Abstract
We develop a computer model for how two different chemical catalysts in solution, A and B, could be driven to form AB complexes, based on the concentration gradients of a substrate or product that they share in common. If A's product is B's substrate, B will be attracted to A, mediated by a common resource that is not otherwise plentiful in the environment. By this simple physicochemical mechanism, chemical reactions could spontaneously associate to become chained together in solution. According to the model, such catalyst self-association processes may resemble other processes of “stochastic innovation,” such as Darwinian evolution in biology, that involve a search among options, a selection among those options, and then a lock-in of that selection. Like Darwinian processes, this simple chemical process exhibits cooperation, competition, innovation, and a preference for consistency. This model may be useful for understanding organizational processes in prebiotic chemistry and for developing new kinds of self-organization in chemically reacting systems.
Keywords: chemical evolution, self-organization, abiogenesis, catalytic chains
There are several examples of what might be called “stochastic innovation,” whereby a biological, physical, or sociological system: (i) searches among viable options, then (ii) selects one or more of those options that is “best” by some metric, then (iii) locks in that selection for the future. In biology, the best-known example is Darwinian evolution (1), where variation is the term that describes the search step, and natural selection is the term that describes steps (i) and (ii). Stochastic innovation occurs in learning and memory and neural development, where the correlated firings of neurons can lead to changes in synaptic strength (2–5), and to the development of the vascular system, where new blood vessels grow toward oxygen-deprived cells (6, 7).
Stochastic innovation appears in other arenas, too. Human beings, businesses, and social organizations evolve through decision-making: they search among the options available to them, make self-serving choices, then remember and act on those decisions in the future. Social insects, like ants, search randomly for food, then lock in the discovery with chemical trails for the rest of the colony (8, 9). Computer models of artificial life and artificial societies show that stochastic innovation can meet apparent goals that were not programmed into them at earlier times (10, 11). The power of stochastic innovation is that it can lead to complex behaviors or organizational structures that are responsive to changes in the environment, even though such processes are unguided, unplanned, and stochastic.
Our interest here is in whether stochastic innovation might also be achievable in chemistry and biochemistry. Can chemical and biochemical reactions be chained together in complex and innovative ways, driven only by simple physicochemical search and selection processes? If so, it may be useful, not only as a tool in chemistry and biochemistry, but also for giving insights into the processes of chemical organization that may have occurred during prebiotic evolution.
Here, we propose a simple model. Our goal is not to explain some existing body of data, because we know of none that pertains. Rather, our goal here is to propose a type of organizing principle that has not been explored before, as far as we know, but that is based on well established physicochemical principles and that can be tested by experiments. Our initial motivation for this work was to understand some puzzles of prebiotic chemistry, where, it could be argued, the field is just as limited by a lack of specific testable models at the moment as it is by a lack of experiments.
Model of Agents and Resources.
We focus here on catalysts, such as enzymes or simple surfaces. We call a catalyst an “agent.” An agent converts a substrate to a product; we label agents alphabetically (see Fig. 1). We call a substrate or product a “resource”; we label resources numerically. We assume that agents are Michaelis–Menten catalysts; i.e., they bind to their substrate before converting the substrate to a product. In our model, resources may be supplied by the external environment. Such environmental resources may vary with time in a random or controlled way by external forces, but, for simplicity, we assume they are uniformly distributed in space.
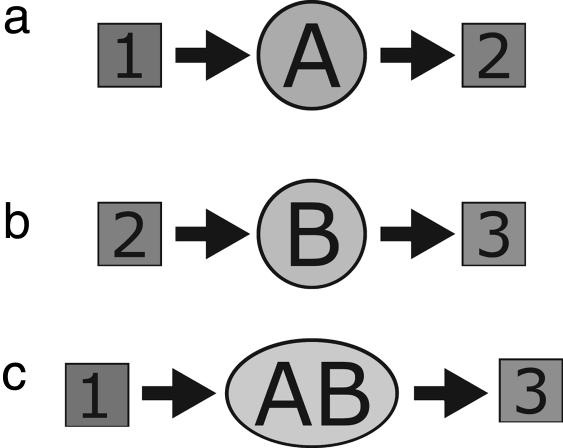
Fig. 1.
