Abstract
Leishmaniasis is a geographically widespread severe disease, with an increasing incidence of two million cases per year and 350 million people from 88 countries at risk. The causative agents are species of Leishmania, a protozoan flagellate. Visceral leishmaniasis, the most severe form of the disease, lethal if untreated, is caused by species of the Leishmania donovani complex. These species are morphologically indistinguishable but have been identified by molecular methods, predominantly multilocus enzyme electrophoresis. We have conducted a multifactorial genetic analysis that includes DNA sequences of protein-coding genes as well as noncoding segments, microsatellites, restriction-fragment length polymorphisms, and randomly amplified polymorphic DNAs, for a total of ≈18,000 characters for each of 25 geographically representative strains. Genotype is strongly correlated with geographical (continental) origin, but not with current taxonomy or clinical outcome. We propose a new taxonomy, in which Leishmania infantum and L. donovani are the only recognized species of the L. donovani complex, and we present an evolutionary hypothesis for the origin and dispersal of the species. The genus Leishmania may have originated in South America, but diversified after migration into Asia. L. donovani and L. infantum diverged ≈1 Mya, with further divergence of infraspecific genetic groups between 0.4 and 0.8 Mya. The prevailing mode of reproduction is clonal, but there is evidence of genetic exchange between strains, particularly in Africa.
Keywords: Leishmania infantum, Leishmaniasis, parasitic protozoa, phylogeny, population genetics
The leishmaniases are a complex of diseases, caused by kinetoplastid flagellates of the genus Leishmania, which include visceral leishmaniasis (VL), the most severe form of the disease, lethal if left untreated, and several forms of cutaneous leishmaniasis (CL), which may be mutilating, disfiguring, or disabling when lesions are multiple. Three hundred fifty million people in 88 countries are at risk. The yearly incidence is 0.5 million cases of VL and 1.5 million cases of CL (1). The number of people suffering from these diseases has increased during the last decade (2).
Leishmaniasis is transmitted by the bite of female phlebotomine sandflies belonging to some 30 species, different throughout the world. Twenty Leishmania species are pathogenic for humans. The causative agents of VL are members of the Leishmania donovani complex, classified into four species: Leishmania archibaldi, Leishmania chagasi, Leishmania donovani, and Leishmania infantum, distinguished by their vectors and reservoir hosts and in pathology (3).
Thousands of Leishmania strains have been typed by multilocus enzyme electrophoresis (MLEE) (4, 5). Their classification has been challenged by studies of L. donovani complex strains, using molecular markers, including coding and noncoding DNA sequences of nuclear or mitochondrial origin, random amplified polymorphic DNA (RAPD), restriction fragment length polymorphism (RFLP), and microsatellites. These studies suggest that the taxonomy of the L. donovani complex needs to be revised. On the basis of the RAPD and DNA sequence analyses, L. chagasi has been synonymized with L. infantum, which is consistent with a recent introduction of L. infantum in the New World (6). Whether L. archibaldi is a valid taxon has been questioned, because markers that distinguish it from L. donovani are not reliable, and other markers do not generate a single clade of L. archibaldi strains (7–13). Some molecular studies have shown that Sudanese strains of L. infantum are genetically indistinguishable from local L. donovani and that all Sudanese strains form a monophyletic genetic group. Thus, the complex would seem to consist of only two valid taxa, L. infantum, which prevails in Europe, North Africa, South and Central America, and L. donovani, which prevails in East-Africa, India, and parts of the Middle East.
Ambiguities concerning a satisfactory taxonomy and a reliable phylogeny emerge from two sources: (i) Leishmania species complexes exhibit limited diversity, and (ii) most studies include insufficient discriminatory markers so that differences are few, usually not exceeding dozens of characters. Therefore, the trees have low information content and are prone to contradictory outcomes.
We have sought to achieve a satisfactory level of resolution, by considering a large data set that includes several kinds of molecular markers applied to 25 strains representative of the L. donovani complex with respect to outstanding issues: 14 L. infantum strains from Europe, 6 L. donovani strains from India and East Africa, and 2 Sudanese L. infantum and 3 L. archibaldi strains, as classified by MLEE. We carry out a multifactorial analysis that includes ≈18,000 characters per strain (≈450,000 characters). Our results favor a taxonomy and phylogeny that are consistent with geographical distribution but not with the MLEE-based taxonomy. Our results confirm that some strains have probably resulted from recent genetic recombination events, even though the prevailing mode of reproduction of Leishmania is clonal.
Results
The 25 strain representatives of the L. donovani complex were selected from the extensive collection in the Montpellier cryobank, by taking into account their zymodeme type [as defined by the set of allozymes encompassed in the Montpellier (MON) classification], associated clinical presentation, geographic origin, and species assignment (Table 1). Our aim was to obtain a reliable phylogeny for the L. donovani complex, to devise a better taxonomy, and to identify clade-specific markers for population genetics analysis of L. infantum in Europe and of the Sudanese/Ethiopian strains, hence the greater representation of these two groups. Two Indian strains (LG9 and LG10) originate from the main endemic region of Bihar in India; the third Indian strain (LG16) had been found by several molecular methods to be part of a Kenyan genetic group (14, 15) and it is used here as representative of this group. For each strain, a total of 18,618 characters was obtained, including microsatellites, DNA sequences of protein-coding genes, noncoding and intergenic regions, RFLPs and RAPDs (Table 2).
Table 1.
