Abstract
The initiation of DNA replication in the budding yeast Saccharomyces cerevisiae occurs in two sequential and mutually exclusive steps. Prereplicative complexes (pre-RCs) containing origin recognition complex (ORC), Cdc6p, and the MCM2–7 proteins assemble only under conditions of low cyclin-dependent kinase (Cdk) activity during G1, whereas origin activation is driven by the increase in Cdk activity at the end of G1. As a first step toward the reconstitution of this two-step process in vitro, we describe a system in which extracts prepared from G1-arrested cells promote sequential assembly of ORC, Cdc6p, and MCM2–7 proteins onto exogenously added origin-containing DNA. This reaction requires an intact ARS consensus sequence and requires ATP for two distinct steps. Extracts from cells arrested in mitosis also can support the binding of ORC but are unable to load either Cdc6p or MCM2–7 proteins. This system should be useful for studying the mechanism and regulation of pre-RC assembly.
Elaboration of the molecular events involved in initiating chromosomal DNA replication in eukaryotic cells would be greatly aided by the establishment of origin-dependent cell-free DNA replication systems. The budding yeast, Saccharomyces cerevisiae, would be ideal for such systems because short, well-defined DNA sequences act as both replicator elements (1–4) (sequences essential for initiation) and origins of bidirectional replication in vivo (5–7). A system has been described in which nuclei isolated from G1-arrested cells initiate replication in an S phase extract (8); however, to date, an efficient, origin-specific cell-free system from budding yeast that initiates DNA replication on exogenously added DNA in vitro has not been described.
The initiation of DNA replication in budding yeast can be considered a two-step process (reviewed in refs. 9 and 10). The first step involves the ordered assembly of prereplicative complexes (pre-RCs) at potential replication origins. Binding of the origin recognition complex (ORC) to the essential ARS consensus sequence (ACS) occurs first and is required for the loading of Cdc6p, which in turn is required for the loading of the MCM2–7 family of proteins. In the second step, origin firing is triggered during the S phase by the action of two protein kinases, Cdc7p with its regulatory subunit Dbf4p and the major cyclin-dependent kinase (Cdk), Cdc28p, with its regulatory subunits, the B-type cyclins (Clbs).
In addition to its positive role in initiation, Cdc28 kinase prevents the reassembly of pre-RCs from S phase until the end of mitosis (11–14). Recent analysis has shown that Cdc28 prevents the accumulation of the MCM proteins in the nucleus (15, 16) and targets Cdc6p for ubiquitin-mediated proteolysis (17–19). This dual role for Cdks in triggering initiation and preventing reinitiation ensures that replication origins cannot initiate more than once in a single cell cycle.
This two-step mechanism for DNA replication has an important implication for biochemical analysis. Because Cdc28 has both positive and negative effects on replication, it might not be possible to assemble pre-RCs and activate replication in a single extract. Therefore, as a step toward a soluble cell free replication system and to gain a deeper understanding of how DNA replication is limited to one round per cell cycle, we have developed a cell-free system for the assembly of pre-RCs that we describe in this paper.
Materials and Methods
Strains and Media.
The yeast strains used were YGP82 (MATa, ura3–52, trp1Δ, prb1–1122, prc1–407, pep4–3, leu2–3,112, nuc1∷LEU2, cdc6∷GAL-CDC6∷TRP1; kindly provided by G. Perkins, Imperial Cancer Research Fund) and YCD2 (MATa, ura3–52, trp1Δ, prb1–1122, prc1–407, pep4–3, leu2–3,112, nuc1∷LEU2, cdc6∷GAL-CDC6∷TRP1, CDC47-MHtag∷URA3; ref. 20). Cells were grown in YP (1% yeast extract, Difco; 2% bacto-peptone, Difco) containing 2% galactose (YP-Gal) or 2% glucose (YPD).
Cell Culture.
Cells were grown to a density of 2 × 107 cells per ml in YP-Gal at 30°C and arrested in G2/M by the addition of nocodazole to a final concentration of 5 μg/ml and incubation for 3.5 h. Cells were then washed, resuspended in YP-Gal containing 10 μg/ml α-factor, and incubated for 2.5 h to arrest in G1. To repress Cdc6p expression, glucose was added 2.5 h after the nocodazole addition to a final concentration of 2% and the culture was incubated for 1 h. Then, the cells were released into YPD containing α-factor as described above.
Cell Extracts.
Because pre-RC components are differentially extracted (21), we made two types of extracts: a high-salt extract and a low-salt extract. ORC and Cdc6p are efficiently solubilized in the high-salt extract. However, Mcm proteins are not efficiently extracted with 300 mM NaCl once they have been loaded onto chromatin. In contrast, when they are not loaded onto chromatin, Mcm proteins are soluble even in low-salt extracts. Therefore, a second low-salt extract was prepared from cells lacking Cdc6p. In the reactions described in this paper, we combined equal amounts of the high-salt and the low-salt extracts to supply ORC, Cdc6p, and Mcm proteins for incubations. This combination helped to decrease the final salt concentration in the loading reaction. We subsequently have found that, although it improves loading efficiency, the low-salt extract is not essential for the loading reaction.