Agents (lettered circles) and resources (numbered squares). (a) Agent of type A converts substrate 1 to product 2. (b) Agent of type B converts substrate 2 to product 3. (c) When agents A and B are complexed together, two reactions are chained together, converting substrate 1s to product 3s.
Fig. 1 shows an example. Agent A converts a substrate 1 to a product 2. Agent B converts substrate 2 to product 3. Key components of our model are the common resources, which are substrates or products that serve in common among different types of agents. For example, in Fig. 1, resource 2 is a common resource because it is both a product of A and a reactant for B. Figs. 1 and 3 also show that if agents A and B come together by some process, then the AB complex is a “machine” that converts 1s to 3s, mediated by the intermediary resource 2s.
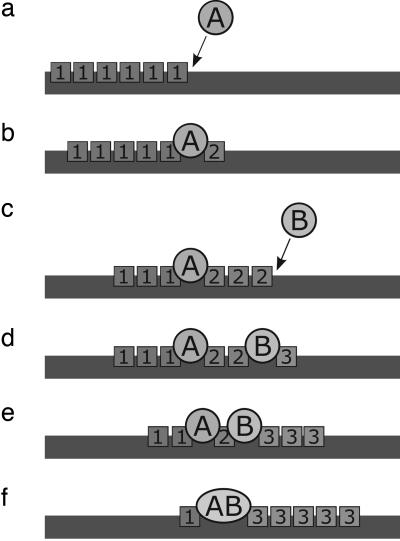
Fig. 3.
Attraction on the lattice. (a) Agent A leaves the bulk and binds to the surface lattice in a region where resource 1s are concentrated. (b) Agent A converts 1s to 2s. (c) Agent B leaves the bulk and binds to the surface lattice where 2s are concentrated, which tends to be near the As that produced them. (d) Agent B produces 3s. (e and f) Agent B associates with A (e), forming a complex, which is now (f) a machine that converts 1s to 3s.
Agent B (Fig. 1) may take up a substrate molecule 2 from either of two sources: either the 2 was produced as the output from a nearby A agent, or the 2 was supplied externally from the environment, if 2s are available from external sources. Because we assume that Bs are Michaelis–Menten catalysts, a B will bind to its substrate, a 2 in this case. Bs will concentrate around 2s simply because Bs flow down their chemical potential gradients, in the same way that solutes in chromatographic mobile phases will seek out and bind to stationary-phase surfaces for which they have affinity.
Principles of Attraction and Shielding.
There are two possibilities for each B agent (Fig. 2): attraction or shielding.
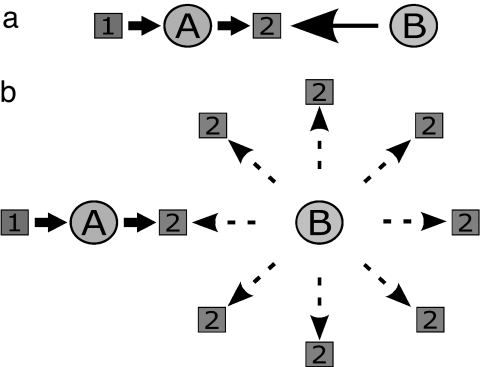
Attraction. As attract Bs through the following indirect mechanism: As produce 2s; 2s are localized near the As; those 2s attract Bs, concentrating the Bs around the As, thus leading to more AB complexation than would have occurred without the intermediary 2s (see Fig. 2a). This enhancement happens when: (i) A agents are present, (ii) 1s (the substrates for As) are plentiful, and (iii) 2s are depleted in the environment (i.e., available only at small or zero concentrations). AB complexation introduces into the system an “innovation,” i.e., an ability to produce 3s from 1s, an ability that does not simply and directly result from the presence of A or B alone (see Fig. 1c). Significant chaining together of agents is an emergent property of our system. In short, AB complexes are driven to form through mutual indirect attraction, mediated by 2s, the common resource, but this attraction occurs only when 2s are depleted from the environment.
