Abstract
Heat shock protein (hsp)90 functions in a complex chaperoning pathway where its activity is modulated by ATP and by interaction with several co-chaperones. One co-chaperone, p23, binds selectively to the ATP-bound state of hsp90. However, the isolated ATP-binding domain of hsp90 does not bind p23. In an effort to identify the p23-binding domain, we have constructed a series of hsp90 deletion mutants fused with glutathione-S-transferase (GST). Full-length GST-hsp90 is able to bind p23, and also, to chaperone assembly of progesterone receptor complexes. Truncations from the C terminus of GST-hsp90 reveal a C-terminal boundary for the p23-binding domain at approximately residue 490. This fragment contains, in order, the ATP-binding domain, a highly charged region, and 203 residues beyond the charged region. p23 binding is unaffected by deletion of the charged region, indicating that two noncontiguous regions of hsp90 are involved in p23 binding. These regions are only effective when hsp90 is in a dimeric state as shown by loss of p23 binding upon removal of GST or as shown by use of FK506-binding protein12-hsp90 constructs that form dimers and bind p23 only in the presence of a bivalent drug. Thus, p23 binding requires an hsp90 dimer with close proximity between N-terminal regions of hsp90 and a conformation specified by ATP.
Heat shock protein (hsp)90 is an abundant and ubiquitous molecular chaperone that is required to assist the conformational maturation of specific targets involved in many key functions of the cell such as cell-cycle regulation and signal transduction. Among hsp90 targets are several protein kinases such as v-Src, Weel, and c-Raf, transcriptional regulators such as p53 and steroid receptors, and two polymerases: that of hepatitis B virus and telomerase (for review, see refs. 1–4). To date, the best known maturation process driven by hsp90 is the assembly of steroid receptors into a high-affinity hormone-binding conformation. This complex, multistep process occurs in an ATP-dependent manner and involves several other chaperones and co-chaperones (4). Three dynamic steps have been observed in this assembly process (5–7). The receptor initially associates with hsp70, assisted by hsp40. This association develops into an intermediate complex in which the co-chaperone, hsp organizing protein (Hop), associates with both hsp70 on the receptor and hsp90, while the protein Hip binds to hsp70. As the receptor complex matures, hsp90 bound to the receptor interacts with p23 and any one of four tetratricopeptide repeat-containing proteins, FK506-binding protein (FKBP)52, FKBP51, cyclophilin-40, or phosphoprotein phosphatase 5. The details of the interactions between these partner proteins are poorly understood and the mechanisms driving the transitions from one complex to another are still unresolved.
hsp90 is a dimeric protein with multiple domains. Several proteins involved in the hsp90 chaperone pathway possess structurally related tetratricopeptide repeat motifs that mediated their interaction with hsp90 and/or hsp70 (8–11). The common tetratricopeptide repeat acceptor site for steroid receptor-associated immunophilins and Hop is located in the dimerization domain of hsp90 near the C terminus (see Fig. 1). The N-terminal domain of hsp90 contains an adenine nucleotide-binding site, which also binds the benzoquinone ansamycin drug, geldanamycin, a potent inhibitor of hsp90 (12–14). The region between these two domains is poorly understood with regard to structure or function. It contains a highly charged region of ≈70 residues and a potential leucine zipper motif. Several studies indicate that hsp90 functions through marked conformational changes involving interdomain interactions. For example, the binding of Hop near the C terminus of hsp90 is suppressed by the binding of ATP and when Hop is bound to hsp90, the binding and hydrolysis of ATP are inhibited (15, 16).
Figure 1.
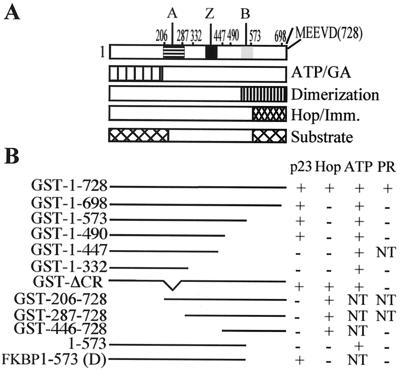
Mutations and structural domains of hsp90. (A) The location of two highly charged regions (A, 206–287 and B, 548–563), a potential leucine zipper region (Z, 378–429), and the conserved MEEVD at the C terminus of hsp90. Regions that bind ATP/geldanamycin (GA) (12–14) or co-chaperones (8–11) plus two domains that have been proposed for substrate binding (29, 30) are also indicated. (B) The hsp90 constructs used in this study are illustrated plus a summary of their activities for binding p23, Hop, and, ATP or for chaperoning PR as active (+), not active (−) or not tested (NT).
The precise role of p23 in the assembly or function of hsp90-chaperoning complexes is still unclear. p23 binds directly to hsp90 and this interaction requires an ATP- and temperature-dependent conformational change in hsp90 (17). In vitro studies with the receptors for glucocorticoids (18) or progesterone (7) show that p23 greatly enhances the efficiency of hsp90 complex assembly and stabilizes the mature complex, but it is not essential for complex formation (18). This latter point is supported by studies in yeast in which deletion of p23 causes only slight effects on cell growth and the functioning of exogenous steroid receptors (19–21). Overexpression of p23 in animal cells enhances the activity of some steroid receptors but suppresses others (22).