Whole-cell extracts were prepared according to the method of Schultz et al. (22) with modifications. Briefly, the cells were washed at 4°C twice with cell wash buffer (20 mM Hepes⋅KOH, pH 7.8/1 M sorbitol) and once with 10 vol of lysis buffer (100 mM Hepes⋅KOH, pH 7.8/0.8 M sorbitol/50 mM potassium glutamate/10 mM MgOAc/2 mM EDTA). The cells were resuspended in 0.25 vol (v/w) of lysis buffer containing 4 mM DTT and 4× protease inhibitors, and frozen by dropping the cell suspension into liquid nitrogen. The 1× protease inhibitors consist of 1 mM 4-(2-aminoethyl)-benzenesulfonyl fluoride hydrochloride (AEBSF), 2 μg/ml aprotinin, 1 mM benzamidine hydrochloride, 10 μg/ml leupeptin, and 1 μg/ml pepstatin A. Afterward, extracts were prepared in the cold room. Dry ice was ground in a coffee mill to make dry ice powder as a coolant, and the frozen yeast beads were broken in the coffee mill in the presence of the dry ice powder. Typically, 5–10 g of the cell pellet was processed at a time. The broken yeast powder was thawed to give a homogenate. To prepare the high-salt extract, potassium glutamate was added to the homogenate to give a final concentration of 300 mM. For the low-salt extract, no potassium glutamate was added. Then, the homogenate was incubated for 30 min and centrifuged in a Sorvall SS-34 rotor at 20,000 rpm for 20 min at 4°C. The supernatant was withdrawn by puncturing the tube wall with a syringe and a needle and clarified by centrifugation in a Beckman SW55 rotor at 55,000 rpm for 1 h at 4°C. The recovered supernatant was aliquotted, frozen in liquid nitrogen, and stored at −70°C as a whole-cell extract. The extracts were stable for at least 3 months at −70°C. The protein concentration of the extract is typically 50–80 mg/ml. Protein concentrations were determined with the Bio-Rad protein assay, using BSA as the standard.
Preparation of ARS1 Beads.
Oligonucleotides for A-rich and T-rich strands of wild-type ARS1 sequence (791–880; ref. 23) and for the T-rich strand labeled with biotin at the 5′ end (Fig. 1B) were chemically synthesized and purified by polyacrylamide urea gel electrophoresis. They were mixed at a ratio of 10, 8, and 2 nmol, respectively, and subsequently annealed, phosphorylated with [γ-32P]ATP, and ligated as described (24). The ligation products corresponding to 5 nmol of annealed single-copy ARS1 DNA were incubated with 10 mg streptavidin-coated paramagnetic beads (Dynabeads M-280 Streptavidin; Dynal, Great Neck, NY) in a 200 μl coupling mixture at room temperature overnight, according to the manufacturer's instructions. The beads were washed three times with 10 mM Hepes⋅KOH (pH 7.6), 1 mM EDTA, and 1 M KOAc and three times with 10 mM Hepes⋅KOH (pH 7.6) and 1 mM EDTA and resuspended in 10 mM Hepes⋅KOH (pH 7.6) and 1 mM EDTA at 80 mg beads per ml. The mutant ARS1 beads were prepared in the same manner, using another set of three oligonucleotides of a mutant form of ARS1 in which an XhoI linker replaced the 8 bp of the A element (858). Typically, ligated DNA immobilized on each mg of beads corresponded to 50 pmol of ARS1 sequence (single copy).
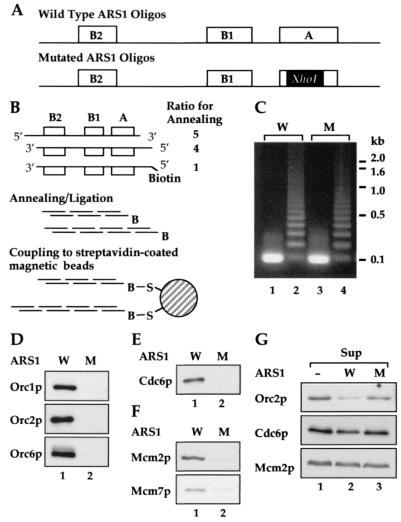
Figure 1.