Shielding. In contrast, when environmental 2s are plentiful and available in all directions, Bs will not selectively migrate toward As (see Fig. 2b). We call this “shielding.” (A more biological example of shielding is chemotaxis. A bacterium will swim toward a point source of food, except if food is uniformly distributed everywhere in space; then the bacterium would not migrate preferentially toward any one single point source. Chemotaxis, of course, is a complex process, but it illustrates how a favored direction of motion can result from simple physicochemical forces that change when environmental resources vary.) In the present model, attraction and shielding are simple consequences of concentration gradients.
Fig. 2.
Attraction and shielding. (a) Attraction. Bs are attracted to 2s, which are produced by As, hence Bs are attracted to As. (b) Shielding. When 2s are plentiful in the environment, Bs are attracted to them in all directions, hence have no special net tendency to associate with As.
Details of the Model.
Here is our model for how stochastic innovation might arise in a system of chemical catalysts. We assume that catalyst agents can adsorb to a surface. The prebiotic origin of life may have involved surfaces, such as minerals or clays, on which reactions took place (12–14).
Our model involves two compartments. First, there is a surface lattice where all of the reactions take place. Second, that surface is in direct contact with a bulk solution just above it, which serves as a chemical potential “bath,” a source of agents and resources for the surface. The surface simply provides a mechanism for trapping the 2s and Bs, slowing their escape from the As, providing the basis for the AB complexation enhancement. There are NA molecules of each agent type in the simulation, and EN (E1, E2, … ) represent the concentrations of that resource in the bulk solution. Fig. 3 shows the steps: (i) 1s attract As from the bulk onto the surface lattice, (ii) As produce 2s, (iii) which then attract Bs, (iv) leading to a machine in which As and Bs are clustered on the surface to produce 3s from 1s. Agents and resources diffuse rapidly throughout the bath, so they can be regarded as being in equilibrium within the bath over the time scale of the processes that happen on the surface. The lattice has Lx × Ly lattice sites; it just serves to coarse-grain the spatial localization. Each lattice site is large enough to be occupied by multiple molecules of different types at the same time.
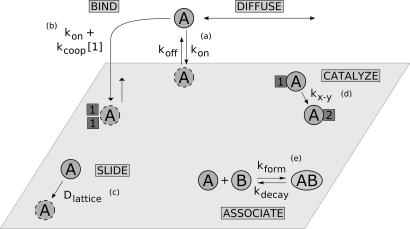
We believe this is the minimal model that captures how catalysts might self-associate in solution. During any given time interval in the simulation, any or all of the following processes may occur (see Fig. 4).
A resource molecule or an agent molecule may drop down from the bulk and associate with the surface lattice, with rate coefficient kon. To keep the model simple, all four types of molecule bind the lattice with the same rate constant.
Any resource or agent molecule on the surface may detach from the surface and be released into the bulk, with rate coefficient koff.
Because of the Michaelis–Menten binding property of each agent, an agent will attach more rapidly to a site where substrate is concentrated, in proportion to the concentration of its substrate at that site, with a rate coefficient kcoop.
An agent molecule A can convert any 1s to 2s on its lattice site with a rate coefficient k1–2.
An agent molecule B can convert any 2s to 3s on its lattice site with coefficient k2–3.
Agents and resources can diffuse laterally on the surface lattice with coefficient Dlattice.
An agent A and agent B on the same lattice site can associate with each other, with rate coefficient kform, forming a new species of agent, the AB complex.
