Abstract
Genes subject to monoallelic expression are expressed from only one of the two alleles either selected at random (random monoallelic genes) or in a parent-of-origin specific manner (imprinted genes). Because high densities of long interspersed nuclear element (LINE)-1 transposon sequence have been implicated in X-inactivation, we asked whether monoallelically expressed autosomal genes are also flanked by high densities of LINE-1 sequence. A statistical analysis of repeat content in the regions surrounding monoallelically and biallelically expressed genes revealed that random monoallelic genes were flanked by significantly higher densities of LINE-1 sequence, evolutionarily more recent and less truncated LINE-1 elements, fewer CpG islands, and fewer base-pairs of short interspersed nuclear elements (SINEs) sequence than biallelically expressed genes. Random monoallelic and imprinted genes were pooled and subjected to a clustering analysis algorithm, which found two clusters on the basis of aforementioned sequence characteristics. Interestingly, these clusters did not follow the random monoallelic vs. imprinted classifications. We infer that chromosomal sequence context plays a role in monoallelic gene expression and may involve the recognition of long repeats or other features. The sequence characteristics that distinguished the high-LINE-1 category were used to identify more than 1,000 additional genes from the human and mouse genomes as candidate genes for monoallelic expression.
Genes expressed from only one allele (monoallelic genes) are either selected at random (random monoallelic genes) or in a parent-of-origin specific manner (imprinted genes). Both types of monoallelic genes reside in chromosomal regions defined by allelic differences in chromatin properties, including replication timing (Table 1). These regions vary in scale from single genes (e.g., IL-2/Il-2), to small gene clusters [e.g., PWS/AS gene region (1)], to the ≈3,000 genes that are silenced during X-inactivation (2).
Table 1. Mouse and human genes that are known to exhibit allelic differences or allelic equivalence.
| Species/gene | Chromosome | Allelic expression | Rep. timing | Reference |
|---|---|---|---|---|
| Random monoallelic | ||||
| Interleukins | ||||
| m | ||||
| Il-2 | 3 | M | A | 39 |
| Il-1a* | 2 | M | - | 42 |
| Il-4 | 11 | M | - | 44 |
| h | ||||
| IL-2 | 4 | M | - | 46 |
| X chromosome | ||||
| m | ||||
| Xist† | X | M | - | 49 |
| h | ||||
| XIST | X | M | A | 51, 52 |
| Immunoglobin loci | ||||
| m | ||||
| Igκ | 6 | M | A | 55 |
| IgL‡ | 16 | M | A | 55 |
| IgH‡ | 12 | M | A | 55 |
| TCRB | 6 | M | A | 55 |
| h | ||||
| Igκ | 2 | M | - | 61 |
| IgL‡ | 22 | M | - | 61 |
| IgH‡ | 14 | M | - | 61 |
| TCRA* | 7 | M | - | 61 |
| TCRB‡ | 14 | M | - | 61 |
| TCRD‡ | 14 | M | - | 61 |
| TCRG‡ | 7 | M | - | 71 |
| Olfactory receptors | ||||
| m | ||||
| Or10 | 14 | M | A | 60 |
| Olfr41‡ | 7 | M | A | 60 |
| Olfr72‡ | 9 | M | A | 60 |
| Ors25‡ | 7 | M | A | 60 |
| Ora16‡ | 2 | M | A | 60 |
| OrM71‡ | 9 | M | A | 60 |
| h | ||||
| OR1E1 | 17 | M | - | 61 |
| OR12D2‡ | 2 | M | - | 61 |
| OR2S2‡ | 9 | M | - | 61 |
| OR51B2‡ | 11 | M | - | 61 |
| NK receptors | ||||
| m | ||||
| Nkg2d* | 6 | M | - | 87 |
| Ly49A*‡ | 6 | M | - | 87 |
| Ly49G2*‡ | Un | M | - | 87 |
| Genes near t complex | ||||
| m | ||||
| Nubp2 | 17 | M | - | 92 |
| Igfals‡ | 17 | M | - | 92 |
| Jsap‡ | 17 | M | - | 92 |
| Imprinted | ||||
| m | ||||
| Tssc3 | 7 | M | - | 37 |
| U2af1-rs1 | 11 | M | - | 40, 41 |
| Peg3 | 7 | M | - | 43 |
| Mas1 | 17 | M | - | 45 |
| Gnas | 2 | M | - | 47 |
| Rasgrf1 | 9 | M | - | 48 |
| Ins1 | 19 | M | - | 50 |
| H19 | 7 | M | A | 41, 53 |
| Snrpn | 7 | M | A | 41, 54 |
| Igf2 | 7 | M | A | 56 |
| m | ||||
| p57(KIP2) | 7 | M | A | 57, 58 |
| Zfp127 | 7 | M | - | 59 |
| Igf2-R | 17 | M | - | 40, 41 |
| Ndn | 7 | M | - | 62 |
| Mash2 | 7 | M | - | 63 |
| h | ||||
| H19 | 11 | M | - | 64 |
| ARHI | 1 | M | - | 66 |
| MAGEL2 | 15 | M | - | 68 |
| PEG10 | 7 | M | - | 69 |
| PLAGL1 | 6 | M | - | 72 |
| ZIM2 | 19 | M | - | 73 |
| ZNF264 | 19 | M | - | 74 |
| TP73 | 1 | M | - | 75 |
| CDKN1C | 11 | M | - | 76 |
| HYMAI | 6 | M | - | 78 |
| NDN | 15 | M | - | 79 |
| NNAT | 20 | M | - | 81 |
| GRB10 | 7 | M | - | 82 |
| ZIM3 | 19 | M | - | 74 |
| GNAS1 | 20 | M | - | 84 |
| DLK1 | 14 | M | - | 85 |
| MEST | 7 | M | - | 86 |
| ZNF215* | 11 | M | - | 88 |
| UBE3A | 15 | M | - | 89 |
| ATP10C | 15 | M | - | 90 |
| IGF2 | 11 | M | - | 41, 91 |
| p57(KIP2) | 11 | M | - | 76 |
| WT1* | 11 | M | - | 93 |
| SNRPN | 15 | M | - | 94 |
| Biallelic | ||||
| m | ||||
| Cebpb | 2 | B | - | 38 |
| Edn3 | 2 | B | - | 38 |
| Chrna4 | 2 | B | - | 38 |
| E2f1 | 2 | B | - | 38 |
| Ntsr | 2 | B | - | 38 |
| Kcnb1 | 2 | B | - | 38 |
| Mc3r | 2 | B | - | 38 |
| C-mpl | 4 | - | S | 39 |
| Tmp | 6 | - | S | 55 |
| Cd2 | 3 | - | S | 55 |
| Rras | 7 | - | S | 41 |
| Alb | 5 | - | S | 41 |
| Myc | 15 | - | S | 60 |
| Pfkl | 10 | - | S | 41 |
| Trp53 | 11 | - | S | 41 |
| h | ||||
| ACTB | 7 | B | - | 65 |
| TSGA14 | 7 | B | - | 67 |
| NAP1L4 | 11 | B | - | 64 |
| IGFBP1 | 7 | B | - | 70 |
| ACHE | 7 | - | S | 41 |
| APOB | 2 | - | S | 41 |
| CD3D | 11 | - | S | 41 |
| MYC | 8 | - | S | 41 |
| TP53 | 17 | - | S | 77 |
| PYGM | 11 | - | S | 41 |
| ERBB2 | 17 | - | S | 80 |
| RB1 | 13 | - | S | 80 |
| RUNX | 21 | - | S | 83 |
m, mouse; h, human; Rep., replication.