Strains of the Leishmania donovani complex investigated in this study
| Code | WHO code | MLEE | MLEE-based species assignment | Country | Type of infection | Heterozygous sites |
|---|---|---|---|---|---|---|
| LG1 | MHOM/FR/78/LEM75 | MON-1 | L. infantum | France | Visceral | 0 |
| LG2 | MHOM/FR/95/LPN114 | MON-1 | L. infantum | France | Visceral | 0 |
| LG3 | MHOM/ES/93/PM1 | MON-1 | L. infantum | Spain | Visceral | 1 |
| LG4 | MHOM/FR/97/LSL29 | MON-1 | L. infantum | France | Cutaneous | 1 |
| LG5 | MHOM/ES/86/BCN16 | MON-1 | L. infantum | Spain | Cutaneous | 1 |
| LG6 | MHOM/PT/00/IMT260 | MON-1 | L. infantum | Portugal | Cutaneous | 0 |
| LG7 | MHOM/FR/96/LEM3249 | MON-29 | L. infantum | France | Cutaneous | 1 |
| LG8 | MHOM/ES/91/LEM2298 | MON-183 | L. infantum | Spain | Visceral | 1 |
| LG9 | MHOM/IN/00/DEVI | MON-2 | L. donovani | India | Visceral | 1 |
| LG10 | MHOM/IN/96/THAK35 | MON-2 | L. donovani | India | Visceral | 0 |
| LG11 | MHOM/ET/72/GEBRE 1 | MON-82 | L. archibaldi | Ethiopia | Visceral | 2 |
| LG12 | MHOM/SD/82/GILANI | MON-30 | L. donovani | Sudan | Visceral | 1 |
| LG13 | MHOM/ET/00/HUSSEN | MON-31 | L. donovani | Ethiopia | Visceral | 0 |
| LG14 | MHOM/FR/80/LEM189 | MON-11 | L. infantum | France | Cutaneous | 0 |
| LG15 | MHOM/MT/85/BUCK | MON-78 | L. infantum | Malta | Cutaneous | 3 |
| LG16 | MHOM/IN/54/SC23 | MON-38 | L. donovani | India | Visceral | 1 |
| LG17 | MCAN/SD/00/LEM3946 | MON-274 | L. donovani | Sudan | Visceral | 12 |
| LG18 | MHOM/SD/62/3S | MON-81 | L. infantum | Sudan | Visceral | 1 |
| LG19 | MHOM/ES/88/LLM175 | MON-198 | L. infantum | Spain | Visceral | 0 |
| LG20 | MHOM/ES/92/LLM373 | MON-199 | L. infantum | Spain | Visceral | 1 |
| LG21 | MHOM/IT/94/ISS1036 | MON-228 | L. infantum | Italy | Visceral | 2 |
| LG22 | MHOM/IT/93/ISS800 | MON-188 | L. infantum | Italy | Visceral | 0 |
| LG23 | MHOM/SD/97/LEM3472 | MON-267 | L. infantum | Sudan | Visceral + PKDL | 1 |
| LG24 | MHOM/SD/97/LEM3429 | MON-257 | L. archibaldi | Sudan | Visceral | 5 |
| LG25 | MHOM/SD/97/LEM3463 | MON-258 | L. archibaldi | Sudan | Visceral | 3 |
Strains isolated from humans [MHOM for Homo sapiens in the World Health Organization (WHO) code], except for LG17, which was originally isolated from a dog (MCAN for Canis). The MON designations refer to different zymodemes, or patterns defined by MLEE. Several MON-1 strains are included because this is the predominant MLEE type, in order to ascertain whether MON-1 is heterogeneous with respect to other markers. LG16 from India is included as representative of a set of strains from Kenya that have been shown, by protein coding genes (14), internal transcribed spacer sequences (10), and microsatellites (15), to be closely related to LG16. PKDL (LG23) refers to a particular syndrome known as ″post-kala azar dermal leishmaniasis.″ Notice the large number of heterozygous sites for LG17 and LG24 (12 and 5, respectively). RAPD markers were not available for strains LG16 to LG25.
Table 2.
Genetic markers for the analysis of 25 strains of the L. donovani complex
| Marker | Characters |
||
|---|---|---|---|
| Total | Variable | Parsimony informative | |
| Fingerprinting markers | |||
| RFLP | 160 | 135 | 111 |
| RAPD* | 787 | 652 | 488 |
| Microsatellites | 116 | 116 | 85 |
| DNA sequences: noncoding | |||
| 3′ histone H1 noncoding region (3′nc-H1) | 766 | 11 | 8 |
| rRNA internal transcribed spacer (ITS) | 1,025 | 2 | 2 |
| 60S acidic ribosomal protein intergenic region (ir-PO) | 995 | 26 | 24 |
| DNA sequences: coding | |||
| Aspartate aminotransferase (asat) | 1,239 | 2 | 2 |
| Fumarate hydratase (fh) | 1,201 | 6 | 1 |
| Glucose-6-phosphate dehydrogenase (g6pdh) | 1,689 | 10 | 7 |
| Glucose-6-phosphate isomerase (gpi) | 1,818 | 15 | 11 |
| Isocitrate dehydrogenase (icd) | 1,308 | 8 | 7 |
| Leishmania-activated C kinase (lack)† | 325 | 1 | 1 |
| LP7 methyl transferase (lp7)† | 405 | 4 | 2 |
| Malic enzyme (me) | 1,644 | 12 | 9 |
| Mannose phosphate isomerase (mpi) | 1,266 | 7 | 4 |
| Nucleoside hydrolase 1 (nh1) | 945 | 6 | 3 |
| Nucleoside hydrolase 2 (nh2) | 1,050 | 3 | 2 |
| 6-phosphogluconate dehydrogenase (pgd) | 1,440 | 21 | 5 |
| Trypanothione reductase (tr)† | 439 | 4 | 3 |
| Combined data set (RAPD data excluded) | 17,831 | 389 | 287 |
| Total | 18,618 | 1,041 | 775 |
*RAPD markers were not available for strains LG16 to LG25.