An AB complex may dissociate, either in the bulk or on the surface, with rate coefficient kdecay. Supporting information (SI) Table 1 and SI Text provide further details and typical values of the parameters (see SI Table 1), as well as a pseudocode implementation (see SI Text). (Source code is available on request.)
Fig. 4.
Processes in the lattice model. (a) Molecules exchange between the bulk solution and bound to a random lattice site. (b) Agents have an additional binding rate at lattice regions with their input resource. (c) Bound molecules move about the surface. (d) Agents convert input resources to output resources at their site. (e) Two agents at a site can form an agent complex.
Results
Exploring Attraction and Shielding.
Our computer simulations show that if an agent A produces 2s at a rate that is faster than the 2s diffuse away, and if the A is bound to the surface, then 2s will be concentrated near the As on the surface. Agents of type B, which use 2s as substrates, explore space stochastically but will be attracted to the 2s, on average, thus binding to surface lattice sites near the most productive As. If there is a mutual affinity of As for Bs, AB complexes will form on the surface.
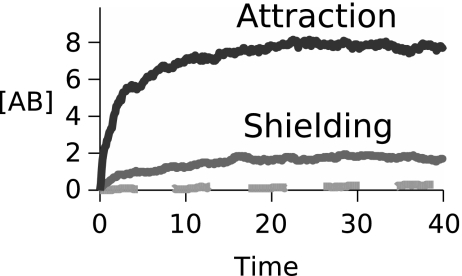
Fig. 5 shows: (i) attraction, the situation in which Bs migrate to As to form complexes, driven by depletion of the common resource, 2; (ii) shielding, the situation in which common resource 2 is plentiful in the environment, so Bs are not selectively attracted to As; and (iii) a control simulation showing that when As produce no common resource 2s, there is essentially no formation of AB complexes, even though there is some intrinsic affinity between the As and Bs. In this model, AB complex formation is a nonequilibrium process; it happens only in the presence of a gradient of 2s concentrated around the As. The complex formation process can result from either highly productive As or the environmental depletion of 2s from the system.
Fig. 5.
Numbers of AB complexes formed vs. time. Dark line: under attraction conditions, no environmental supply of resource 2 (E1 = 104, E2 = 0). Light line: under shielding conditions, with resource 2 provided by the environment (E1 = 104, E2 = 103). Dashed line: control experiment; no 2s are available because the environment has none, and As are unproductive because of the absence of 1s (E1 = 0, E2 = 0). (Time is in units of 1,000 simulation units for this and following plots.)
The rate of complex formation: increases with the productivity of the agents and decreases with the degree of environmental shielding. AB complexation is largest when As are productive (requiring that bulk 1s are plentiful). AB complexation is reduced by shielding (when environmental 2s are plentiful) (see SI Fig. 8).
Some Properties of the Model: Cooperation, Competition, Consistency, and Innovation.
Our agents cooperate with each other. When the environment provides no substrate for catalyst B, B will migrate toward A via its substrates. The model shows a driving force for innovation: when the common resource is depleted, the system evolves an ability to create 3s directly from 1s, thus creating two chemical reactions chained together where there were only two isolated reactions before.
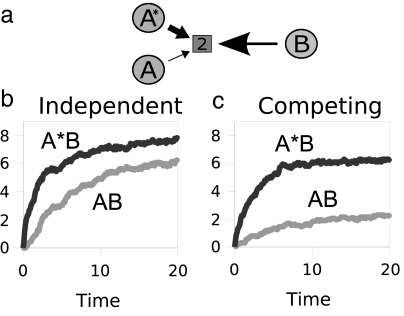
Our model also exhibits competition. Fig. 6 shows a version of the model in which As convert 1s to 2s and Bs convert 2s to 3s, as before, but now there is an additional agent, labeled A*. A* is a superior version of A. A* converts 1s to 2s faster than As catalyze the same conversion. Fig. 6 b and c show that complex formation increases with As productivity. For comparison, Fig. 6b just shows independent experiments of a single A interacting with B: curve [A*B] shows what happens when the A produces 2s rapidly, and curve [AB] shows what happens when the A produces 2s more slowly. In contrast, Fig. 6c shows A and A* together in the same solution, competing for Bs. A* outcompetes A to associate with the Bs. Comparing Fig. 6c with Fig. 6b shows a nonadditivity because of the competition for the same parameters: When A and A* compete in the same solution, the “rich get richer.” Within our simple chemical model system, this competition resembles Darwinian selection, except that our metric of “success” is AB complex formation, whereas the metric of success in biological systems is survival.