Monoallelically expressed in a subset of cells.
Paternally imprinted in some tissues.
Excluded from the analyses that involve individual representatives from each gene family (see supporting information).
On the basis of the inability of X-inactivation to spread into many autosomal regions in X/autosome translocations, it was proposed that way stations located throughout the X chromosome are required for gene silencing to spread (3, 4). The discovery that inactivation correlates with high densities of long interspersed nuclear elements (LINEs) in mice led to the hypothesis that LINE elements are way stations (5). An analysis of human genome sequence supported this hypothesis (6). LINEs are mostly truncated descendants of 6- to 7-kb (7) transposons enriched in chromosomal G bands (8) and in L1 and L2 isochores (genomic DNA fractions in Cs2SO4 density gradients) (9). LINE elements make up 14.5% and 17.6% of mouse and human autosomes and 28.9% and 31.0% of mouse and human X chromosomal sequence (E.A. and Y.M., unpublished data). Other abundant repeats throughout the genomes are the <350-bp short interspersed nuclear elements (SINEs), enriched in chromosomal R bands (8) and H3 and H4 isochores (9) and the retrovirus-derived long terminal repeats (LTRs). A host-defense mechanism (10, 11) is thought to suppress the multiplication of intact elements by the formation of heterochromatin over their sequence (12–15).
The high density of LINE-1 elements around X-inactivated genes raises the possibility that monoallelically expressed autosomal genes are also flanked by high densities of repetitive sequence. Here we show that monoallelic autosomal genes display significantly higher densities of LINE-1 sequence, less truncated LINE-1 elements, and fewer CpG islands and SINE elements in their flanking regions. We find evidence that imprinted and random monoallelic genes each separate into two statistically significant distinct groups. We identify >1,000 additional genes that share the characteristics of one of these groups but are not currently known to exhibit allelic differences in gene expression or chromatin structure.
Methods
Data. We identified 33 random monoallelically expressed (20 mouse, 13 human) genes; 39 imprinted (15 mouse, 24 human) genes; and 28 (15 mouse, 13 human) genes that we designated as being biallelically expressed (Table 1). Genes were assigned to the biallelically expressed category on the basis of gene expression data and/or evidence of synchronous replication timing. All monoallelic genes that have been assayed for replication timing are asynchronously replicating (Table 1 and ref. 16). We consequently assumed that synchronously replicated genes are biallelically expressed. However, we consider it plausible that biallelic genes that are replicated asynchronously exist. The presence of biallelically expressed genes within the aforementioned monoallelic gene set would bias the results of the current study toward the null hypothesis of no difference between the monoallelic and biallelic groups.
The majority of known autosomal monoallelic genes belong to gene families. A single representative from each of these gene families was selected (Table 1). In addition, 150 genes (75 mouse, 75 human) were randomly selected from the National Center for Biotechnology Information Reference Sequence (RefSeq) genes. The location of each gene and the sequence characteristics surrounding it were determined by using the University of California, Santa Cruz (UCSC) Genome Browser (http://genome.cse.ucsc.edu), February 2002 and June 2002 builds for mouse and human, respectively. Repeat information was determined by using the repeatmasker (A. F. A. Smit and P. Green, http://ftp.genome.washington.edu/RM/RepeatMasker.html) track of the UCSC Genome Browser. See Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org, for a more detailed description of the procedures used to obtain data.
Sequence Analysis. A set of 68 covariates that described each gene and the sequence in the region surrounding it, defined as 100 kb to the 3′ and 5′ sides of the gene, was written. Definitions of covariates are in Supporting Methods.
Statistical Methods. Univariate differences in covariates were tested across categorical groupings by using the Kruskal–Wallis test (17). Distributions of covariates across categorical groupings were visualized with boxplots.
To facilitate unsupervised learning, an intrinsic dissimilarity measure was constructed with a random-forest analysis of the covariates describing the 200-kb regions flanking the genes (18–20). We visualized the data via 3D isotonic multidimensional scaling (MDS) plots (21). The Partitioning Around Medoids (PAM) algorithm (22) was used to group the genes into k clusters on the basis of their Euclidean distances in the MDS plot. To evaluate which k best described the data, we used the prediction strength (23). To describe the differences between the resulting clusters in terms of the covariates, we chose a cutoff for each covariate that minimized the average impurity [as measured by the Gini index (24)] over the resulting two subsets (genes with covariate values larger vs. smaller than the cutoff). The most important dichotomized covariates were used in Perl scripts to identify RefSeq genes that share the characteristics of the high LINE-1 cluster.