†These three genes have not been used in standard MLEE studies or for exploring the genetic structure of the parasites.
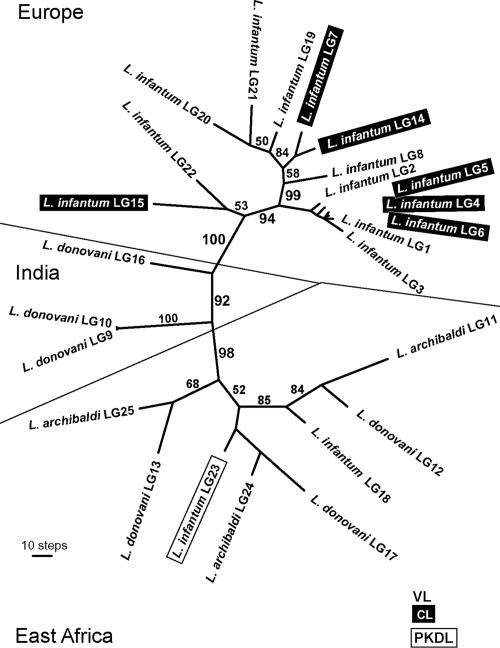
The data sets were analyzed separately and in various combinations. Results of the combined analysis of all characters (RAPDs excluded) are shown in Fig. 1. Sixteen equally parsimonious trees were obtained, but all trees showed quite similar topologies to the one in the figure. The most notable feature of all trees is a definite geographical clustering, reflecting an extremely strong correlation between genetic diversity and geographic origin. The basal nodes of the tree, i.e., those defining the main geographical clusters and their relationships, are all statistically reliable (bootstrap values of 92–100; see Fig. 1). If the RAPD data are included, the basal bootstrap values decrease, probably because of the incompleteness of the RAPD (data for 10 strains were not available) and their lack of consistent codominant inheritance (because of null alleles), their anonymous nature, different asymmetrical transformation probabilities, and possible GC priming bias (16). Nevertheless, when RAPDs are analyzed separately by an appropriate distance method [e.g., NeiLi or UpHolt as implemented in Phylogenetic Analysis Using Parsimony (PAUP), or programs specially designed to process RAPD data, such as FreeTree (17)], the same geographical relationships seen in Fig. 1 are obtained (data not shown). When the complete data set (including RAPDs) was analyzed by using mean or total distances, the topology resembled that shown in Fig. 1, except for the clustering of the Sudanese strains LG17 and LG25 together with the Indian strains LG9 and LG10 (data not shown). However, the resolution of basal nodes and the bootstrap support of the combined distance trees are significantly lower than in the case of parsimony. We have also analyzed separately individual data sets (DNA sequences, RFLPs and RAPDs) and obtained the same geographical associations shown in Fig. 1, but with reduced statistical support and resolution, because of lower information content. We compiled a concatenated sequence from our DNA data, but this placed the Indian/Kenyan strain LG16 at the root of the European clade. The rest of the overall geographical groupings were similar to those shown in Fig. 1.
Fig. 1.
Unrooted maximum-parsimony tree (one of 16 equally parsimonious trees) inferred from the combined data set of DNA sequences, microsatellites, and RFLP data (17,831 characters, 287 parsimony informative characters). All characters are given equal weight; numbers above branches are percent bootstrap values based on 1,000 replicates. Length of the tree is 773 steps. VL, visceral leishmaniasis; CL, cutaneous leishmaniasis; PKDL, post-kala azar dermal leishmaniasis. (Scale bar, 10 steps.)
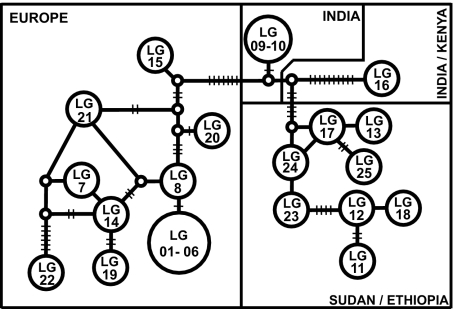
To get additional insight into the relationships among the strains, we analyzed our data set, using the coalescent-based statistical parsimony network approach. To simplify the analysis of these diploid organisms, heterozygous positions revealed by sequencing of the protein-coding genes were incorporated into the data set as ambiguous (degenerate code) and were thus treated as missing data. The exclusion of heterozygotes resulted in 19 different haplotypes. The concatenated network (Fig. 2) again reveals a strong correlation between molecular data and geographic origin. As reflected by the number of mutational steps (short cross lines), the parsimony network places the Indian and Indian/Kenyan strains between the African and the European strains, as was the case in the unrooted phylogeny of Fig. 1, about equally distant from the two sets.
Fig. 2.
Statistical parsimony network (TCS software, Version 1.21) based on a concatenated data set of 10 enzyme-coding genes (12,694 nt, 37 parsimony informative characters). Circles with strain designation represent different haplotypes. The short crosslines represent mutational steps between haplotypes. Heterozygous positions were not considered for the network construction.
Heterozygosity for the single-copy protein-coding genes is generally lower within the European clade than the African clade (Table 1) and varies among genes, with highest heterozygosity observed for the genes encoding fumarate hydratase, glucose-6-phosphate isomerase, and isocitrate dehydrogenase. One Sudanese strain (LG17) is extremely heterozygous (see Table 1).