Fig. 6.
Competition. (a) Agent B can associate with either agent A or agent A*, a superior producer of resource 2. (b) Complex formation increases as the productivity of As increase. (c) Agent A competes with a superagent A* (for A*, kx-y = 0.1 and for A, kx-y = 0.001). Competition enhances the difference between A*B and AB complex formation.
Our model shows that consistency has value, exhibiting “tortoise and hare” behavior. One type of agent, At, the tortoise, is a slow consistent producer of 2s. The other type of agent, Ah, the hare, is highly productive, but only in short bursts. Even though the time-averaged productivity in converting 1s to 2s is identical here for these two types of agents, the tortoise wins. The tortoises form complexes with Bs at a faster rate than the hares form complexes with Bs. Thus, sustained consistency is more effective for complex formation than high-activity-burst behavior. (See SI Fig. 9.)
Other Functional Hierarchies.
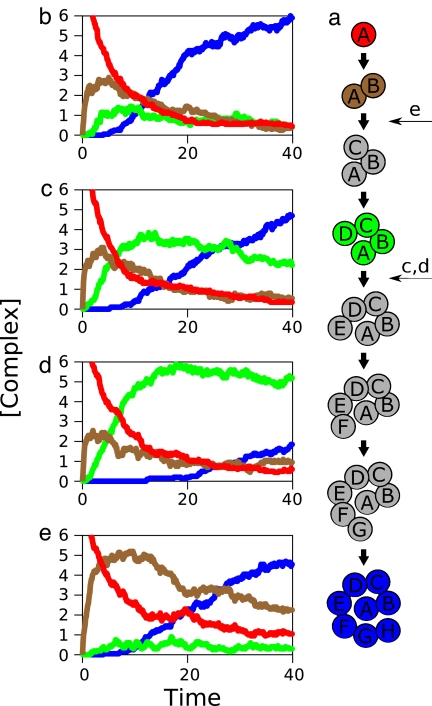
This model indicates how more complex chains of catalytic activity could be formed. Fig. 7a shows a linear series of catalytic agents that become chained together to convert 1s to 2s to 3s to 4s, etc. Fig. 7b shows the time dynamics of formation, when the environment is simply providing 1s. It shows that the final endstate machine, a concatenation of catalysts A, B, C, D, E, F, G, and H, grows monotonically populated, and that no intermediate smaller machine is ever substantially populated during the time course of development. Thus, multiple catalysts can be driven together, potentially into a variety of topological arrangements, including metabolic chains, networks, and cycles.
Fig. 7.
Formation of multiagent complexes. (a) Eight agents assemble sequentially to form a catalytic chain, using only the input resource for agent A. (b) When there is no environmental shielding, the full chain assembles rapidly, with no stable intermediates. (c) The presence of environment resources (E5 = 100) shields the step between agents D and E, resulting in longer-lived minor intermediates. (d) A 10-fold stronger environmental supply (E5 = 1,000) results in the stable dominance of the A-D intermediate chain over the complete chain. (e) The presence of environment resources (E3 = 100) at an earlier stage, between B-C, leads to an initially stronger, but ultimately minor, intermediate chain.