All statistical analyses were conducted by using the freely available software package r (25), which can be downloaded from http://cran.r-project.org/. See Supporting Methods for a more detailed description of the statistical procedures used.
Results
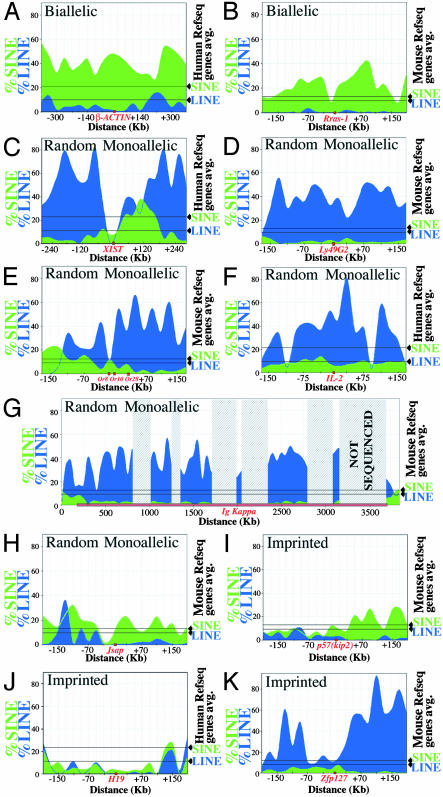
LINEs Are Significantly Less Truncated and More Abundant Around Random Monoallelically Expressed Genes. We asked whether monoallelically expressed autosomal genes are flanked by high densities of repetitive sequence. The repeatmasker track of the UCSC genome browser was used to compare the contributions of repetitive sequence elements in regions flanking known mouse and human random monoallelic, imprinted, and biallelically expressed genes. Visual inspection revealed that human and mouse genes known to be biallelically expressed were flanked by less LINE-1 sequence (Fig. 1 A and B), as compared with gene averages calculated from all RefSeq genes over a 200-kb flanking region (see Methods). In contrast, most (Fig. 1 C–G) but not all (Fig. 1H) of the random monoallelic mouse and human genes examined were flanked by far larger proportions of LINE sequence than the gene average. Most imprinted genes examined did not display above-average LINE-1 sequence (Fig. 1 I and J), although some did (Fig. 1K).
Fig. 1.
LINE-1 and SINE sequence abundance in regions flanking biallelically expressed, random monoallelic, and imprinted genes. Percent LINE-1 and SINE were calculated as the quotient of base pairs of repeats to total base pairs as described (see supporting information). Graphs for percent LINE-1 and SINE sequence are superimposed, with the higher value behind the lower value. Green, SINE sequence content; blue, LINE-1 sequence content; red box, gene; stippled red line, Ig gene region; gray box, region for which sequence is not available. The overall LINE-1 and SINE averages for 200-kb regions flanking all available mouse or human RefSeq genes (see Methods) are marked to the right of each plot. LINE-1 average, blue letters; SINE average, green letters. Biallelically expressed genes: human β-ACTIN (A) and mouse Rras-1 (B). Random monoallelically expressed genes: human XIST (C); mouse Ly49G2 (D); mouse olfactory gene cluster containing Or8, Or10, and Or28 (E); human IL-2 (F); mouse Ig κ gene region (G); mouse Jsap (H). Imprinted genes: mouse p57 (kip2) (I); human H19 (J); mouse Zfp127 (K).
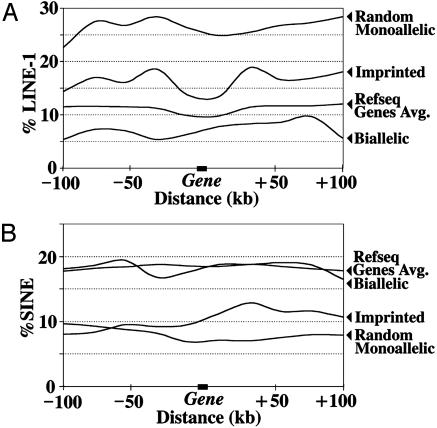
To determine whether these results were representative of all known monoallelic genes and to obtain more detailed description of flanking sequence, 33 random monoallelic, 39 imprinted, and 28 biallelically expressed genes were identified in the literature (Table 1) and the 200-kb flanking sequences compared. Both random monoallelic and imprinted genes were flanked by larger proportions of LINE-1 sequence than the genome average, with the proportion of LINE-1 sequence being highest for random monoallelic genes (Fig. 2A). Biallelically expressed genes displayed lower than average LINE-1 sequence (Fig. 2 A). The proportion of LINE-1 sequence was roughly constant over the 200-kb regions averaged for all available RefSeq genes except in the ≈20-kb region flanking the gene where it was noticeably lower (Fig. 2 A). Finally, both random monoallelic and imprinted genes were flanked by lower-than-average amounts of SINE sequence (Fig. 2B). These LINE and SINE sequence characteristics were maintained throughout clusters of random monoallelic genes but did not continue as one considered sequences progressively further away from monoallelic genes (Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 2.
Average LINE-1 and SINE sequence abundance in regions flanking genes that are or are not subject to monoallelic expression. Percent LINE-1 or SINE sequence in sequential 20-kb sequence windows proceeding 100 kb upstream of the 5′ end of the gene, or extending 100 kb downstream from the 3′ end of the gene, are shown. Densities were calculated as described in Methods. (A) LINE-1 sequence averages. (B) SINE sequence averages.