The important result apparent in Figs. 1 and 2 is that the genetic configuration of the L. donovani complex strains is determined, first and foremost, indeed almost exclusively, by geographic origin, independently of taxonomic designation or clinical pleomorphism. Thus, all eight East African strains constitute a common clade, supported with 98% bootstrap, independently of whether they have been designated in the past as L. infantum, L. donovani or L. archibaldi (Fig. 1). The three Indian L. donovani strains (LG9, LG10, and LG16) are paraphyletic, which is consistent with them being members of two different, yet related, genetic groups. Finally, all of the European L. infantum strains invariably group together, forming two subclades: a larger and well supported subclade (94% bootstrap) composed of 12 strains from Spain, Portugal, France, and Italy; and a smaller subclade that comprises two strains from Malta and Italy (LG15 and LG22), which are set apart from all other European strains by all character sets. There is good support for the monophyly of the MON-1 type in Europe (which includes up to 90% of all typed L. infantum strains): All six MON-1 strains cluster together as the crown group in Fig. 1, although three are associated with VL and the other three with the cutaneous form of the disease.
We have explored the genetic structure of the L. donovani complex populations based on the sequence of the 10 enzyme-coding genes (Table 3). The ratio of nonsynonymous to synonymous substitutions is nearly equal for European, Indian, and Indian/Kenyan clades, but strongly biased toward synonymous substitutions in Africa, suggesting selection against amino acid replacements. Although not significant, the Tajima and Fu Li selection tests show a similar trend, i.e., negative values for European and positive values for African strains. None of these values is statistically significant due, at least in part, to the small numbers of strains involved. Table 3 shows that nucleotide diversity (π or θ) is low for all strains, somewhat lower in Europe than in Africa, although the difference is not significant. Haplotype diversity (HD) is highest in Africa, where it has the maximum possible value of 1, as we would expect, because each of the eight strains has a unique combination for the sequenced enzyme-coding genes as pointed out above and shown in Fig. 2. Haplotype diversity is lower in Europe (HD = 0.835), where six (LG1-LG6) of 14 strains share the same MLEE type (MON-1, Table 1) and indeed the same haplotype for the 10 combined enzyme-coding genes.
Table 3.
Genetic diversity in the L. donovani complex
| Complex | HD | π | Θ | NS/S | D (Tajima) | D* (Fu Li) |
|---|---|---|---|---|---|---|
| Geographical | ||||||
| Europe (n = 14) | 0.835 | 0.00042 | 0.00052 | 10/11 | −0.83724 (n.s.) | −1.07384 (n.s.) |
| Africa (n = 8) | 1.000 | 0.00075 | 0.00064 | 3/18 | 0.94878 (n.s.) | 0.73184 (n.s.) |
| India, India/Kenya (n = 3) | 0.667 | 0.00074 | 0.00074 | 6/8 | n/a | n/a |
| All (n = 25) | 0.947 | 0.000114 | 0.000119 | 21/36 | −0.17323 (n.s.) | −0.60186 (n.s.) |
| Clinical | ||||||
| Visceral (n = 18) | 0.977 | 0.00125 | 0.00126 | 20/35 | 0.02794 (n.s.) | −0.41053 (n.s.) |
| Cutaneous (n = 6) | 0.800 | 0.00042 | 0.00045 | 6/7 | −0.17323 (n.s.) | −0.60186 (n.s.) |
HD is haplotype diversity, π and Θ measure polymorphism per site, and NS/S is the ratio of nonsynonymous to synonymous substitutions. n.s., not significant.
Table 3 also gives the population structure parameters for the strains as grouped according to the form of the disease they cause (VL or CL), regardless of geographic origin. The diversity values (π, θ, and HD) are lower for strains causing CL than for those causing VL. These differences may reflect that our strains causing the mild form of the disease are from Europe and thus correlate with geographic origin, which might account for the observed pattern, although the small number of strains does not allow any generally significant conclusions.
The Kst and Fst indices (Table 4) reveal a high level of divergence between the populations. Estimates of the gene flow (Nm) obtained from Fst statistics are very low, showing little if any gene flow between continents, although they show lesser isolation between Africa and India than between any of these two and Europe. Some markers may be specific for particular genetic groups; for example, for enzyme-coding genes, allele 3 of pgd and allele 3 of gpi (14) have only been found in Indian MON-2 strains, whereas alleles 6 and 7 of nh1 and alleles 4 and 5 of pgd have only been found in Kenyan strains.
Table 4.
Geographic divergence (KST and FST) and gene flow (Nm) between populations of L. donovani
| Geographic divergence | Kst | Fst | Nm (Fst) |
|---|---|---|---|
| Europe–India, India/Kenya | 0.34510 | 0.58345 | 0.18 |
| Europe–Africa | 0.49762 | 0.65304 | 0.13 |
| India, India/Kenya–Africa | 0.26358 | 0.45204 | 0.30 |
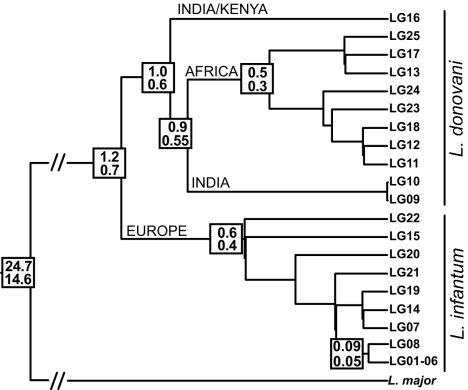
To explore evolution of the L. donovani complex, we have calibrated it against Leishmania major, on the basis of genes encoding the glycosomal form of gapdh and the large subunit of RNA polymerase II (rpoII), for which DNA sequences from other kinetoplastid parasites are available (18). The estimated times of divergence between L. major and the L. donovani complex are 14.6 (rpoII) and 24.7 (gapdh) Mya (Figs. 3 and 4 and Table 5). These estimates were obtained by the penalized likelihood method and truncated Newton algorithm as implemented in the r8s software (19), because the data sets for either gene significantly depart from the clock-like model. Figs. 3 and 4 and Table 5 also give the estimated time of divergence between taxa or groups of strains within the L. donovani complex. These estimates were obtained by the Langley-Fitch method (20), because the clock-like model could not be rejected on the basis of the 10 enzyme-coding genes in our data set (12, 14). The data suggest that possibly somewhere in central Asia, ≈1.2–0.7 Mya, the ancestral population of the L. donovani complex diverged into two separate clades, L. donovani and L. infantum. The Indian/Kenyan L. donovani subclade seems to represent an early offshoot of the L. donovani clade (1.0–0.6 Mya), followed by the Indian subclade. Between 0.6 and 0.3 Mya, further diversification occurred within L. infantum and Sudanese/Ethiopian L. donovani (Table 5).