Previous efforts to identify the site of p23 binding on hsp90 have not been successful. Mutations in the ATP-binding domain show that this domain is essential for p23 binding (23, 24). However, the isolated domain does not bind p23 (24), and several deletion mutations near the C terminus of hsp90 also have been shown to eliminate p23 binding (25). These studies indicate that p23 binding depends on conformation and could involve several regions of hsp90.
In the present study, we show that p23 binding can be localized to a fragment of hsp90, but binding involves two noncontiguous regions and requires the dimeric structure of hsp90.
Materials and Methods
Plasmids and Constructions.
N- and C-terminally truncated mutants were made by PCR amplification using the chicken hsp90α cDNA, in the pSP72 vector (26) or pSVK3 (27) as template. Amplified fragments were cloned into the procaryotic expression vector pGEX in frame with glutathione-S-transferase (GST, Pharmacia). The FKBP12-hsp90–1-573 fusion protein (FKBP1–573) was prepared by PCR using vector pC4Fv1E containing FKBP12 sequence (furnished by ARIAD Pharmaceuticals, Cambridge, MA) and pET-23 (Novagen) containing full-length chicken hsp90α as templates. After amplification, the fusion sequence was cloned into pET-23. All DNA sequences were confirmed by automated sequencing.
Protein Expression and Purification.
The hsp90 was overexpressed in Escherichia coli and purified from cell lysates by DEAE-cellulose and heparin-agarose column chromatography and Mono Q FPLC as described previously (24). The preparation was >99% pure as assessed by densitometry of SDS/PAGE gels. hsp70, Ydj1, Hop, and p23 were expressed and purified as described previously (16).
GST Fusion Protein Purification.
The expression plasmids for GST-hsp90 and mutants were transformed into E. coli strain UT5600 or BL21(DE3). Cells were sonicated in lysis buffer [10 mM Tris/100 mM KCl, pH 7.5/10 mM monothioglycerol/0.05% Nonidet P-40/4 mM EDTA and the protease inhibitors 0.1 mM leupeptin/0.1 mg/ml bacitracin/77 μg/ml aprotinin/1.5 μm of pepstatin/1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride] and cleared by ultracentrifugation (100,000 × g for 30 min at 4°C). Lysates were loaded on gluthatione-Sepharose. After exhaustive washing, the retained proteins were eluted from the resin with the same buffer containing 30 mM glutathione, pH 7.5. The eluted proteins were then loaded on a Mono Q FPLC column (Pharmacia) and eluted with a linear gradient of 0–1 M KCl. Finally, the peak fractions containing the proteins were dialyzed in 10 mM Tris⋅HCl, 0.1 mM EDTA, 1 mM DTT, and 50 mM KCl, pH 7.5 and concentrated by ultrafiltration. All of the purification steps were carried at 4°C. In some cases, GST was removed from fusion proteins by thrombin digestion. Incubations at 30° for 3 h were performed with 50 units of thrombin/mg of protein in 10 mM Tris, 50 mM KCl, and 2 mM DTT, pH 7.5. GST was removed by adsorption to glutathione-Sepharose and the remaining protein was fractionated on Superdex 200.
Size Exclusion Chromatography.
A Superdex 200 column (10/10, Pharmacia-Biotech) was equilibrated with Tris 20 mM, pH 7.5, 100 mM KCl, 2 mM DTT, and 1 mM EDTA. Elution was performed by using the same buffer at a flow rate of 0.5 ml/min and proteins were detected by the UV A280. The column was calibrated with thyroglubulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), BSA (67 kDa), and chymotrypsinogen A (25 kDa) (Pharmacia Biotech).
p23-Binding Assay.
The binding of p23 to hsp90 or mutants was measured by combining the hsp90 proteins with 5 μg of p23 in a final volume of 200 μl as described previously (17). The binding buffer was 10 mM Tris⋅HCl, pH 7.5, 50 mM KCl, 2 mM DTT, 20 mM Na2MoO4, 0.01% Nonidet P-40, 8 mM MgCl, and 4 mM ATP, and an ATP regeneration system (10 mM phosphocreatine/7 units of creatine phosphokinase). After incubation for 30–60 min at 30°C, the reactions were chilled on ice and added to anti-p23 antibody JJ3 (28) bound to Protein A-Sepharose (20 μl) for 40 min. The resin was resuspended every 5–10 min. The resin pellets were then washed five times with 1-ml volume of binding buffer. Bound proteins were eluted from the antibody resin and resolved by SDS/PAGE (28).
Hop-Binding Assay.