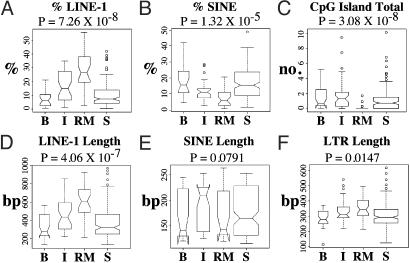
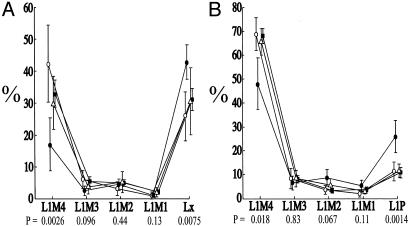
The 200-kb flanking regions for these genes and 150 randomly chosen (75 mouse, 75 human) genes were subjected to more detailed statistical analyses that confirmed the significance of these trends and yielded additional information. LINE-1 and SINE elements, respectively, constituted significantly higher and lower proportions of flanking sequence in random monoallelic genes compared with biallelic and randomly sampled genes (Fig. 3 A and B). LTR and LINE-2 elements did not display significant differences in abundance (not shown). LINE-1 elements were significantly longer around random monoallelic genes than around biallelic genes (Fig. 3D), whereas SINE length was about the same between random monoallelic and biallelic genes (Fig. 3E). Random monoallelic genes were also flanked by significantly fewer CpG islands (Fig. 3C) and significantly lower GC content (not shown). One possible explanation for these findings is that, in both mice and humans, gene conversion among members of each of the gene families (natural killer receptors, olfactory receptors, Ig genes, and T cell receptors) had fortuitously reduced the genomic diversity of the sample set in favor of high densities of flanking LINE-1 sequence. To address this possibility, the analysis was repeated by using a gene set that included only one randomly selected representative from each of these gene families. This analysis also yielded statistically significant differences between the mono- and biallelic gene categories in all of the aforementioned major distinguishing sequence characteristics (Fig. 7, which is published as supporting information on the PNAS web site). Finally, it was found that the LINE-1 elements flanking random monoallelic genes consisted of higher proportions of the evolutionarily more recent mouse-specific (Fig. 4A) and primate-specific (Fig. 4B) LINE-1 subtypes.
Fig. 3.
Boxplots of select covariates. B, biallelically expressed genes; I, imprinted genes; RM, random monoallelically expressed genes; S, random sampling of 150 genes. The width of each box plot reflects the sample size. P values are determined by the Kruskal–Wallis test comparing B, I, and RM genes.
Fig. 4.
Distribution of LINE-1 ages between groups of genes. Plotted are the L1 subfamily mean percentages and their 95% confidence intervals. Below each LINE-1 type are P values across the corresponding values for the biallelic (white circle), imprinted (triangle), and random monoallelic (black circle) genes (Kruskal–Wallis test). Average (see Methods) for a random sample of 150 genes (squares). Mean percentages were calculated as the quotient of subfamily elements and total L1 elements. (A) Five mouse subfamilies are listed from the most ancient (L1M4) to the most recent (Lx). (B) Five human L1 subfamilies listed left to right from the evolutionarily most ancient (L1M4) repeats to the most recent (L1P). See supporting information for more details.
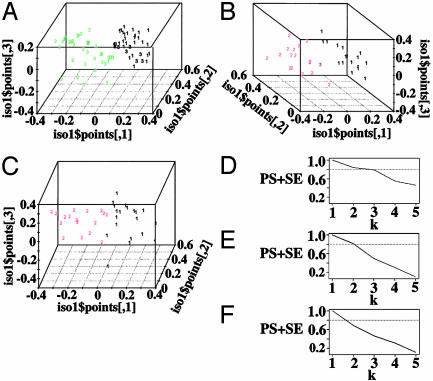
High variability among the statistically significant features that distinguished monoallelic genes from biallelic and randomly sampled genes (Figs. 1 and 3) prompted an investigation into whether the monoallelic genes consisted of more than one category. Combined imprinted and random monoallelic mouse and human genes were clustered on the basis of an intrinsic dissimilarity measure, constructed from 51 covariates describing the 200-kb flanking regions (see Methods). The clustering algorithm found that the most significant separation of genes was by the percentage of LINE-1 sequence (%LINE-1) or the percentage of SINE sequence (%SINE) followed closely by separation according to species, mouse or human (not shown). Clustering according to species (Fig. 5 A and D) was assumed not to pertain to monoallelic gene expression, and the procedure was repeated for the genes from each species.
Fig. 5.
Isotonic multidimensional scaling plots (A–C) and prediction strength plots (D–F) of the 39 imprinted and 33 randomly inactivated genes in Table 1. (A) Mouse (green) and human (black) genes. (B) Mouse genes: black, cluster 1; red, cluster 2. (C) Human genes: black, cluster 1; red, cluster 2. We estimated the number of gene clusters using the prediction strength criterion: the number of clusters is estimated as the largest k such that prediction strength (PS)+SE is >0.8 (dashed line). Prediction strengths plus their standard errors for different choices of the number of clusters (k) are shown. (D) Mouse and human genes. (E) Mouse genes. (F) Human genes.
Strong evidence was found for two distinct clusters among mouse genes (Fig. 5 B and E). The splitting algorithm (see Methods) found that %LINE-1 perfectly characterized the two clusters. All of the genes in one cluster had %LINE-1 >15.5%; the other, <15.5%. The number of Lx and L1P LINE-1 subtypes (higher, respectively, in the mouse and human high LINE-1 clusters), %SINE, and number of CpG islands (both lower in the high LINE-1 cluster) also characterized the two clusters well (Table 2, which is published as supporting information on the PNAS web site). Note that the statistical significance of the mouse data analysis may be reduced by errors in the assembly of the mouse genome sequence. Also note that the proportion of random monoallelic vs. imprinted genes was not statistically significantly different between the clusters (P = 0.13). There was also some evidence for a similar binary clustering of human monoallelic genes (Fig. 5C and Table 2), although the prediction strength criterion for two clusters was not met (Fig. 5F). The number of primate-specific LINE-1 elements best classified the two clusters, with all 17 of the genes with ≤13 primate-specific LINE-1 elements falling in a high LINE-1 cluster and all but three of the remaining genes falling in the other. Again, the proportion of random monoallelic vs. imprinted genes was not statistically significantly different between the clusters (P = 0.09).