Fig. 3.
Maximum-likelihood γ-corrected tree constructed under the clock model, used for divergence time estimates (Mya). Numbers at nodes denote age inferred by the clock-like Langley-Fitch method (r8s software) with calibration points inferred from the gapdh and rpoII data sets (upper and lower values).
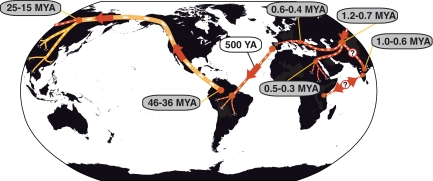
Fig. 4.
Origin and dispersal of Leishmania. A predecessor of L. donovani group and L. major would have evolved from monoxenous parasites of insects in South America ≈46–36 Mya and moved to Asia via the Bering land bridge (yellow line). The ancestor of the L. donovani complex diverged from other Leishmania species ≈14–24 Mya (red line). This predecessor arrived in central Asia and ≈1 Mya diverged into European L. infantum, African L. donovani and Indian/Kenyan L. donovani. L. infantum was later introduced in South America by European settlers, whereas L. donovani, represented here by strain LG16, would have been transferred by immigrants/slaves from India to Kenya and/or vice versa. Time estimates based on our data are in gray; those in white are taken from published literature.
Table 5.
Time of divergence between different species or populations
| Comparison | gapdh | rpoII |
|---|---|---|
| L. major and L. donovani s.l. | 24.73 | 14.6 |
| L. donovani s.s. and L. infantum | 1.18 | 0.73 |
| L. infantum | 0.63 | 0.39 |
| L. infantum(origin of MON-1) | 0.09 | 0.05 |
| India/Kenya (LG16) and the rest of L. donovani | 0.99 | 0.59 |
| Indian L. donovani(LG9/10) and African strains | 0.89 | 0.55 |
| African L. donovani | 0.52 | 0.32 |
The age of divergence between the populations analyzed in our study (rows 2–6) is inferred from the 10 enzyme-coding genes data set, calibrated against the timing of the split between L. major and L. donovani (row 1) as estimated by either gapdh and rpoii (see text for further details).
Discussion
The leishmaniases are geographically widespread severe diseases with an incidence of two million cases per year and 350 million people at risk. The incidence is increasing worldwide (2), likely because of increased travel and population migration, such as immunologically naïve and malnourished refugee populations into endemic areas in the Sudan, and the movement of infected people into nonendemic regions (21). Global warming and other environmental factors may also be contributing to the increased incidence.
Several trends have emerged from dozens of analyses. They include a partial correlation between genetic diversity and geographic origin, lack of identified association between genotypes and predictable clinical outcome, some flexibility in host specificity, hybrid genotypes and mixed infections of strains assigned to different species, and an incomplete correlation between genetic markers and phenotyping by MLEE, the present gold standard for the identification of Leishmania species (8, 10–12, 14, 15, 22, 23). Although the genetic groups may be robust in such trees, the basal nodes are usually not well supported, probably because of genetic recombination. Consequently, there has been as yet no clear view of the evolution of the L. donovani complex. The limitations are evident within L. infantum and its most common MLEE profile, MON-1. Discriminatory markers have been elusive for different species or genetic groups within the L. donovani complex.
The MLEE-based taxonomy of the L. donovani complex has been challenged by investigation of other molecular markers. We have sought to evaluate the current taxonomy of the L. donovani complex by joint consideration of the molecular data available, which include in our extended analysis ≈18,000 characters for each of 25 strains representative of the geographical distribution and clinical significance of this species complex. We also seek to elucidate the evolutionary history of the genus Leishmania, a parasitic protozoan of great public health significance. Our analysis represents one of the most extensive attempts to examine intra- and interspecific genetic diversity in a group of protists.
It has been proposed that the genus Leishmania first appeared either in the Old World (24, 25) or the New World (26–28). The New World origin is supported by the high genetic diversity of neotropical Leishmania species and by combined amino acid, DNA, and RNA polymerase-based trees, which root in America (data not shown). This claim has received support by the description of a monoxenic insect flagellate from Costa Rica that branches at the root of the Leishmania clade (29). Our results, based on the extensive data sets for gapdh and rpoII and calibrated by reference to the T. brucei–T. cruzi split dated at 100 Mya (30), are consistent with that interpretation. We propose that the ancestor of the New World leishmaniases evolved in South America in the Paleocene or Eocene, ≈46–36 Mya (Fig. 4) and then migrated via the Bering land bridge to Asia. The Leishmania lineage would have, then, dispersed through Central and/or Southeast Asia during the Miocene, 24–14 Mya (31), where a major diversification gave rise to Leishmania aethiopica, L. major, Leishmania gerbilli, Leishmania turanica, Leishmania tropica, and the L. donovani complex (26, 32). L. infantum would have split from the early L. donovani lineage ≈1 Mya, and L. donovani soon thereafter invaded India and Africa. Closing the circle, after 500 years ago, MON-1 European strains were transferred to South America, represented by the species formerly designated L. chagasi, considered synonymous with L. infantum (6). The two main reservoir hosts of the L. donovani complex are humans and canids whose historical movements likely have influenced the distribution of L. donovani and L. infantum.