A cell-free system for the assembly of pre-RCs. (A–C) Preparation of ARS1 beads. (A) Complementary oligonucleotides for ARS1 beads. The positions of the A, B1, and B2 elements are indicated by open boxes. In the mutated ARS1 oligo DNA, 8 bp of the A element was replaced with an 8-bp XhoI linker sequence. (B) Preparation of ARS1 beads. See Materials and Methods for details. (C) Aliquots of annealed wild-type (W) and mutant (M) oligonucleotides, before (1 and 3) and after (2 and 4) the ligation, were run on an agarose gel and stained with ethidium bromide. (D–F) Assembly of pre-RC components on ARS1 beads. Whole-cell extracts prepared from YGP82 cells expressing Cdc6p were incubated with either wild-type (W) or mutant (M) ARS1 beads. (D) ORC loading. (E) Cdc6p loading. (F) Mcm loading. To examine the loading of Mcm7p, whole-cell extracts were prepared from YCD2 cells, and Mcm7p tagged with c-myc epitope was detected with the 9E10 antibody. (G) The levels of pre-RC components in reaction mixtures. After the incubation, samples were taken from the supernatants separated from the wild-type (ARS1 W) or mutant (ARS1 M) beads. A reaction mixture lacking ARS1 beads was constructed separately, and a sample was taken without an incubation (ARS1 −). The levels of pre-RC components were detected by immunoblotting (Sup).
Loading Assay.
For the standard assembly reaction, 40 μl reaction buffer [50 mM Hepes⋅KOH, pH 7.6/625 mM sorbitol/20 mM MgOAc/0.125 mM EDTA/5 mM EGTA/2 mM DTT/6 mM ATP/ATP regenerating system/2× protease inhibitors/1.5 mg/ml poly(dI-dC)⋅poly(dI-dC)/2.5 mg/ml sonicated pBluescript KS+] containing either wild-type or mutant ARS1 beads (20 pmol ARS1 per reaction) was mixed with 20 μl high-salt extract and 20 μl low-salt extract to give a final volume of 80 μl. The ATP regenerating system consisted of 40 mM creatine phosphate and 16 units of creatine phosphokinase. For the titration, increasing amounts of the extracts were added as indicated in the figures. The final volume of the reaction mixture was adjusted to 80 μl by adding dilution buffer (50 mM Hepes⋅KOH, pH 7.8/87.5 mM potassium glutamate/400 mM sorbitol/5 mM MgOAc/1 mM EDTA). The reaction mixture was incubated at 24°C for 8 min, and the reaction was stopped by the addition of 9 vol of ice-cold wash buffer (50 mM Hepes⋅KOH, pH 7.6/75 mM potassium glutamate/1 mM EGTA/5 mM MgOAc/30% sucrose/0.1% Triton X-100/1 mM DTT/1× protease inhibitors). The beads were isolated from the supernatant with a magnetic separator at 4°C, rinsed once with 800 μl wash buffer, and boiled in Laemmli's SDS sample buffer. A sample was taken from the supernatant and boiled in SDS sample buffer.
Immunoblotting.
Immunoblots were performed as described (18). Orc1p, Orc2p, Orc6p, Cdc6p, and Mcm2p were detected with the polyclonal antibody JDI51 (A. Schepers and J.F.X.D., unpublished results) at a dilution of 1:250, the polyclonal JAB12 at 1:500 (25), the monoclonal SB49 (a gift from B. Stillman, Cold Spring Harbor Laboratory) at 1:1,000, the monoclonal 9H/85 at 5 μg/ml (26), and the polyclonal sc6680 (Santa Cruz Biotechnology) at 1:2,000. Mcm7p tagged with c-myc epitope was detected by using the monoclonal 9E10.
Results
A Cell-Free System for Pre-RC Formation.
We have chosen one of the best-characterized budding yeast replication origins, ARS1, to use for the development of a cell-free pre-RC assembly system. The core ARS1 origin is a 193-bp segment composed of multiple sequence elements known as A, B1, B2, and B3 (23). The A and B1 elements serve as the recognition sequence both in vitro and in vivo for ORC (25, 27, 28). The A element is essential for both ORC binding and origin function in vivo, and the B1 element is required for high-affinity ORC binding both in vivo and in vitro and is very important for origin function. ORC remains bound to these sequences throughout the cell cycle in vivo. The function of the B2 element in replication is unknown; however, the pre-RC assembles over it in vivo (29), and MCM loading in vivo is reduced in the B2 mutant (30). DNA replication initiates between the B1 and B2 elements in vivo (31, 32). The B3 element, which has an auxiliary role in origin function in vivo, serves as the binding site for ARS binding factor 1 (ABF1) both in vivo and in vitro (29, 33, 34).
The A, B1, and B2 elements comprise a reasonably efficient ARS (23) in the absence of B3 and can be synthesized in vitro as an oligonucleotide of approximately 100 nt. Double-stranded ARS1 oligonucleotides containing a small fraction of 5′-biotinylated oligonucleotide were ligated together (Fig. 1 A–C) and attached to streptavidin-coated paramagnetic beads. As a control, oligonucleotides containing a mutant form of ARS1 in which an XhoI linker replaces essential sequences in the A element were prepared in an identical manner. This mutant form of ARS1 abolishes origin function in vivo, ORC binding in vivo and in vitro, and pre-RC assembly in vivo (23, 25, 27, 28, 35, 36). The strategy for preparation of ARS1-containing beads is outlined in Fig. 1 A and B and is described in further detail in Materials and Methods.