Search for Genes That Are Candidates for Monoallelic Expression. The mouse and human genomes were searched for genes that share the characteristics of their respective high LINE-1 groups. Our goal was to obtain a list of monoallelic candidate genes that contains a low number of false positives rather than a comprehensive list of monoallelic genes. Therefore, we chose monoallelic gene predictors with high specificity and sacrificed on their sensitivity. A mouse monoallelic gene predictor of >19% LINE-1 sequence, <5.5% SINE sequence, <10% L1M4 LINE-1 sequence, and <540 CpG island base pairs yielded 526 candidate mouse monoallelic genes (Table 3, which is published as supporting information on the PNAS web site). A human monoallelic gene predictor of >18% LINE-1 sequence, <12% SINE sequence, >13.5%L1P LINE-1 sequence, <75% L1M4 LINE-1 sequence, and <470 CpG island base pairs produced 896 candidate human monoallelic genes (Table 4, which is published as supporting information on the PNAS web site). The high specificity of the predictor came at the cost of lower sensitivity: only 14 of the 18 genes in the mouse high LINE-1 gene cluster satisfied all conditions of the mouse predictor, and only 13 of 20 genes in the human high LINE-1 gene cluster satisfied all conditions. Importantly, none of the known biallelic genes were misclassified as monoallelic genes. Most of the candidate monoallelic genes are not known to be subject to monoallelic expression.
Discussion
We have shown that a majority of the known random monoallelically expressed autosomal genes are located in LINE-1 dense genomic regions. A view increasingly gaining support is that, during X-inactivation, LINE-1 elements (5, 6) act as way stations (3, 4) that promote heterochromatin to spread throughout the X chromosome. The current study further implicates LINE-1 elements in establishing nonequivalent chromatin structures and monoallelic expression at a subset of autosomal genes. These genes, plus a minority of imprinted genes, reside in a distinct chromosomal context characterized by a significantly higher density of L1 sequence, evolutionarily more recent and less truncated LINE elements, fewer CpG islands, and less SINE sequence than biallelic and other monoallelic genes. The exclusion of SINE elements from the flanking regions of imprinted genes had been previously reported (26).
The LINE sequences that helped distinguish monoallelic genes were primarily the relatively evolutionarily recent rodent- and primate-specific varieties (6, 27), suggesting that these LINE elements accumulated in parallel during eutherian radiation. The results of the clustering analysis revealed that only a subset of monoallelic genes is distinguished by high density of LINE sequence. Because animals presumably had functioning olfactory and immune responses before the radiation, we propose that the monoallelic expression of olfactory receptors, Igs, and other genes predates the participation of LINE elements, and that a subset of these genes subsequently adopted a strategy for monoallelic expression that involves LINE elements. The spread of X-inactivation has similarly been hypothesized to predate a role for LINE elements (6).
Trans-interactions have been reported between the alleles of various imprinted genes (28–32) and between the two X chromosomes (S. Diaz-Perez, G. Csankovszki, M. Blanco, V. Gallegos-Garcia, J. Pehrson, R. Jaenisch & Y.M., unpublished work) (33). To explain these interactions, it was proposed that imprinted autosomal gene regions (29) and X chromosomes (34) homologously pair. Support for this model comes from a 3D fluorescence in situ hybridization analysis that showed that the two copies of the imprinted PWS/AS and Ins2/Igf-2/H19 gene clusters homologously associate (35). We propose that random monoallelic genes homologously pair and that LINE, but not SINE, elements function as substrates for homologous pairing (pairing stations). We further propose that a different foundation for homologous pairing exists around the monoallelic genes that are flanked by low LINE densities.
Candidate genes for monoallelic expression included genes for which monoallelic expression has been predicted or is otherwise expected. Pheromone, vomeronasal, and olfactory receptor genes were recovered. Pheromone and vomeronasal receptors are expected to be monoallelically expressed, like the olfactory receptors, to aid in distinguishing airborne chemicals. The list also included protocaherins, which have been predicted to be monoallelically expressed (W. Dreyer, personal communication) within specific area codes of the body (36). Numerous genes involved in the immune response were also identified. Several immune system genes are already known to be subject to monoallelic expression. This regulation is thought to allow for tight regulation of the gene products (e.g., IL2) or to provide specificity of response to antigens (e.g., Igs and T cell receptor genes). Finally, a large number of genes were identified for which no reason for monoallelic expression could be determined.
Supplementary Material
Acknowledgments
We thank Christina Jamieson and Laura Gammill for helpful discussions and critical feedback. We also thank Allen Day and Rebecca Mar for help in the early stages of this project. E.A. and F.T. were supported by National Science Foundation—Integrative Graduate Education and Research Traineeship Program Bioinformatics Award 9987641. E.A. was also supported by a Vigre grant. Y.M. was supported in part by National Institutes of Health Grants GM6100701 and HD041451-02.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: LINEs, long interspersed nuclear elements; SINEs, short interspersed nuclear elements; RefSeq, Reference Sequence.