Studies with microsatellites (10, 15) and enzyme-coding genes (14) indicate that strains from Kenya not included in our sample are closely related to the Indian strain LG16, which can thus be considered as representative of a genetic group that includes Kenyan L. donovani, here named the India/Kenya group. The same studies and another microsatellite analysis (8) suggest that MON-2 Indian strains (here represented by LG9 and LG10) are distinct from the India/Kenya group but somewhat more related to these than to other L. donovani-complex genetic groups. This may have been due to the introduction of the Indian strains to Africa by Indian immigrants (8, 33) or, conversely, by the slave trade from Africa to India (34, 35). The strains in our analysis suggest that India may have been invaded earlier than Africa, which might imply two different colonizations of Africa, one to Sudan and one to Kenya (10, 14). Equally plausible is that Leishmania may have bypassed India before reaching Africa, and that the populations evolved separately in Kenya and Sudan: relatively recently introduced aggressive strains of Kenyan origin would then have repeatedly swept throughout the Indian subcontinent.
The scenario we propose disagrees with the commonly accepted origin of the L. donovani complex in the Sudan, because Sudanese strains are a recent branch of L. donovani (5, 35, 36). An African origin cannot be discounted, because the tree rooted with L. major reveals that LG16 is at the basis of the L. donovani s.s. clade, with L. infantum as a sister group (Fig. 3). However, it is possible that intermediary strains are missing; for example, little-studied Asian strains.
Figs. 1 and 2 show considerable genetic divergence among European L. infantum strains, although less extensive than in Africa. There is no strict correlation between genetic make-up and country of origin, but this is not unexpected, given the mobility of humans in this region. Several molecular markers have been found that discriminate among MON-1 strains, the most prevalent MLEE type (12, 14, 37, 38). Identification of such markers is crucial for better understanding the population structure and potential spread of the virulent VL strains within Europe. In our phylogenies (Figs. 1 and 2), different clinical outcomes associated with the same MLEE profile (MON-1) are intermingled, which suggests that the host may have an important role in determining the outcome of the disease. An alternative possibility is that some strains may have independently lost the potential to visceralize, particularly those with an MLEE profile that has been isolated from cutaneous cases, such as MON-29. Visceralization in humans is probably an early character of the L. donovani complex, given that the vast majority of strains and all genetic groups of the complex cause VL. It seems unlikely that a pathogenicity island, gene rearrangement, or other pathogenic genome change, responsible for severe visceral disease, would have occurred independently among these strains.
The highly heterozygous strain from the Sudan (LG17), the only canine strain in our study, is likely a product of a recent genetic cross between strains. The presence of several zymodemes in the same host in the Sudan (39) might account for a high frequency of genetic exchange among Leishmania strains in this region, even if their prevailing mode of reproduction is clonal. Multiple heterozygous sites and the alleles shared with homozygous strains indicate that such heterozygosity is due not to recurrent mutation but to genetic exchange. Strain LG17 likely represents a robust putative hybrid within the L. donovani complex and thus deserves a more extensive characterization. More generally, the East African strains exhibit high levels of heterozygosity (Table 1), which suggests that genetic exchange may be more frequent there than among European strains. Genetic hybrids of distinct Leishmania species, L. infantum and L. major, have been isolated from Portuguese immunocompromised patients (40). In Toxoplasma gondii, genetic exchange among predominantly clonal populations has a dramatic impact on pathogenicity (41, 42). In Leishmania, an increased frequency of recombination among the Sudanese strains might account for the emergence of virulent strains that cause high human mortality in this region.
Figs. 1 and 2 suggest a taxonomy different from the classification based on MLEE. It is clear that genetic make-up and, therefore, phylogeny associate with geography rather than with MLEE phylogenies or clinical effects (visceral versus cutaneous). We propose that (i) the monophyletic set of European strains, whether agents of VL or CL, be classified as L. infantum (which would include the already-considered-synonymous L. chagasi); and (ii) the East African strains all be classified as L. donovani s.s. This taxon would include East African strains previously classified as L. archibaldi and L. infantum (see Fig. 1), which are genetically more similar to other East African strains (L. donovani) than the European L. infantum strains are to one another. Currently, L. infantum has been defined as having a MLEE got (asat) 100 phenotype by MON typing. This classification makes L. donovani paraphyletic (intermingled with L. infantum; see East African strains in Fig. 1). The inclusion of the Sudan/Ethiopia strains within a single taxon, L. donovani, is supported by a 98% bootstrap. So, our analysis supports the previous proposal that only strains with phenotypes for got (asat) of 100 and mdh of 100 or 104 (but not 112) (ref. 14) be classified as L. infantum. Figs. 1 and 2 show two sets of strains that are quite different from the rest and from each other: India (LG9 and LG10) and India/Kenya (LG16), each with strong bootstrap support. These strains may be retained for now within L. donovani s.s. Inconsistent species definitions are not unusual in other Leishmania species, for example Leishmania killicki in relation to L. tropica or Leishmania peruviana in relation to Leishmania braziliensis.
We have shown that large data sets may yield well defined species and phylogenies. Star phylogenies, in particular, a common situation, may be similarly resolved. The combination of multiple data sets may help to identify clade-specific markers, which could in turn resolve phylogenies of large strain collections.
Materials and Methods
Data Sets.
The markers for each strain are: (i) RFLP of genes gp63 and cpb; (ii) RAPD from 52 primers (15 strains only); (iii) repeat number in microsatellites; (iv) three noncoding regions, 3′nc-H1, ITS, and ir-PO; (v) 10 protein-coding genes used for MLEE; (vi) three more genes: lack, lp7, and tr. Characters per strain: 18,618, 1,041 of which are variable.