To ensure that Cdk activity is low in our extracts, cells were arrested at Start with α-factor before cell lysis. We used a protease-deficient (pep4, prb1, prc1) and nuclease-deficient (nuc1) strain in which the endogenous CDC6 gene was replaced by a copy of CDC6 under the control of the inducible GAL1,10 promoter. We used this strain because Cdc6p, an essential pre-RC component, is normally present at very low levels in α-factor-arrested cells (26, 37). By growing cells in galactose before α-factor arrest, we can induce high levels of Cdc6p in these cells. We were unable to detect any proteins in crude extracts that tightly associated with Cdc6p (unpublished observations), thus we reasoned that high Cdc6p levels might help to promote protein–protein interactions required for pre-RC formation. Because GAL-CDC6 is the only copy of the CDC6 gene, we also can use this strain to prepare extracts from cells lacking Cdc6p, allowing us to examine the requirement for Cdc6p in the cell-free system.
To begin to examine pre-RC assembly, wild-type or mutant ARS1 beads were incubated with yeast extracts, as described in Materials and Methods, at 24°C in the presence of nonspecific competitor DNA. Under these conditions, the loading of pre-RC components is reasonably quick, reaching a maximum in approximately 10 min. Reactions then were diluted 10-fold with wash buffer, and beads were separated from the extracts with a magnet. The beads were rinsed, and the proteins associated with the beads were analyzed by immunoblotting. As shown in Fig. 1D, Orc1p, Orc2p, and Orc6p were clearly detected on wild-type (W) ARS1 beads, whereas no signal was observed on mutant (M) ARS1 beads (lanes 1 and 2). Thus, ORC is loaded in an origin-dependent reaction, consistent with its known properties as a sequence-specific DNA binding protein. Fig. 1E shows that Cdc6p also is loaded specifically onto the wild-type ARS1 beads in these extracts. Because there is no loading of Cdc6p onto the ACS mutant ARS1 beads, the loading onto wild-type ARS1 beads suggests that ORC is required for the Cdc6p loading in these extracts. Finally, Fig. 1F shows that two proteins from the MCM family, Mcm2p and Mcm7p, were also bound to the wild type but not to the mutant ARS1 beads. Therefore, these extracts can support the assembly of the known pre-RC components, ORC, Cdc6p, and the MCM2–7 proteins, onto DNA in an origin-dependent reaction.
After the incubation with ARS1 beads, we examined the levels of the individual pre-RC components remaining in the supernatants. Fig. 1G shows that Orc2p is efficiently depleted from the supernatant after incubation with wild-type but not with mutant ARS1 sequence (lanes 2 and 3), indicating efficient binding of ORC to wild-type ARS1 beads. Based on the amount of Orc2p in cells (25) and the amount of ARS1 DNA in the reactions, we estimate that only approximately 1% of the total amount of ARS1 DNA in these reactions is bound by ORC. Thus, at present, we cannot examine pre-RCs assembled in vitro by DNase1 footprinting, because footprinting techniques require near-saturation of the DNA template. In contrast to ORC, neither Cdc6p nor Mcm2p was depleted from the extracts after incubation. Because Cdc6p is overexpressed in these extracts, it is perhaps not surprising that Cdc6p is not depleted from the extracts. However, that the Mcm2p levels are not decreased in the supernatants suggests that the loading of MCM proteins in these extracts is not as efficient as the loading of ORC. It is possible that MCM loading may require some additional unidentified factor present in limiting amounts in the extracts. Alternatively, some aspect of the loading reaction may require further optimization. Regardless, these experiments demonstrate the assembly of budding yeast pre-RC components on a purified DNA template. This is also a description of an in vitro MCM loading reaction requiring a specific DNA sequence.
Ordered Assembly of Pre-RC Components.