References
- 1.Nicholls, R. D. & Knepper, J. L. (2001) Annu. Rev. Genom. Hum. Genet. 2, 153–175. [DOI] [PubMed] [Google Scholar]
- 2.Lyon, M. F. (1961) Nature 190, 372–373. [DOI] [PubMed] [Google Scholar]
- 3.Riggs, A. D. (1990) Aust. J. Zool. 37, 419–441. [Google Scholar]
- 4.Gartler, S. M. & Riggs, A. D. (1983) Annu. Rev. Genet. 17, 155–190. [DOI] [PubMed] [Google Scholar]
- 5.Lyon, M. F. (1998) Cytogenet. Cell Genet. 80, 133–137. [DOI] [PubMed] [Google Scholar]
- 6.Bailey, J. A., Carrel, L., Chakravarti, A. & Eichler, E. E. (2000) Proc. Natl. Acad. Sci. USA 97, 6634–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smit, A. F., Toth, G., Riggs, A. D. & Jurka, J. (1995) J. Mol. Biol. 246, 401–407. [DOI] [PubMed] [Google Scholar]
- 8.Holmquist, G. P. (1989) J. Mol. Evol. 28, 469–486. [DOI] [PubMed] [Google Scholar]
- 9.Pavlicek, A., Jabbari, K., Paces, J., Paces, V., Hejnar, J. V. & Bernardi, G. (2001) Gene 276, 39–45. [DOI] [PubMed] [Google Scholar]
- 10.Yoder, J. A., Walsh, C. P. & Bestor, T. H. (1997) Trends Genet. 13, 335–340. [DOI] [PubMed] [Google Scholar]
- 11.Bestor, T. H. (1998) Epigenetics 187–199.
- 12.Dorer, D. R. & Henikoff, S. (1994) Cell 77, 993–1002. [DOI] [PubMed] [Google Scholar]
- 13.Garrick, D., Fiering, S., Martin, D. I. & Whitelaw, E. (1998) Nat. Genet. 18, 56–59. [DOI] [PubMed] [Google Scholar]
- 14.Pal-Bhadra, M., Bhadra, U. & Birchler, J. A. (1997) Cell 90, 479–490. [DOI] [PubMed] [Google Scholar]
- 15.Matzke, M., Matzke, A. J. M. & Schied, O. M. (1994) in Homologous Recombination and Gene Silencing in Plants, ed. Paszkowski, J. (Kluwer, Dordrecht, The Netherlands), pp. 271–307.
- 16.Issa, M., Blank, C. E. & Atherton, G. W. (1969) Cytogenetics 8, 219–237. [DOI] [PubMed] [Google Scholar]
- 17.Kruskal, J. B. (1964) Psychometrika 29, 1–27. [Google Scholar]
- 18.Breiman, L. (1999) Manual on Setting Up and Understanding Random Forests http://stat-www.berkeley.edu/users/breiman/rf.htm.
- 19.Breiman, L. (2001) Machine Learn. 45, 5–32. [Google Scholar]
- 20.Shi, T. & Horvath, S. (2003) in Proceedings of the 7th Joint Conference on Information Sciences, ed. Chiu, D. K. Y. (Association for Intelligent Machinery, Durham, NC), in press.
- 21.Shepard, R. N. (1962) Psychometrika 27, 219–246. [Google Scholar]
- 22.Kaufman, L. & Rousseeuw, P. J. (1990) Finding Groups in Data: An Introduction to Cluster Analysis (Wiley, New York).
- 23.Tibshirani, R., Walther, G., Botstein, D. & Brown, P. (2001) Technical Report http://www-stat.stanford.edu/~timb/ftp/predstr.pdf.
- 24.Breiman, L., Friedman, J. H., Olshen, R. A. & Stone, C. J. (1984) Classification and Regression Trees (Wadsworth, Belmont, CA).
- 25.Ihaka, R. & Gentleman, R. (1996) J. Comput. Graph. Stat. 5, 299–314. [Google Scholar]
- 26.Greally, J. M. (2002) Proc. Natl. Acad. Sci. USA 99, 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smit, A. F. (1996) Curr. Opin. Genet. Dev. 6, 743–748. [DOI] [PubMed] [Google Scholar]
- 28.Hu, J.-F., Vu, T. H. & Hoffman, A. R. (1997) J. Biol. Chem. 272, 20715–20720. [DOI] [PubMed] [Google Scholar]
- 29.LaSalle, J. M. & Lalande, M. (1995) Nat. Genet. 9, 386–394. [DOI] [PubMed] [Google Scholar]
- 30.Shemer, R., Birger, Y., Dean, W. L., Reik, W., Riggs, A. D. & Razin, A. (1996) Proc. Natl. Acad. Sci. USA 93, 6371–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duvillie, B., Bucchini, D., Tang, T., Jami, J. & Paldi, A. (1998) Genomics 47, 52–57. [DOI] [PubMed] [Google Scholar]
- 32.Tsai, J.-Y. & Silver, L. M. (1991) Genetics 129, 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marahrens, Y., Loring, J. & Jaenisch, R. (1998) Cell 92, 657–664. [DOI] [PubMed] [Google Scholar]
- 34.Marahrens, Y. (1999) Genes Dev. 13, 2624–2632. [DOI] [PubMed] [Google Scholar]
- 35.LaSalle, J. M. & Lalande, M. (1996) Science 272, 725–728. [DOI] [PubMed] [Google Scholar]
- 36.Dreyer, W. J. & Roman-Dreyer, J. (1999) Genetica 107, 249–259. [PubMed] [Google Scholar]
- 37.Qian, N., Frank, D., O'Keefe, D., Dao, D., Zhao, L., Yuan, L., Wang, Q. & Keating, M. (1997) Hum. Mol. Genet. 6, 2021–2029. [DOI] [PubMed] [Google Scholar]