The binding of Hop to hsp90 or hsp90 mutants was measured by combining 5 μg of Hop with 10 μg of hsp90 in 200 μl of 10 mM Tris⋅HCl, pH 7.5, 100 mM KCl, 2 mM DTT, 0.01% Nonidet P-40 (16). The mixture was incubated for 30 min at 30°C, chilled on ice, and adsorbed to anti-Hop antibody F5 bound to Protein A-Sepharose for 40 min. The resin pellets were then washed five times with 1 ml of binding buffer. Bound proteins were resolved by SDS/PAGE.
ATP-Sepharose Binding.
ATP-Sepharose binding was performed as described previously (12) using γ-phosphate-linked ATP (Upstate Biotechnology).
Progesterone Receptor Complex Assembly.
Progesterone receptor (PR) from chick oviduct was adsorbed onto PR22 antibody-Protein A-Sepharose and was assembled into complexes as described by Kosano et al. (7). The incubation contained ≈0.05 μM PR plus 1.4 μM hsp70, 0.8 μM hsp90 dimer, 0.2 μM Ydj-1, and 0.08 μM Hop plus 2.6 μM p23. The samples also contained 20 mM Tris, pH 7.5, 5 mM MgCl2, 2 mM DTT, 0.01% Nonidet P-40, 50 mM KCl, and 5 mM ATP. After incubation for 30 min at 30°C, 0.1 μM [3H] progesterone was added for incubation on ice for 4 h. The complexes were then isolated and assessed for bound progesterone and for protein composition.
Results
The hsp90 constructs used in this study are illustrated in Fig. 1 along with a summary of their binding activities. Most of these were prepared as N-terminally fused proteins with GST or with FKBP12. The proteins were overexpressed in E. coli and purified.
GST-hsp90 Is an Active Chaperone.
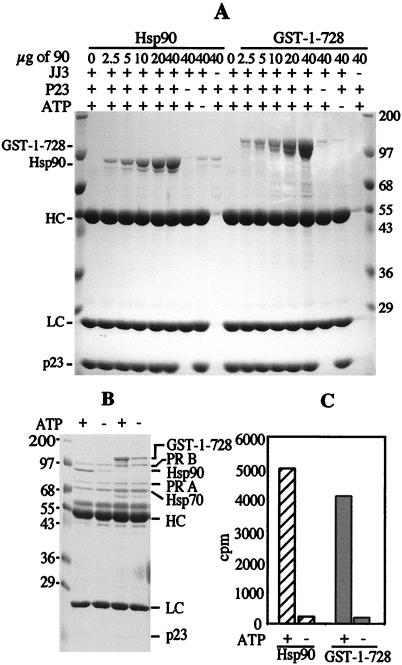
It was of initial interest to see if the fusion of GST to the amino terminus of hsp90 altered the activity of this protein. GST exists as a stable dimer (31–33) and would, therefore, join hsp90 monomers in a region that is not known to have subunit interactions. We compared the binding of p23 to hsp90 and to GST-1–728 as shown in Fig. 2A. p23 was combined with hsp90 or GST-1–728 under conditions that promote binding (see Materials and Methods). Complexes were assessed by immunoprecipitation by using an antibody to p23 followed by SDS/PAGE. GST-1–728 binds p23 in an ATP-dependent manner that is almost identical to that of hsp90. The two hsp90 proteins appear to have similar affinities for p23 and the ATP concentration dependencies for these interactions are also comparable (results not shown). We also tested the ability of GST-1–728 to bind and chaperone the PR. This is a more demanding assay that requires the additional proteins hsp40 (Ydj1), hsp70 and Hop (7). Under proper conditions, the monomeric PR forms a complex with these proteins that promotes the hormone-binding activity of the receptor. The two receptor forms, PR-A and PR-B, appear to behave identically in complex formation (34). As shown in Fig. 2 B and C, GST-1–728 is active in complex formation and in achieving the hormone-binding state, even though its activity is somewhat less than wild-type hsp90. These results demonstrate that hsp90 functions well even when the amino-terminal ends are restricted by GST.
Figure 2.
hsp90 and GST-1–728 have comparable p23-binding and PR assembly activities. (A) p23 (5 μg) was incubated with increasing concentrations of GST-1–728 or hsp90 in the presence of 2 mM ATP and the regeneration system as described in Materials and Methods. The proteins in complexes were resolved by SDS/PAGE. The position of Mr markers plus antibody heavy chain (HC) and light chain (LC) are indicated. The samples without hsp90 contained 40 μg of GST. (B and C) Progesterone receptor complexes were assembled in vitro in the presence of 5 mM ATP, 2 μg of Ydj 1, 20 μg of hsp70, 5 μg of Hop, 5 μg of p23, and either 20 μg of hsp90 or 26 μg of GST-1–728. Proteins in complex with PR were assessed by SDS/PAGE (B) and hormone-binding activity was measured (C).
The C-Terminal Domains Are not Required for p23 Binding.