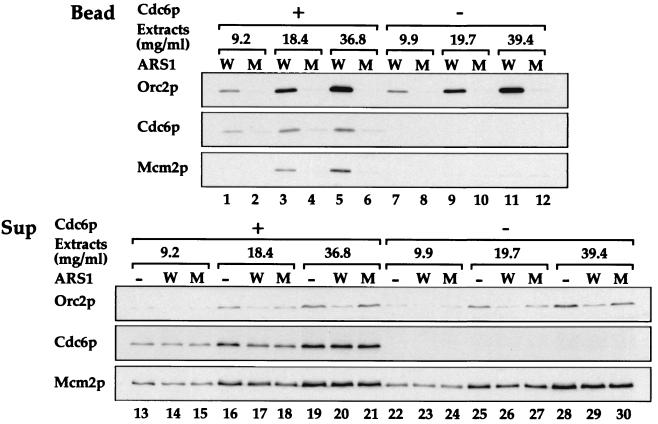
To begin to examine the mechanism of pre-RC assembly in vitro, we prepared extracts from G1-arrested cells under conditions of Cdc6p expression or repression. Fig. 2 shows a titration of extract using either wild-type or mutant ARS1 beads. This experiment shows that extracts either containing or lacking Cdc6p supported similar levels of Orc2p binding to wild-type but not mutant ARS1 beads (lanes 1–12). Moreover, ORC is efficiently depleted from the supernatants from the wild-type bead in both sets of reactions (lanes 13–30). This depletion is consistent with the fact that ORC is bound to ARSs before Cdc6p expression in vivo and remains bound to origins in G1, even in the absence of Cdc6p expression (38). Thus, in our assay, Cdc6p does not appear to affect either the affinity or specificity of ORC binding. Lanes 13–30 also show the presence (lanes 13–21) or absence (lanes 22–30) of Cdc6p from the two extracts. In the presence of Cdc6p, Mcm2p was preferentially loaded onto wild-type ARS1 beads; however, in the absence of Cdc6p, this preferential loading of Mcm2p was lost. Thus, the specific loading of MCM proteins in this cell-free system, like pre-RC assembly in vivo, depends on the Cdc6p. These results demonstrate that in this cell-free system, extracts from G1-arrested cells support the ordered assembly of pre-RC components, including ORC, Cdc6p, and MCM proteins, on a short, well defined specific DNA sequence.
Figure 2.
Cdc6p-dependent protein loading on ARS1 beads. Whole-cell extracts were prepared from YGP82 cells in which Cdc6p was expressed (Cdc6p +) or repressed (Cdc6p −). Wild-type (W) or mutant (M) ARS1 beads were incubated with increasing amounts of the extracts, as indicated by their final concentrations in the reaction mixtures (Extracts). The levels of ORC, Cdc6p, and Mcm proteins associated with the beads (Bead) or in the supernatants (Sup) were detected by immunoblotting as described in Fig. 1.
Two Requirements for ATP in Pre-RC Assembly.
ORC requires a nucleoside triphosphate (preferably ATP) to bind ARS DNA in vitro (27, 39). There are at least two ATP binding sites in ORC, one on Orc1p and one on Orc5p, and mutation of one of these (Orc1p) prevents ORC binding in vitro and is nonfunctional in vivo (39). In addition, Cdc6p and the MCM proteins all have potential nucleotide binding sites, and mutation of the nucleotide binding site of Cdc6p prevents its function in vivo (40–43). These observations led us to test the requirement for nucleotides in pre-RC assembly in vitro.
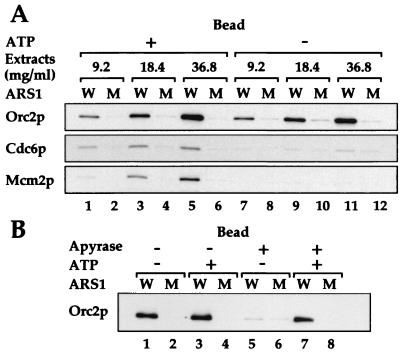
The extract titrations in Fig. 3A (lanes 1–6) show that, under standard reaction conditions containing ATP and an ATP regenerating system, ORC, Cdc6p, and Mcm2p bound specifically to wild-type ARS1 beads and not mutant ARS1 beads. In the absence of exogenously added ATP and the ATP regenerating system, specific ORC binding was undiminished (lanes 7–12). However, the omission of ATP and the ATP regenerating system almost completely eliminated the loading of both Cdc6p and Mcm2p (lanes 7–12). This elimination of loading demonstrates that ATP is required for pre-RC assembly in vitro. In addition, this experiment shows that, under conditions in which ORC can efficiently bind to its target sequence in vitro, ATP is required for the recruitment of Cdc6p and MCM proteins to the pre-RC. Because Cdc6p binds before the MCM2–7 complex, this result indicates that the association of Cdc6p with ORC requires ATP.
Figure 3.
ATP requirement for protein assembly on ARS1 beads. (A) ARS 1 beads were incubated with whole-cell extracts prepared from YGP82 cells expressing Cdc6p either in the presence of (ATP +) or in the absence of (ATP −) exogenously added ATP and the ATP regenerating system. Proteins associating with the beads (Bead) or remaining in the supernatants (Sup) were detected by immunoblotting as before. (B) Loading reactions were carried out either with or without exogenously added ATP and the ATP regenerating system (ATP + or −). To the indicated reactions (Apyrase +), 5 units of apyrase was added. After incubation with ARS1 beads, Orc2p associating with the beads (Bead) and remaining in the supernatants (Sup) was detected.
The observed binding of ORC in the absence of added ATP was unexpected because ORC clearly requires ATP to bind to ARS1 DNA in vitro (27, 39). We reasoned that endogenous nucleotides present in the extracts might be enough to support ORC binding (but not Cdc6p loading) in the absence of added ATP. To deplete endogenous nucleotides, therefore, we treated extracts with apyrase, an enzyme capable of hydrolyzing ATP to ADP and, ultimately, to AMP. Fig. 3B shows that, in the absence of added ATP, ORC binding was greatly reduced in the apyrase-treated reactions (lanes 5 and 6). This effect was not a nonspecific effect of apyrase because it could be prevented by the addition of ATP and ATP regenerating system to the reactions (Fig. 3B, lanes 7 and 8). These results indicate that pre-RC assembly in vitro requires ATP for at least two distinct steps, ORC binding and Cdc6p recruitment.