- 38.Williamson, C. M., Dutton, E. R., Abbott, C. M., Beechey, C. V., Ball, S. T. & Peters, J. (1995) Genet. Res. 65, 83–93. [DOI] [PubMed] [Google Scholar]
- 39.Hollander, G. A., Zuklys, S., Morel, C., Mizoguchi, E., Mobisson, K., Simpson, S., Terhorst, C., Wishart, W., Golan, D. E., Bhan, A. K., et al. (1998) Science 279, 2118–2121. [DOI] [PubMed] [Google Scholar]
- 40.Barlow, D. P., Stoger, R., Herrmann, B. G., Saito, K. & Schweifer, N. (1991) Nature 349, 84–87. [DOI] [PubMed] [Google Scholar]
- 41.Kitsberg, D., Selig, S., Brandeis, M., Simon, I., Keshet, I., Driscoll, D. J., Nicholls, R. D. & Cedar, H. (1993) Nature 1993, 459–463. [DOI] [PubMed] [Google Scholar]
- 42.Verweij, C. L., Bayley, J. P., Bakker, A. & Kaijzel, E. L. (2001) Adv. Exp. Med. Biol. 495, 129–139. [DOI] [PubMed] [Google Scholar]
- 43.Kuroiwa, Y., Kaneko-Ishino, T., Kagitani, F., Kohda, T., Li, L. L., Tada, M., Suzuki, R., Yokoyama, M., Shiroishi, T., Wakana, S., et al. (1996) Nat. Genet. 12, 186–190. [DOI] [PubMed] [Google Scholar]
- 44.Bix, M. & Locksley, R. M. (1998) Science 281, 1352–1354. [DOI] [PubMed] [Google Scholar]
- 45.Villar, A. J. & Pedersen, R. A. (1994) Nat. Genet. 8, 373–379. [DOI] [PubMed] [Google Scholar]
- 46.Matesanz, F., Delgado, C., Fresno, M. & Alcina, A. (2000) Eur. J. Immunol. 30, 3516–3521. [DOI] [PubMed] [Google Scholar]
- 47.Williamson, C. M., Schofield, J., Dutton, E. R., Seymour, A., Beechey, C. V., Edwards, Y. H. & Peters, J. (1996) Genomics 36, 280–287. [DOI] [PubMed] [Google Scholar]
- 48.Plass, C., Shibata, H., Kalcheva, I., Mullins, L., Kotelevtseva, N., Mullins, J., Kato, R., Sasaki, H. & Hirotsune, S. (1996) Nat. Genet. 14, 106–109. [DOI] [PubMed] [Google Scholar]
- 49.Brockdorff, N., Ashworth, A., Kay, G. F., Cooper, P., Smith, S., McCabe, V. M., Norris, D. P., Penny, G. D., Patel, D. & Rastan, S. (1991) Nature 351, 329–331. [DOI] [PubMed] [Google Scholar]
- 50.Giddings, S. J., King, C. D., Harman, K. W., Flood, J. F. & Carnaghi, L. R. (1994) Nat. Genet. 6, 310–313. [DOI] [PubMed] [Google Scholar]
- 51.Brown, C. J., Ballabio, A., Rupert, J. L., Lafreniere, R. G., Grompe, M., Tonlorenzi, R. & Willard, H. F. (1991) Nature 349, 38–44. [DOI] [PubMed] [Google Scholar]
- 52.Gartler, S. M., Goldstein, L., Tyler-Freer, S. E. & Hansen, R. S. (1999) Hum. Mol. Genet. 8, 1085–1089. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, Y. & Tycko, B. (1992) Nat. Genet. 1, 40–44. [DOI] [PubMed] [Google Scholar]
- 54.Leff, S. E., Brannan, C. I., Reed, M. L., Ozcelik, T., Francke, U., Copeland, N. G. & Jenkins, N. A. (1992) Nat. Genet. 2, 259–264. [DOI] [PubMed] [Google Scholar]
- 55.Mostoslavsky, R., Singh, N., Tenzen, T., Goldmit, M., Gabay, C., Elizur, S., Qi, P., Reubinoff, B. E., Chess, A., Cedar, H., et al. (2001) Nature 414, 221–225. [DOI] [PubMed] [Google Scholar]
- 56.DeChiara, T. M., Robertson, E. J. & Efstratiadis, A. (1991) Cell 64, 849–859. [DOI] [PubMed] [Google Scholar]
- 57.Kagotani, K., Takebayashi, S., Kohda, A., Taguchi, H., Paulsen, M., Walter, J., Reik, W. & Okumura, K. (2002) Biosci. Biotechnol. Biochem. 66, 1046–1051. [DOI] [PubMed] [Google Scholar]
- 58.Hatada, I. & Mukai, T. (1995) Nat. Genet. 11, 204–206. [DOI] [PubMed] [Google Scholar]
- 59.Jong, M. T., Carey, A. H., Caldwell, K. A., Lau, M. H., Handel, M. A., Driscoll, D. J., Stewart, C. L., Rinchick, E. M. & Nicholls, R. D. (1999) Hum. Mol. Genet. 8, 795–803. [DOI] [PubMed] [Google Scholar]
- 60.Chess, A., Simon, I., Cedar, H. & Axel, R. (1994) Cell 78, 823–834. [DOI] [PubMed] [Google Scholar]
- 61.Chess, A. (1998) Science 279, 2067–2068. [DOI] [PubMed] [Google Scholar]
- 62.Watrin, F., Roeckel, N., Lacroix, L., Mignon, C., Mattei, M. G., Disteche, C. & Muscatelli, F. (1997) Eur. J. Hum. Genet. 5, 324–332. [PubMed] [Google Scholar]
- 63.Guillemot, F., Caspary, T., Tilghman, S. M., Copeland, N. G., Gilbert, D. J., Jenkins, N. A. & Nagy, A. (1995) Nat. Genet. 9, 235–242. [DOI] [PubMed] [Google Scholar]
- 64.Onyango, P., Jiang, S., Uejima, H., Shamblott, M. J., Gearhart, J. D., Cui, H. & Feinberg, A. P. (2002) Proc. Natl. Acad. Sci. USA 99, 10599–10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, G., Taneja, K. L., Singer, R. H. & Green, M. R. (1994) Nature 372, 809–812. [DOI] [PubMed] [Google Scholar]