Previous studies by Chen et al. (25) showed that p23 binding is markedly reduced by mutations at the C terminus and within the dimerization domain even though ATP binding near the N terminus is also required. Obermann et al. (23) reported that removal of 103 residues from the C terminus abolished the interaction of hsp90 with p23 in a yeast two-hybrid system. However, when tested as GST fusion proteins, we found that deletions from the C terminus were tolerated as shown in Fig. 3A. The C-terminal deletion mutant, GST-1–698, lacking 30 amino-acids of hsp90α was able to bind p23. This interaction had the same requirement for ATP as did wild-type. Therefore, the conserved MEEVD sequence motif was not essential for this activity.
Figure 3.
Effects of hsp90 deletions on p23 binding. (A) GST-1–728 and GST-1–698 were compared for p23 binding in the presence or absence of 2 mM ATP. Background measurements were assessed by omitting p23 or the p23 antibody JJ3. (B) hsp90, GST-1–728, and GST-1–573 were assessed for p23 binding in the presence or absence of ATP, adenosine 5′-diphosphate, or geldanamycin (GA). (C) The GST-hsp90 constructs indicated were compared for p23 binding in the presence or absence of ATP. In all cases, the hsp90 constructs from p23 complexes were resolved by SDS/PAGE and stained with Coomassie Blue.
A series of C-terminal truncations was tested to determine the boundary of the p23-binding region. The predominant dimerization domain of hsp90 is near the C terminus (35) and GST-1–573 lacks most of this domain (Fig. 1). However, this mutant clearly binds p23 (Fig. 3B). This interaction was supported by ATP, but not by ADP, and it was blocked by addition of the hsp90 inhibitor, geldanamycin, as shown previously for full-length hsp90 (17). Further truncation to GST-1–490 did not alter p23 binding. However, the additional deletion of 43 residues (GST-1–447) completely abolished p23 binding (Fig. 3C). These 43 residues are not sufficient for p23 binding because GST-446–728 has no p23-binding activity (Fig. 3C). However, this mutant was able to interact with Hop, indicating that it was produced as a functional fragment (Fig. 1 and results not shown). Thus, the C-terminal boundary of the p23-binding domain lies between residues 447 and 490 of hsp90.
p23 Binding Requires Two Separate Regions of hsp90.
GST-1–490 contains the ATP-binding domain followed by a highly charged region plus 203 residues beyond this region. Previous studies indicated that the isolated ATP-binding domain (residues 1–221) is unable to bind p23 (12). Fig. 3C shows that a larger fragment, GST-1–332, containing the charged domain, is also unable to bind p23. This fragment was fully capable of binding to ATP-Sepharose supporting the integrity of its structure (Fig. 1 and results not shown).
We next tested the significance of the charged domain in the p23 interaction. This domain has been shown to improve the binding of denatured proteins to an N-terminal fragment of hsp90 in vitro (36); however, hsp90 lacking the charged domain is able to maintain viability in yeast lacking endogenous hsp90 (37). Because the charged domain lies between two regions needed for p23 binding, we expected it to be important for this interaction. However, when this region, residues 206–287, was deleted from full-length hsp90 (GSTΔCR), normal binding of p23 was observed (Fig. 3C). Thus, the charged region is not part of the p23-binding site nor is it needed for proper conformation of this site.
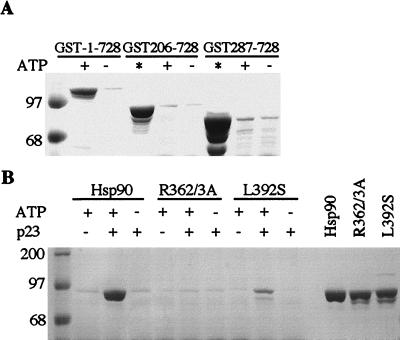
It is possible that deletion of the ATP-binding domain would allow the binding of p23 to region 287–490 in an unregulated fashion. This possibility was tested by using two constructs; GST-206–728, which contains the charged region, and GST-287–728, lacking the charged region. Neither of these proteins was able to bind p23 (Fig. 4A), even though they both bound well to Hop (Fig. 1 and results not shown). Because the close proximity of GST might interfere with p23 binding to these fragments, they were also tested after proteolytic removal of the GST. Again, no p23 binding was observed (not shown). These results show that p23 binding cannot be confined to residues 287–490 but also requires direct or indirect (conformational) contributions from the ATP-binding domain.
Figure 4.
Binding of hsp90 mutants to p23. (A) GST-206–728 and GST-287–728 were assessed for p23 binding in the presence or absence of ATP. GST-1–728 was included as a positive control and the total protein assayed for each mutant is shown in lanes labeled (*) (B) p23 Binding is shown using hsp90 or the two full-length hsp90 mutants, R362/3A and L392S. The total protein assayed is shown in the last three lanes.
As a further test for the importance of region 287–490 in p23 binding, two point mutations were made in this region of the full-length hsp90 protein. In one case, R362 and R363 were both changed to alanine, and in the other, L392 was changed to serine. These residues were selected because they exist in two highly conserved regions of the hsp90 sequence (38). L392 is in a region proposed to have a leucine-zipper structure that may be involved in protein interactions (39). Both of these mutants co-migrate with wild-type hsp90 on gel filtration and they are able to bind Hop (not shown). As shown in Fig. 4B, R362/3A lost the ability to bind p23. L392S bound p23 but appears to be somewhat less active than hsp90wt. Thus, region 287–490 clearly participates in p23 binding when using full-length hsp90 in the absence of GST.