Phylogenetic Analysis.
Nonsequence data were incorporated into a 0/1 matrix representing absence/presence. Amino acid sequences were aligned by using the Megaling package (DNA Star, Madison, WI) and back-translated to nucleotides. The data sets were analyzed separately and in various combinations. A concatenated data set combining all data except RAPDs (16) was analyzed by using maximum parsimony as implemented in PAUP software, Version 4.0b10 (43) with characters equally weighted. Particular data sets and combinations (DNA sequences and fingerprints) were analyzed by maximum parsimony, maximum likelihood, minimum evolution, and LogDet-paralinear distances (trees not shown). The statistical parsimony network was constructed with TCS software, Version 1.21 (44). Heterozygous positions were considered as missing data.
DNA Polymorphism and Genetic Diversity.
Polymorphism, neutrality indices (45, 46), Fst, Kst, and Nm (47) were computed with DnaSP software, Version 4.01 (48).
Divergence Time.
The reference point was the split Trypanosoma brucei/Trypanosoma cruzi, estimated at 100 Mya (30), assuming similar rates of evolution in Leishmania. We estimated first the L. major–L. donovani split, using gapdh and rpoII, for which kinetoplastid data exist (18); next, we estimated the L. donovani complex, using the concatenated data set of 10 genes (12, 14), rooted with L. major. All estimates used penalized likelihood, the Langley Fitch method, and the truncated Newton algorithm as in the r8s software (19). The clock model was tested with PAUP 4.0b10 and Likelihood Ratio software (49).
Acknowledgments
This work was supported by European Community Grant QLK2-CT-2001-01810, Grant Agency of the Czech Academy of Sciences Grant Z60220518, and Ministry of Education of the Czech Republic Grants 2B06129 and LC07032.
Abbreviations
- VL
visceral leishmaniasis
- CL
cutaneous leishmaniasis
- MLEE
multilocus enzyme electrophoresis
- MON
Montpellier
- RFLP
restriction fragment length polymorphisms
- RAPD
randomly amplified polymorphic DNA.
Footnotes
The authors declare no conflict of interest.
References
- 1.Desjeux P. Trans R Soc Trop Med Hyg. 2001;95:239–243. doi: 10.1016/s0035-9203(01)90223-8. [DOI] [PubMed] [Google Scholar]
- 2.Dujardin JC. Trends Parasitol. 2006;22:4–6. doi: 10.1016/j.pt.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Lainson R, Shaw JJ. In: The Leishmaniases in Biology and Medicine. Peters W, Killick-Kendrick R, editors. London: Academic; 1987. pp. 1–120. [Google Scholar]
- 4.Rioux J-A, Lanotte G, Serres E, Pratlong F, Bastien P, Perieres J. Ann Parasitol Hum Comp. 1990;65:111–125. doi: 10.1051/parasite/1990653111. [DOI] [PubMed] [Google Scholar]
- 5.Pratlong F, Dereure J, Bucheton B, El-Safi S, Dessein A, Lanotte G, Dedet JP. Parasitology. 2001;122:599–605. doi: 10.1017/s0031182001007867. [DOI] [PubMed] [Google Scholar]
- 6.Mauricio IL, Stothard JR, Miles MA. Parasitol Today. 2000;16:188–189. doi: 10.1016/s0169-4758(00)01637-9. [DOI] [PubMed] [Google Scholar]
- 7.Lewin S, Schönian G, El Tai N, Oskam L, Bastien P, Presber W. Int J Parasitol. 2002;32:1267–1276. doi: 10.1016/s0020-7519(02)00091-7. [DOI] [PubMed] [Google Scholar]
- 8.Jamjoon MB, Ashford RW, Bates PA, Chance ML, Kemp SJ, Watts PC, Noyes HA. Parasitology. 2004;129:1–11. doi: 10.1017/s0031182004005955. [DOI] [PubMed] [Google Scholar]
- 9.Mauricio IL, Stothard JR, Miles MA. Parasitology. 2004;128:263–267. doi: 10.1017/s0031182003004578. [DOI] [PubMed] [Google Scholar]
- 10.Kuhls K, Mauricio IL, Pratlong F, Presber W, Schönian G. Microbes Infect. 2005;7:1224–1234. doi: 10.1016/j.micinf.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Quispe-Tintaya KW, Laurent T, Decuypere S, Hide H, Banuls AL, De Doncker S, Rijal S, Canavate C, Camping L, Dujardin JC. J Infect Dis. 2005;192:685–692. doi: 10.1086/432077. [DOI] [PubMed] [Google Scholar]
- 12.Zemanová E, Jirků M, Mauricio IL, Horák A, Miles MA, Lukeš J. Int J Parasitol. 2007;37:149–160. doi: 10.1016/j.ijpara.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Mauricio I, Gaunt MW, Stothard JR, Miles MA. Int J Parasitol. 2007;37:565–576. doi: 10.1016/j.ijpara.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 14.Mauricio IL, Yeo M, Baghaei M, Doto D, Silk R, Pratlong F, Zemanová E, Dedet J- P, Lukeš J, Miles MA. Int J Parasitol. 2006;36:757–769. doi: 10.1016/j.ijpara.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Kuhls K, Keilonat L, Ochsenreiter S, Schaar M, Schweynoch C, Presber W, Schönian G. Microbes Infect. 2007;9:334–343. doi: 10.1016/j.micinf.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Backeljau T, De Bruyn L, De Wolf H, Jordaens K, Van Dongen S, Verhagen R, Winnepenninckx B. Cladistics. 1995;11:119–130. doi: 10.1111/j.1096-0031.1995.tb00083.x. [DOI] [PubMed] [Google Scholar]
