemistry. 2001;40:387–394. doi: 10.1021/bi001680l. [DOI] [PubMed] [Google Scholar]
- 43.von Grotthuss M, Plewczynski D, Ginalski K, Rychlewski L, Shakhnovich EI. PDB-UF: database of predicted enzymatic functions for unannotated protein structures from structural genomics. BMC Bioinformatics. 2006;7:53. doi: 10.1186/1471-2105-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plewczynski D, Pas J, von Grotthuss M, Rychlewski L. 3D-Hit: fast structural comparison of proteins. Appl. Bioinformatics. 2002;1:223–225. [PubMed] [Google Scholar]
- 45.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrison A, Pearl F, Mott R, Thornton J, Orengo C. Quantifying the similarities within fold space. J. Mol. Biol. 2002;323:909–926. doi: 10.1016/s0022-2836(02)00992-0. [DOI] [PubMed] [Google Scholar]
- 47.Grishin NV. Fold change in evolution of protein structures. J. Struct. Biol. 2001;134:167–185. doi: 10.1006/jsbi.2001.4335. [DOI] [PubMed] [Google Scholar]
- 48.Grishin NV. KH domain: one motif, two folds. Nucleic Acids Res. 2001;29:638–643. doi: 10.1093/nar/29.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krishna SS, Grishin NV. Structural drift: a possible path to protein fold change. Bioinformatics. 2005;21:1308–1310. doi: 10.1093/bioinformatics/bti227. [DOI] [PubMed] [Google Scholar]
- 50.Reeves GA, Dallman TJ, Redfern O, Akpor A, Orengo CA. Structural diversity of domain superfamilies in the CATH database. J. Mol. Biol. 2006;360:725–741. doi: 10.1016/j.jmb.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 51.Gong W, O'Gara M, Blumenthal RM, Cheng X. Structure of pvu II DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res. 1997;25:2702–2715. doi: 10.1093/nar/25.14.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scavetta RD, Thomas CB, Walsh MA, Szegedi S, Joachimiak A, Gumport RI, Churchill ME. Structure of RsrI methyltransferase, a member of the N6-adenine beta class of DNA methyltransferases. Nucleic Acids Res. 2000;28:3950–3961. doi: 10.1093/nar/28.20.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shindyalov IN, Bourne PE. An alternative view of protein fold space. Proteins. 2000;38:247–260. [PubMed] [Google Scholar]
- 54.Szustakowski JD, Kasif S, Weng Z. Less is more: towards an optimal universal description of protein folds. Bioinformatics. 2005;21 Suppl 2:ii66–ii71. doi: 10.1093/bioinformatics/bti1111. [DOI] [PubMed] [Google Scholar]
- 55.Swindells MB, Orengo CA, Jones DT, Hutchinson EG, Thornton JM. Contemporary approaches to protein structure classification. Bioessays. 1998;20:884–891. doi: 10.1002/(SICI)1521-1878(199811)20:11<884::AID-BIES3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.