A Dimer Structure Is Required for p23 Binding.
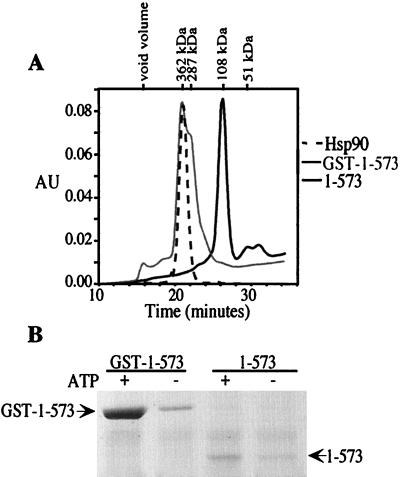
To test the influence of the GST dimer on p23 binding to hsp90 fragments, GST was cleaved by thrombin digestion and removed by adsorption to glutathione-Sepharose. This was first done by using GST-1–698, and surprisingly, most of the p23-binding activity was lost. When the preparation was analyzed by gel filtration, it showed heterogeneity consistent with a mixture of monomeric and dimeric forms (not shown). To more clearly illustrate the importance of the dimer, fragment 1–573 was obtained after thrombin cleavage. This fragment appears to be monomeric (Fig. 5A), and it has no p23-binding activity (Fig. 5B). Thus, only the dimeric form of hsp90 is able to bind p23.
Figure 5.
GST-dependent binding of 1–573 to p23. Fragment 1–573 was prepared by cleavage of GST (see Materials and Methods). (A) Size exclusion chromatography on Superdex 200 was used to compare the sizes of hsp90, GST-1–573, and 1–573. (B) The binding of p23 to GST-1–573 and to 1–573 is shown plus and minus ATP. Equivalent amounts of protein were used for the binding assay.
Finally, to demonstrate unambiguously the requirement for dimerization, a fusion protein was prepared with FKBP12 attached to the N terminus of hsp90–1-573. This exists in monomeric form, but it can be induced to form dimers through interaction of the FKBP12 drug-binding sites with the bivalent drug AP20187 (ARIAD) (40, 41). Purified FKBP-1–573 (Fig. 6A) was combined with the drug at various ratios and the extent of dimer formation was assessed by native gel electrophoresis (Fig. 6B). Optimal dimerization occurred with a drug:protein ratio of approximately 1:2 and diminished when the drug was in excess. The ability of these protein forms to bind p23 is shown in Fig. 6C. Monomeric FKBP-1–573 was unable to bind p23, but substantial binding was achieved with drug addition at ratios that promote dimerization, confirming the dimer requirement for this hsp90 activity. p23 binding is optimal with a drug:protein ratio of 1:4, slightly less than the optimum for dimerization. The reason for this is unknown.
Figure 6.
Dimer-dependent binding of p23. (A) SDS/PAGE analysis of purified 1–573 and FKBP-1–573 proteins. (B) Dimerization of FKBP-1–573 is dependent on the proportion of added bivalent drug. FKBP-1–573 was incubated for 1 h at 30° in the presence of the bivalent drug AP20187 (closely related to AP1903; ref. 38). drug/FKBP-1–573 ratios were varied between 0 and 2, and monomers (M) and dimers (D) were resolved by native PAGE. (C) p23 Binding correlates with drug-induced dimerization. Samples were prepared with FKBP-1–573 plus various ratios of AP20187 as in B and tested for p23-binding activity. Shown is the amount of FKBP-1–573 bound to p23 and resolved by SDS/PAGE and Western blotting with an antibody to hsp90.
Discussion
Earlier attempts to define the site of p23 binding on hsp90 were not successful, but suggested that multiple regions may be involved (23, 25). The difficulties of these studies are explained by the present results. p23 binding is a complex process that requires (i) the binding of ATP at the nucleotide-binding domain, (ii) communication between the nucleotide-binding domain and a noncontiguous downstream domain, and (iii) a conformation that is expressed only by hsp90 in a dimeric state. Thus, the interaction of p23 with hsp90 can be altered by mutations that influence ATP binding, the downstream domain or dimer formation and, consequently, p23 binding is a stringent test for the integrity of hsp90 quaternary structure.