- 17.Hampl V, Pavlíček A, Flegr J. Int J Syst Evol Microbiol. 2001;51:731–735. doi: 10.1099/00207713-51-3-731. [DOI] [PubMed] [Google Scholar]
- 18.Simpson AGB, Stevens JR, Lukeš J. Trends Parasitol. 2006;22:168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Sanderson MJ. Bioinform Appl Note. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 20.Langley CH, Fitch WM. J Mol Evol. 1974;3:161–177. doi: 10.1007/BF01797451. [DOI] [PubMed] [Google Scholar]
- 21.Desjeux P. Comp Immunol Microbiol Infect Dis. 2004;27:305–318. doi: 10.1016/j.cimid.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 22.Quispe-Tintaya KW, Ying X, Dedet J-P, Rijal S, De Bolle X, Dujardin JC. J Infect Dis. 2004;189:1035–1043. doi: 10.1086/382049. [DOI] [PubMed] [Google Scholar]
- 23.Waki K, Dutta S, Ray D, Kolli BK, Akman L, Kawazu S-I, Lin C-P, Chang K-P. Eukaryot Cell. 2007;6:198–210. doi: 10.1128/EC.00282-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerr SF. Mem Inst Osw Cruz. 2000;95:75–80. doi: 10.1590/s0074-02762000000100011. [DOI] [PubMed] [Google Scholar]
- 25.Momen H, Cupolillo E. Mem Inst Oswaldo Cruz. 2000;95:583–588. doi: 10.1590/s0074-02762000000400023. [DOI] [PubMed] [Google Scholar]
- 26.Noyes HA, Arana BA, Chance ML, Maignon R. J Euk Microbiol. 1997;44:511–517. doi: 10.1111/j.1550-7408.1997.tb05732.x. [DOI] [PubMed] [Google Scholar]
- 27.Noyes HA. Mem Inst Oswaldo Cruz. 1998;93:657–661. doi: 10.1590/s0074-02761998000500017. [DOI] [PubMed] [Google Scholar]
- 28.Stevens JR, Noyes HA, Schofield CJ, Gibson W. Adv Parasitol. 2001;48:1–56. doi: 10.1016/s0065-308x(01)48003-1. [DOI] [PubMed] [Google Scholar]
- 29.Yurchenko VA, Lukeš J, Jirků M, Zeledón R, Maslov DA. Parasitology. 2006;133:537–546. doi: 10.1017/S0031182006000746. [DOI] [PubMed] [Google Scholar]
- 30.Stevens JR, Rambaut A. Infect Genet Evol. 2001;1:143–150. doi: 10.1016/s1567-1348(01)00018-1. [DOI] [PubMed] [Google Scholar]
- 31.Fernandes AP, Nelson K, Beverley SM. Proc Natl Acad Sci USA. 1993;90:11608–11612. doi: 10.1073/pnas.90.24.11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Croan DG, Morrison DA, Ellis JT. Mol Biochem Parasitol. 1997;89:149–159. doi: 10.1016/s0166-6851(97)00111-4. [DOI] [PubMed] [Google Scholar]
- 33.Sang DK, Pratlong F, Ashford RW. Trans R Soc Trop Med Hyg. 1992;86:621–622. doi: 10.1016/0035-9203(92)90153-4. [DOI] [PubMed] [Google Scholar]
- 34.Ashford RW. Int J Parasitol. 2000;30:1269–1281. doi: 10.1016/s0020-7519(00)00136-3. [DOI] [PubMed] [Google Scholar]
- 35.Ibrahim ME, Barker DC. Inf Gen Evol. 2001;1:61–68. doi: 10.1016/s1567-1348(01)00009-0. [DOI] [PubMed] [Google Scholar]
- 36.Ashford RW, Seaman J, Schorscher J, Pratlong F. Trans R Soc Trop Med Hyg. 1992;86:379–380. doi: 10.1016/0035-9203(92)90229-6. [DOI] [PubMed] [Google Scholar]
- 37.Ochsenreither S, Kuhls K, Schaar M, Presber W, Schönian G. J Clin Microbiol. 2006;44:495–503. doi: 10.1128/JCM.44.2.495-503.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Botilde Y, Laurent T, Quispe-Tintaya W, Chicharro C, Canavate C, Cruz I, Kuhls K, Schönian G, Dujardin JC. Infect Genet Evol. 2006;6:440–446. doi: 10.1016/j.meegid.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 39.Dereure J, El-Safi SH, Bucheton B, Boni M, Kheir MM, Davoust B, Pratlong F, Feugier E, Lambert M, Dessein A, et al. Microbes Infect. 2003;5:1103–1108. doi: 10.1016/j.micinf.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 40.Ravel C, Cortes S, Pratlong F, Morio F, Dedet J-P, Campino L. Int J Parasit. 2006;36:1383–1388. doi: 10.1016/j.ijpara.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 41.Grigg ME, Bonnefoy S, Hehl AB, Suzuki Y, Boothroyd JC. Science. 2001;294:161–165. doi: 10.1126/science.1061888. [DOI] [PubMed] [Google Scholar]
- 42.Boyle JP, Rajasekar B, Saei JPJ, Ajoka JW, Berriman M, Paulsen I, Roos DS, Sibley LD, White MW, Boothroyd J. Proc Natl Acad Sci USA. 2006;103:10514–10519. doi: 10.1073/pnas.0510319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods) Sunderland, MA: Sinauer; 2003. Version 4. [Google Scholar]
- 44.Clement M, Posada D, Crandall KA. Mol Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 45.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu XY, Li W-H. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nei M. Proc Natl Acad Sci USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 49.Felsenstein J. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]