Mitotic Extracts Are Defective in Pre-RC Assembly.
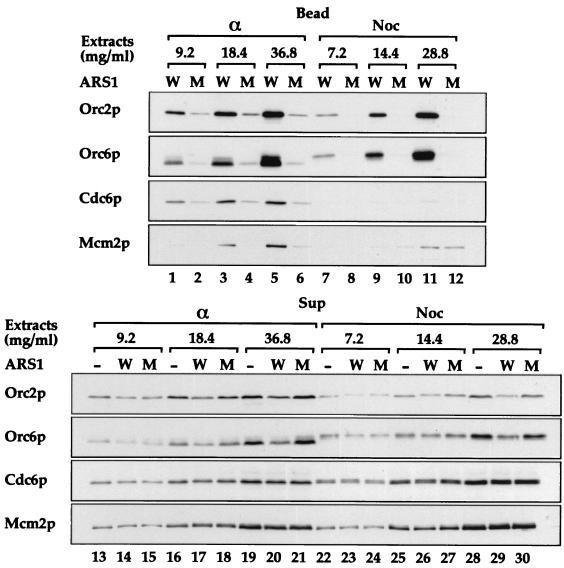
Cells arrested in mitosis with nocodazole are incapable of assembling pre-RCs in vivo, even when Cdc6p is expressed at high levels (12, 26, 38). Therefore, we were interested in determining whether extracts prepared from such nocodazole-arrested cells expressing Cdc6p were capable of pre-RC assembly in vitro. Fig. 4 (lanes 1–12) shows that extracts from both nocodazole and α-factor-arrested cells can efficiently load ORC in a sequence-specific manner, consistent with the observation that ORC binds ARSs in vivo through the cell cycle. Strikingly, whereas extracts from α-factor-arrested cells efficiently loaded Cdc6p and Mcm2p in addition to ORC (lanes 1–6), the extracts from nocodazole-arrested cells were unable to load either Cdc6p or Mcm2p onto wild-type ARS1 beads (lanes 7–12). Fig. 4 (lanes 13–30) shows that ORC, Cdc6p, and Mcm2p were present in nocodazole extracts at levels similar to those in the α-factor extracts. We note that both Cdc6p and Orc6p in the extracts from nocodazole-arrested cells appear as slower migrating forms due to phosphorylation by Cdc28p, suggesting that the inability of Cdc6p to load in these extracts may reflect inhibition by Cdc28p.
Figure 4.
Pre-RC assembly can occur in extracts of G1 cells but is blocked in extracts of G2/M cells. Whole-cell extracts prepared from Cdc6p-expressing cells (YGP82), arrested either in G1 phase with α-factor (α) or in G2/M with nocodazole (Noc), were incubated with wild-type (W) or mutant (M) ARS1 beads. The levels of ORC, Cdc6p, and Mcm2p associated with the beads (Bead) or present in the supernatants (Sup) were determined as above.
Discussion
We have described a cell-free system that supports the ordered assembly of a full complement of pre-RC components from yeast extracts onto exogenously added origin-containing DNA. This is an origin-dependent in vitro pre-RC assembly system. Cdc6p is required for MCM loading but not for the loading of ORC, which is consistent with the sequential loading of ORC, Cdc6p, and the MCM2–7 complex. Moreover, pre-RC assembly requires ATP and occurs with extracts from G1-arrested but not G2/M-arrested cells. Thus, this cell-free system recapitulates many features of pre-RC assembly in vivo.
Our results suggest that nucleotides play at least two roles in pre-RC assembly. The first role is to promote ORC binding. As shown by Bell and Stillman (27), the binding of purified ORC to ARS DNA requires the presence of ATP. The fact that depletion of all nucleoside diphosphates and triphosphates from the reactions by apyrase treatment of extracts prevents the loading of ORC is consistent with this conclusion. When ATP was omitted from reactions but extracts were not treated with apyrase, ORC was still able to bind to origin DNA, but neither Cdc6p nor the MCM2–7 proteins were loaded. The ability of ORC to bind its cognate sequence in the absence of exogenously added ATP suggests either (i) that low levels of ATP in the extract are sufficient to support ORC binding but not pre-RC assembly or (ii) a nucleotide other than ATP present in the extract can support ORC binding but not pre-RC assembly.