The structure and biochemistry of GST are well known, and this protein has been used in other studies as an agent for dimer formation (31–33, 42, 43). At the start of this study, we anticipated the need to remove GST from our constructs once they had been expressed and purified. When free in solution, hsp90 is dimerized through contacts near the C terminus, leaving the N-termini distant from one another (35). However, electron microscopy studies show a relatively linear or open hsp90 dimer in untreated samples that is converted to a circularized molecule by treatment with heat or ATP, suggesting new interactions near the N-termini (44). Also, an N-terminal fragment of yeast hsp90 has been crystallized as a dimer but with only a small region of dimer contacts that may or may not be of physiological importance (14). Based on this structure, it has been proposed that the N-terminal region of hsp90 can open and close in a clamp-like mechanism to interact with target proteins (45). Thus, several lines of evidence suggest that flexibility within N-terminal domains may be important in the functioning of hsp90. We were, therefore, surprised to find that restriction of the N terminus by GST had little effect on the ATP-dependent binding of p23 or on the more complex chaperoning of the progesterone receptor. We also have tested GST-hsp90 constructs in an assay for the folding of firefly luciferase which involves hsp70, hsp40, Hop and hsp90 (16, 46). As with PR chaperoning, activity was observed with full-length GST-1–728, but not with deletion mutants of hsp90.
The dimerization requirement for p23 binding is shown by the GST-hsp90 constructs, and more rigorously by the FKBP12-hsp90 proteins. However, the close proximity of N-terminal domains induced in these fusion proteins is not sufficient for p23 binding. Conformational changes induced by ATP binding remain essential for p23 interaction. This is likely to involve conformational changes that lead to additional dimer contacts between the hsp90 monomers near the ATP-binding domain. p23 binding may facilitate and stabilize this conformational state of hsp90.
Our results show that the charged region of hsp90 is a discrete domain that can be removed without altering the conformational interactions needed for p23 binding. This indicates that the charged region is not a functional bridge between domains involved in p23 binding. However, it is essential for the chaperoning activity of hsp90 as tested in the progesterone receptor assay (Fig. 1) and the luciferase assay (46). On the other hand, this region is lacking in bacterial hsp90 (HtpG) (38) and in the mitochondrial homolog, TRAP-1 (47). Also, yeast viability is maintained by hsp90 lacking the charged region (37), indicating that it is needed for some hsp90 functions but not others.
A model to illustrate the functional states of hsp90 is shown in Fig. 7. For simplicity, hsp70 and hsp40 are omitted even though they appear to be important for presentation of target proteins to hsp90. Major dimer contacts exist for hsp90 near the C terminus (35), but additional dimer contacts are proposed near the ATP-binding domain that are enhanced by ATP and bring this domain in close proximity with a downstream domain (compare Fig. 7 A and B). The charged domain is shown as an independent structure that does not obscure interaction between the ATP-binding domain and a downstream region. This interaction, which occurs in the presence of ATP, is needed for p23 binding. The open conformation (Fig. 7C) is observed with free, purified hsp90, but this may be an inactive state which binds and hydrolyzes ATP poorly. Conversion to state A or B may occur through interactions with either co-chaperones or target proteins, or both. The sites of target protein binding are still uncertain and are not indicated in the model. Both N-terminal and C-terminal hsp90 fragments have been shown to interact with denatured proteins and peptides (29, 30); however, these fragments are inactive in more complex hsp90 chaperone assays.
Figure 7.
A model for the conversion of hsp90 through three conformational states. States A and B show differential interactions in the absence and presence of ATP. State C is an open nonfunctional conformation of hsp90.
In two ways, hsp90 appears to bind steroid receptors (4–7); first in an intermediate complex where the receptor is inactive and hsp90 is bound to Hop, but not ATP. This complex is converted to one in which the receptor gains hormone-binding activity, hsp90 binds p23 and ATP, and Hop is replaced by an immunophilin (IP). ATP and p23 may act cooperatively to induce conformational changes that transform the C-terminal domain to one that favors the binding of immunophilins over Hop. The ATPase activity of hsp90 is inhibited by Hop, but not by the immunophilin CyP-40 (Cpr6) (15). Thus, ATP hydrolysis can occur from state B, perhaps resulting in dissociation of hsp90 from its substrate and co-chaperone. The two states of hsp90 (Fig. 7 A and B) involve conformational transitions in hsp90 and the target protein and appear to require every domain of hsp90, perhaps including two sites for target protein binding. Further studies on the regulation of nucleotide binding and hydrolysis and the influence of co-chaperones are key aspects for understanding these transitions and the mechanism of hsp90 action.
Acknowledgments
We thank X. Meng and J. Devin-Leclerc for assistance in the preparation of GST constructs and Sara Felts and Sherry Linander for assistance in manuscript preparation. FKBP12 plasmid and the drug AP20187 were kindly provided by ARIAD Pharmaceuticals. This work was supported by National Institutes of Health Grants DK 46249 and HD 90140 (part of the Specialized Cooperative Center Program in Reproduction Research) to D.T. and by Association pour la Recherche sur le Cancer and Ligue Contre le Cancer (Indre) grants to M.G.C.
Abbreviations
- hsp
heat shock protein
- GST
glutathione-S-transferase
- FKBP
FK506-binding protein
- Hop
hsp organizing protein
- PR
progesterone receptor
Note Added in Proof.