- 66.Yu, Y., Xu, F., Peng, H., Fang, X., Zhao, S., Li, Y., Cuevas, B., Kuo, W. L. & Bast, R. C. J. (1999) Proc. Natl. Acad. Sci. USA 96, 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamada, T., Kayashima, T., Yamasaki, K., Ohta, T. & Kishino, T. (2002) Gene 288, 57–63. [DOI] [PubMed] [Google Scholar]
- 68.Lee, S., Kozlov, S., Hernandez, L., Chamberlain, S. J., Brannan, C. I. & Wevrick, R. (2000) Hum. Mol. Genet. 9, 1813–1819. [DOI] [PubMed] [Google Scholar]
- 69.Ono, R., Kobayashi, S., Wagatsuma, H., Aisaka, K., Kohda, T., Kaneko-Ishino, T. & Ishino, F. (2001) Genomics 73, 232–237. [DOI] [PubMed] [Google Scholar]
- 70.Eggermann, K., Wollmann, H. A., Binder, G., Kaiser, P. & Eggermann, T. (1999) Ann. Genet. 42, 117–121. [PubMed] [Google Scholar]
- 71.Heilig, J. S. & Tonegawa, S. (1987) Proc. Natl. Acad. Sci. USA 22, 8070–8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kamiya, M., Judson, H., Okazaki, Y., Kusakabe, M., Bonthron, D. T. & Hayashizaki, Y. (2000) Hum. Mol. Genet. 9, 453–460. [DOI] [PubMed] [Google Scholar]
- 73.Kim, J., Bergmann, A. & Stubbs, L. (2000) Genomics 64, 114–118. [DOI] [PubMed] [Google Scholar]
- 74.Kim, J., Bergmann, A., Wehri, E., Lu, X. & Stubbs, L. (2001) Genomics 77, 91–98. [DOI] [PubMed] [Google Scholar]
- 75.Kaghad, M., Bonnet, H., Yang, A., Creancier, L., Biscan, J. C., Valent, A., Minty, A. & Caput, D. (1997) Cell 90, 809–819. [DOI] [PubMed] [Google Scholar]
- 76.Matsuoka, S., Thompson, J. S., Edwards, M. C., Bartletta, J. M., Elledge, S. J. & Feinberg, A. P. (1996) Proc. Natl. Acad. Sci. USA 93, 3026–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amiel, A., Litmanovich, T., Gaber, E., Lishner, M., Avivi, L. & Fejgin, M. D. (1997) Hum. Genet. 101, 219–222. [DOI] [PubMed] [Google Scholar]
- 78.Arima, T., Drewell, R. A., Oshimura, M., Wake, N. & Surani, M. A. (2000) Genomics 67, 248–255. [DOI] [PubMed] [Google Scholar]
- 79.Jay, P., Rougeulle, C., Massacrier, A., Moncla, A., Mattei, M. G., Malzac, P., Roeckel, N., Taviaux, S., Lefranc, J. L., Lalande, M., et al. (1997) Nat. Genet. 17, 357–361. [DOI] [PubMed] [Google Scholar]
- 80.Amiel, A., Korenstein, A., Gaber, E. & Avivi, L. (1999) Eur. J. Hum. Genet. 7, 223–230. [DOI] [PubMed] [Google Scholar]
- 81.Evans, H. K., Wylie, A. A., Murphy, S. K. & Jirtle, R. L. (2001) Genomics 77, 99–104. [DOI] [PubMed] [Google Scholar]
- 82.Blagitko, N., Mergenthaler, S., Schulz, U., Wollmann, H. A. & Kalscheuer, V. M. (2000) Hum. Mol. Genet. 9, 1587–1595. [DOI] [PubMed] [Google Scholar]
- 83.Dotan, Z. A., Dotan, A., Litmanovitch, T., Ramon, J. & Avivi, L. (2000) Genes Chromosomes Cancer 27, 270–277. [PubMed] [Google Scholar]
- 84.Hayward, B. E., Kamiya, M., Strain, L., Moran, V. & Bonthron, D. T. (1998) Proc. Natl. Acad. Sci. USA 95, 10038–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kobayashi, S., Wagatsuma, H., Ono, R., Ichikawa, H., Kaneko-Ishino, T. & Ishino, F. (2000) Genes Cells 5, 1029–1037. [DOI] [PubMed] [Google Scholar]
- 86.Nishita, Y., Yoshida, I., Sado, T. & Takagi, N. (1996) Genomics 36, 539–542. [DOI] [PubMed] [Google Scholar]
- 87.Held, W. & Kunz, B. (1998) Eur. J. Immunol. 28, 2407–2416. [DOI] [PubMed] [Google Scholar]
- 88.Alders, M. A. & Ryan, A. (2000) Am. J. Hum. Genet. 66, 1473–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rougeulle, C., Glatt, H. & Lalande, M. (1997) Nat. Genet. 17, 14–15. [DOI] [PubMed] [Google Scholar]
- 90.Meguro, M., Kashiwagi, A., Mitsuya, K., Nakao, M. & Oshimura, M. (2001) Nat. Genet. 28, 19–20. [DOI] [PubMed] [Google Scholar]
- 91.Giannoukakis, N., Deal, C., Paquette, J., Goodyer, C. G. & Polychronakos, C. (1993) Nat. Genet. 4, 98–101. [DOI] [PubMed] [Google Scholar]
- 92.Sano, Y., Shimada, T., Nakasima, H., Nicholson, R. H., Eliason, J., Kocarek, T. A. & Ko, M. S. H. (2001) Genome Res. 11, 1833–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jinno, Y., Yun, K., Nishiwaki, K., Kubota, T., Ogawa, O., Reeve, A. E. & Niikawa, N. (1994) Nat. Genet. 6, 305–309. [DOI] [PubMed] [Google Scholar]
- 94.Reed, M. L. & Leff, S. E. (1994) Nat. Genet. 6, 163–167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.