The second role for nucleotides appears to be in the recruitment of Cdc6p to origins. This requirement could be seen when ATP and the ATP regenerating system were omitted from the loading reaction (Fig. 3). Under these conditions ORC binds efficiently to DNA, but Cdc6p and the MCM proteins are not loaded. Given the ordered assembly of the complex, this suggests that the step in which Cdc6 binds to ORC cannot occur in the absence of ATP. At present we do not know whether it is ORC, Cdc6p, and/or some other factor that must bind ATP to load Cdc6. However, we note that previous genetic analysis has suggested that mutants in the Walker A motif of Cdc6p that should prevent ATP binding act as null mutants (41–43) and do not interfere with the function of wild-type Cdc6p, even when overexpressed (41). From this we previously proposed that ATP binding by Cdc6p is required for productive interaction with ORC. Consistent with this prediction, while this paper was under consideration, Mizushima et al. (44) described an ATP-dependent interaction between ORC and Cdc6 and showed that the Walker A motif cdc6 mutant was defective in preventing oligomerization of ORC in vitro.
Extracts from cells arrested in mitosis can load ORC but are unable to load either Cdc6p or the MCM complex. These extracts may lack unidentified essential factor(s) required for pre-RC assembly. We note that our previous analysis has shown that new protein synthesis is required for pre-RC assembly after nocodazole arrest (13). Furthermore, one or more of the known pre-RC components may be present but inactive in these mitotic extracts. Inhibition of Cdc28p activity in cells arrested in mitosis can drive the assembly of pre-RCs in those cells, suggesting that Cdc28p may inactivate one or more pre-RC component. Indeed, the accumulation of MCM proteins in the nucleus is prevented by Cdc28p (15, 16). Moreover, the Cdc6 protein is normally only present during G1 because it is targeted for rapid degradation in response to phosphorylation by Cdc28p (17–19). Because the pre-RC assembly system described here is soluble and does not require nuclear formation and because Cdc6 is overexpressed and present at high levels even in the mitotic extracts, neither of these phenomena can explain the inability of the mitotic extracts to assemble pre-RCs. This suggests that there is at least one additional step during pre-RC assembly that is blocked by Cdc28p. By fractionating G1 and mitotic extracts into individual, essential components, it should be possible to determine which pre-RC components are inactive in the mitotic extract. Previous work has shown that Cdc6p overexpressed from the GAL1,10 promoter could be detected as associating with replication origins in vivo by chromatin immunoprecipitation (CHIP) during G2 (36). Although this does not seem consistent with our results, which show that Cdc6 loading does not occur in extracts from nocodazole-arrested cells, we note that even in these CHIP experiments, the association of Cdc6p with origins in G2 was probably less efficient than association in G1 because it was described as being “slower” than the association in G1 (36). We also suggest that overexpression of Cdc6, which can act as a Cdc28 inhibitor in vitro and in vivo (20, 45), in this experiment may, in fact, contribute to its own loading by lowering CDK activity.
ORC, Cdc6p, and MCM proteins are the best-characterized pre-RC components; however, it is possible that there are additional pre-RC components or that there are other factors required for assembly of the complex but are not components themselves. For example, the fission yeast cdt1 protein and a Xenopus Cdt1 homologue have been shown to be required for MCM loading (46, 47). It will be interesting to see whether related proteins also are required in budding yeast. Other budding yeast proteins such as Mcm10 also may be pre-RC components (48). Using MCM loading as an end point, it should be possible to fractionate the G1 extracts to identify any additional factors involved in pre-RC assembly.
The cell-free system described in this paper should be useful for elucidating the molecular mechanism of pre-RC assembly and for understanding how pre-RC is regulated during the cell cycle. In addition, it represents a first step toward establishing origin-dependent initiation of DNA replication in soluble extracts.
Acknowledgments
We thank José Antonio Tercero, Elizabeth Noton, and Karim Labib for insightful comments on the manuscript; Hiroyuki Yamano for helpful discussions; Gordon Perkins for the yeast strain; Bruce Stillman for the Orc6p antibody; the Imperial Cancer Research Fund peptide synthesis laboratory for α-factor; and the Imperial Cancer Research Fund oligonucleotide synthesis service for oligonucleotides. T.S. was supported by the Toyobo Biotechnology Foundation.