Prodromou et al. (48) have shown evidence for dimer interactions within the N-terminal half of hsp90 that relate to ATPase activity and the binding of p23.
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.220430297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.220430297
References
- 1.Caplan A J. Trends Cell Biol. 1999;9:262–268. doi: 10.1016/s0962-8924(99)01580-9. [DOI] [PubMed] [Google Scholar]
- 2.Buchner J. Trends Biochem Sci. 1999;24:136–141. doi: 10.1016/s0968-0004(99)01373-0. [DOI] [PubMed] [Google Scholar]
- 3.Mayer M P, Bukau B. Curr Biol. 1999;9:R322–R325. doi: 10.1016/s0960-9822(99)80203-6. [DOI] [PubMed] [Google Scholar]
- 4.Pratt W B. Proc Soc Exp Biol Med. 1998;217:420–434. doi: 10.3181/00379727-217-44252. [DOI] [PubMed] [Google Scholar]
- 5.Smith D F. Mol Endocrinol. 1993;7:1418–1429. doi: 10.1210/mend.7.11.7906860. [DOI] [PubMed] [Google Scholar]
- 6.Dittmar K D, Banach M, Galigniana M D, Pratt W B. J Biol Chem. 1998;273:7358–7366. doi: 10.1074/jbc.273.13.7358. [DOI] [PubMed] [Google Scholar]
- 7.Kosano H, Stensgard B, Charlesworth M C, McMahon N, Toft D O. J Biol Chem. 1998;273:32973–32979. doi: 10.1074/jbc.273.49.32973. [DOI] [PubMed] [Google Scholar]
- 8.Owens-Grillo J K, Hoffmann K, Hutchison K A, Yem A W, Diebel M R, Jr, Handschumacher R E, Pratt W B. J Biol Chem. 1995;270:20479–20484. doi: 10.1074/jbc.270.35.20479. [DOI] [PubMed] [Google Scholar]
- 9.Smith D F. Biol Chem. 1998;379:283–288. doi: 10.1515/bchm.1998.379.3.283. [DOI] [PubMed] [Google Scholar]
- 10.Carrello A, Ingley E, Minchin R F, Tsai S, Ratajczak T. J Biol Chem. 1999;274:2682–2689. doi: 10.1074/jbc.274.5.2682. [DOI] [PubMed] [Google Scholar]
- 11.Young J C, Obermann W M J, Hartl F U. J Biol Chem. 1998;273:18007–18010. doi: 10.1074/jbc.273.29.18007. [DOI] [PubMed] [Google Scholar]
- 12.Grenert J P, Sullivan W P, Fadden P, Haystead T A J, Clark J, Mimnaugh E, Krutzsch H, Ochel H-J, Schulte T W, Sausville E, et al. J Biol Chem. 1997;272:23843–23850. doi: 10.1074/jbc.272.38.23843. [DOI] [PubMed] [Google Scholar]
- 13.Stebbins C E, Russo A A, Schneider C, Rosen N, Hartl F U, Pavletich N P. Cell. 1997;89:239–250. doi: 10.1016/s0092-8674(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 14.Prodromou C, Roe S M, O'Brien R, Landbury J E, Piper P W, Pearl L H. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 15.Prodromou C, Siligardi G, O'Brien R, Woolfson D N, Regan L, Panaretou B, Ladbury J E, Piper P W, Pearl L H. EMBO J. 1999;18:754–762. doi: 10.1093/emboj/18.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson B D, Schumacher R J, Ross E D, Toft D O. J Biol Chem. 1998;273:3679–3686. doi: 10.1074/jbc.273.6.3679. [DOI] [PubMed] [Google Scholar]
- 17.Sullivan W, Stensgard B, Caucutt G, Bartha B, McMahon N, Alnemri E S, Litwack G, Toft D O. J Biol Chem. 1997;272:8007–8012. doi: 10.1074/jbc.272.12.8007. [DOI] [PubMed] [Google Scholar]
- 18.Dittmar K D, Demady D R, Stancato L F, Krishna P, Pratt W B. J Biol Chem. 1997;272:21213–21220. doi: 10.1074/jbc.272.34.21213. [DOI] [PubMed] [Google Scholar]
- 19.Bohen S P. Mol Cell Biol. 1998;18:3330–3339. doi: 10.1128/mcb.18.6.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang Y, Fliss A E, Rao J, Caplan A J. Mol Cell Biol. 1998;18:3727–3734. doi: 10.1128/mcb.18.7.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knoblauch R, Garabedian M J. Mol Cell Biol. 1999;19:3748–3759. doi: 10.1128/mcb.19.5.3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freeman B C, Felts S J, Toft D O, Yamamoto K R. Genes Dev. 2000;14:422–434. [PMC free article] [PubMed] [Google Scholar]
- 23.Obermann W M J, Sondermann H, Russo A A, Pavletich N P, Hartl F U. J Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grenert J P, Johnson B D, Toft D O. J Biol Chem. 1999;274:17525–17533. doi: 10.1074/jbc.274.25.17525. [DOI] [PubMed] [Google Scholar]