Abbreviations
- pre-RC
prereplicative complex
- ORC
origin recognition complex
- Cdk
cyclin-dependent kinase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Struhl K, Stinchcomb D T, Scherer S, Davis R W. Proc Natl Acad Sci USA. 1979;76:1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stinchcomb D T, Struhl K, Davis R W. Nature (London) 1979;282:39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- 3.Kingsman A J, Clarke L, Mortimer R K, Carbon J. Gene. 1979;7:141–152. doi: 10.1016/0378-1119(79)90029-5. [DOI] [PubMed] [Google Scholar]
- 4.Hsiao C L, Carbon J. Proc Natl Acad Sci USA. 1979;76:3829–3833. doi: 10.1073/pnas.76.8.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewer B J, Fangman W L. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 6.Huberman J A, Spotila L D, Nawotka K A, El Assouli S M, Davis L R. Cell. 1987;51:473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- 7.Huberman J A, Zhu J G, Davis L R, Newlon C S. Nucleic Acids Res. 1988;16:6373–6384. doi: 10.1093/nar/16.14.6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasero P, Braguglia D, Gasser S M. Genes Dev. 1997;11:1504–1518. doi: 10.1101/gad.11.12.1504. [DOI] [PubMed] [Google Scholar]
- 9.Diffley J F X. Genes Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- 10.Newlon C S. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- 11.Dahmann C, Diffley J F X, Nasmyth K A. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- 12.Piatti S, Bohm T, Cocker J H, Diffley J F X, Nasmyth K. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 13.Noton E A, Diffley J F X. Mol Cell. 2000;5:85–95. doi: 10.1016/s1097-2765(00)80405-0. [DOI] [PubMed] [Google Scholar]
- 14.Detweiler C S, Li J J. Proc Natl Acad Sci USA. 1998;95:2384–2389. doi: 10.1073/pnas.95.5.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labib K, Diffley J F X, Kearsey S E. Nat Cell Biol. 1999;1:415–422. doi: 10.1038/15649. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen V Q, Co C, Irie K, Li J J. Curr Biol. 2000;10:195–205. doi: 10.1016/s0960-9822(00)00337-7. [DOI] [PubMed] [Google Scholar]
- 17.Elsasser S, Chi Y, Yang P, Campbell J L. Mol Biol Cell. 1999;10:3263–3277. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drury L S, Perkins G, Diffley J F X. Curr Biol. 2000;10:231–240. doi: 10.1016/s0960-9822(00)00355-9. [DOI] [PubMed] [Google Scholar]
- 19.Calzada A, Sanchez M, Sanchez E, Bueno A. J Biol Chem. 2000;275:9734–9741. doi: 10.1074/jbc.275.13.9734. [DOI] [PubMed] [Google Scholar]
- 20.Desdouets C, Santocanale C, Drury L S, Perkins G, Foiani M, Plevani P, Diffley J F X. EMBO J. 1998;17:4139–4146. doi: 10.1093/emboj/17.14.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Donovan S, Harwood J, Drury L S, Diffley J F X. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schultz M C, Hockman D J, Harkness T A, Garinther W I, Altheim B A. Proc Natl Acad Sci USA. 1997;94:9034–9039. doi: 10.1073/pnas.94.17.9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marahrens Y, Stillman B. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 24.Kadonaga J T, Tjian R. Proc Natl Acad Sci USA. 1986;83:5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowley A, Cocker J H, Harwood J, Diffley J F X. EMBO J. 1995;14:2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drury L S, Perkins G, Diffley J F X. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bell S P, Stillman B. Nature (London) 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 28.Rao H, Stillman B. Proc Natl Acad Sci USA. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diffley J F X, Cocker J H, Dowell S J, Rowley A. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- 30.Zou L, Stillman B. Mol Cell Biol. 2000;20:3086–3096. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bielinsky A K, Gerbi S A. Science. 1998;279:95–98. doi: 10.1126/science.279.5347.95. [DOI] [PubMed] [Google Scholar]
- 32.Bielinsky A K, Gerbi S A. Mol Cell. 1999;3:477–486. doi: 10.1016/s1097-2765(00)80475-x. [DOI] [PubMed] [Google Scholar]
- 33.Diffley J F, Stillman B. Proc Natl Acad Sci USA. 1988;85:2120–2124. doi: 10.1073/pnas.85.7.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diffley J F X, Cocker J H. Nature (London) 1992;357:169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- 35.Aparicio O M, Weinstein D M, Bell S P. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka T, Knapp D, Nasmyth K. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 37.Piatti S, Lengauer C, Nasmyth K. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cocker J H, Piatti S, Santocanale C, Nasmyth K, Diffley J F X. Nature (London) 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 39.Klemm R D, Austin R J, Bell S P. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 40.Koonin E V. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perkins G, Diffley J F X. Mol Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- 42.Weinreich M, Liang C, Stillman B. Proc Natl Acad Sci USA. 1999;96:441–446. doi: 10.1073/pnas.96.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B, Feng L, Huang S H, Reynolds C P, Wu L, Jong A Y. J Biol Chem. 1999;274:8291–8298. doi: 10.1074/jbc.274.12.8291. [DOI] [PubMed] [Google Scholar]
- 44.Mizushima T, Takahashi N, Stillman B. Genes Dev. 2000;14:1631–1641. [PMC free article] [PubMed] [Google Scholar]
- 45.Elsasser S, Lou F, Wang B, Campbell J L, Jong A. Mol Biol Cell. 1996;7:1723–1735. doi: 10.1091/mbc.7.11.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maiorano D, Moreau J, Mechali M. Nature (London) 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- 47.Nishitani H, Lygerou Z, Nishimoto T, Nurse P. Nature (London) 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- 48.Homesley L, Lei M, Kawasaki Y, Sawyer S, Christensen T, Tye B K. Genes Dev. 2000;14:913–926. [PMC free article] [PubMed] [Google Scholar]