- 25.Chen S, Sullivan W P, Toft D O, Smith D F. Cell Stress Chaperones. 1998;3:118–129. doi: 10.1379/1466-1268(1998)003<0118:diopat>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang K I, Devin J, Cadepond F, Jibard N, Guiochon-Mantel A, Baulieu E E, Catelli M G. Proc Natl Acad Sci USA. 1994;91:340–344. doi: 10.1073/pnas.91.1.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meng X, Devin J, Sullivan W P, Toft D, Baulieu E-E, Catelli M G. J Cell Sci. 1996;109:1677–1687. doi: 10.1242/jcs.109.7.1677. [DOI] [PubMed] [Google Scholar]
- 28.Johnson J L, Toft D O. J Biol Chem. 1994;269:24989–24993. [PubMed] [Google Scholar]
- 29.Young J C, Schneider C, Hartl F U. FEBS Lett. 1997;418:139–143. doi: 10.1016/s0014-5793(97)01363-x. [DOI] [PubMed] [Google Scholar]
- 30.Scheibel T, Weikl T, Buchner J. Proc Natl Acad Sci USA. 1998;95:1495–1499. doi: 10.1073/pnas.95.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McTigue M A, Williams D R, Tainer J A. J Mol Biol. 1995;246:21–27. doi: 10.1006/jmbi.1994.0061. [DOI] [PubMed] [Google Scholar]
- 32.Tudyka T, Skerra A. Protein Sci. 1997;6:2180–2187. doi: 10.1002/pro.5560061012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaplan W, Hüsler P, Klump H, Erhardt J, Sluis-Cremer N, Dirr H. Protein Sci. 1997;6:399–406. doi: 10.1002/pro.5560060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith D F, Faber L E, Toft D O. J Biol Chem. 1990;265:3996–4003. [PubMed] [Google Scholar]
- 35.Nemoto T, Ohara-Nemoto Y, Ota M, Takagl T, Yokoyama K. Eur J Biochem. 1995;233:1–8. doi: 10.1111/j.1432-1033.1995.001_1.x. [DOI] [PubMed] [Google Scholar]
- 36.Scheibel T, Siegmund H I, Jaenicke R, Ganz P, Lilie H, Buchner J. Proc Natl Acad Sci USA. 1999;96:1297–1302. doi: 10.1073/pnas.96.4.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Louvion J-F, Warth R, Picard D. Proc Natl Acad Sci USA. 1996;93:13937–13942. doi: 10.1073/pnas.93.24.13937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta R S. Mol Biol Evol. 1995;12:1063–1073. doi: 10.1093/oxfordjournals.molbev.a040281. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz J A, Mizukami H, Skafar D F. FEBS Lett. 1993;315:109–113. doi: 10.1016/0014-5793(93)81144-o. [DOI] [PubMed] [Google Scholar]
- 40.Amara J F, Clackson T, Rivera V M, Guo T, Keenan T, Natesan S, Pollock R, Yang W, Courage N L, Holt D A, et al. Proc Natl Acad Sci USA. 1997;94:10618–10623. doi: 10.1073/pnas.94.20.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clackson T, Yang W, Rozamus L W, Hatada M, Amara J F, Rollins C T, Stevenson L F, Magari S R, Wood S A, Courage N L, et al. Proc Natl Acad Sci USA. 1998;95:10437–10442. doi: 10.1073/pnas.95.18.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nemoto T, Ota M, Ohara-Nemoto Y, Kaneko M. Anal Biochem. 1995;227:396–399. doi: 10.1006/abio.1995.1300. [DOI] [PubMed] [Google Scholar]
- 43.Zabel U, Häusler C, Weeger M, Schmidt H H H W. J Biol Chem. 1999;274:18149–18152. doi: 10.1074/jbc.274.26.18149. [DOI] [PubMed] [Google Scholar]
- 44.Maruya M, Sameshima M, Nemoto T, Yahara I. J Mol Biol. 1999;285:903–907. doi: 10.1006/jmbi.1998.2349. [DOI] [PubMed] [Google Scholar]
- 45.Pearl L H, Prodromou C. Curr Opin Struct Biol. 2000;10:46–51. doi: 10.1016/s0959-440x(99)00047-0. [DOI] [PubMed] [Google Scholar]
- 46.Johnson, B. D., Chadli, A., Felts, S. J., Bouhouche, I., Catelli, M. G. & Toft, D. O. (2000) J. Biol. Chem., in press. [DOI] [PubMed]
- 47.Felts S J, Owen B A L, Nguyen P M, Trepel J, Donner D B, Toft D O. J Biol Chem. 2000;275:3305–3312. doi: 10.1074/jbc.275.5.3305. [DOI] [PubMed] [Google Scholar]
- 48.Prodromou C, Panaretou B, Chohan S, Siligardi G, O'Brien R, Ladbury J E, Roe S M, Piper P W, Pearl L H. EMBO J. 2000;19:4383–4392. doi: 10.1093/emboj/19.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]