These results bear on an idea that has been called “irreducible complexity.” It has been argued that complex biological and prebiotic chemical systems could not have arisen by simple physicochemical processes, because there would have been no selective advantage for each of the putative incremental changes along the way (15). In that view, what good is half an eye? An organism would not be served by anything less than a full eye, so intermediate structures would not have imparted enough value to survive natural selection. In that view, “irreducible” refers to a system that would fail to function if any one component is removed, and irreducible complexity refers to the idea that such systems require design and could not be developed by stochastic innovation. The counterargument, seen in computer simulations, for example (10), has been that stochastic innovation works differently: evolution doesn't “know” the final end-goal in advance, but finds it through a random search in indirect, incremental steps.
Fig. 7 shows our model version of an irreducible system (a chain of reactions that convert 1s ultimately into 9s; it would fail to do so if any step is removed). This system ultimately converts 1s into 9s through a multistep process in which no step is dispensable. Yet, this machine arises in our model from simple physicochemical processes that have not involved any sort of “design” in advance to achieve this particular goal of producing 9s. In the model, this chaining together of chemical reactions is driven by blind physicochemical forces.
Discussion
Evolutionary “Gaps.”
In evolution, there are sometimes evolutionary gaps in the fossil record, situations in which lifeforms X and Y or features X and Y are known but where there is no evidence of the steps in between. Fig. 7b shows how such gaps arise in our simple machine. In short, the intermediate states are unstable. The steps are downhill. One evolutionary step leads to the next, quickly followed by the next, and so on, without pausing. In the evolutionary metaphor, half an eye never appears as a stable state because such a state is quickly driven by even stronger evolutionary forces to form a complete eye, maybe for a different purpose than the half-eye. Such two-state transitions are also common in protein folding, for example, where the denatured state is followed in time by a partly structured state that is immediately followed by an even more structured state, etc., until the molecule becomes fully folded into the native structure. At the earliest stages of folding, the protein does not know that it is headed toward the native state; it is just seeking a situation that is marginally better than its previous state.
The alternative is to have no evolutionary gap. Fig. 7 c–e shows how that alternative situation arises in our simple model, for a different set of parameters. In these cases, intermediate states are stable and populated during the evolution of the full molecular machine. The system can stall at intermediate points. In both situations (stable intermediates or two-state behavior), this simple model of chemical association resembles behaviors observed in Darwinian biological systems: Evolutionary gaps sometimes appear, and sometimes they don't.
Implications for Prebiotic Chemistry.
There have been two general models for prebiotic evolution: genetics-first (GF) or metabolism-first (MF) (16–18). In GF, some genetic machinery, or capability for self-reproduction, is presumed to arise at an early stage; for example, using RNA molecules (19, 20). Biology then evolves from that point. Once genetic machinery exists, there are many plausible models for the later stages, based on hypercycles (21, 22), and/or based on the many powerful RNA and protein evolution experiments that have been performed (23–25). The challenge in accepting GF as a first step, however, is that it is complex and requires the joint emergence of catalysis, compartmentation, and heritability, all at the same time (26). de Duve has noted (16) that GF “is accepted much less for its likelihood than for the lack of an alternative.”
In the MF model (14, 16, 27), chemical reactions become chained together evolutionarily before the appearance of genetic machinery. Although the requirements for MF are, in theory, more elementary than for GF, a key question about MF is how catalysts might become organized on their own, in the absence of a genetic system (28, 29).
We believe the present model of stochastic innovation based on attraction and shielding among chemical catalysts provides a plausible mechanism by which simple metabolic chains and cycles of reactions could have come together, perhaps at least long enough for a genetic system to then emerge. Of course, an important virtue of ultimately having a genetic system is that it provides much longer term “memory” for the “lock-in” step (step 3 in the Introduction) than does nongenetic propagation, where memory is merely provided by a ratio of off-rates to resource fluctuation times.
A key distinction between stochastic innovation, explored here, and design-based innovation, in which a complex system is engineered and constructed by a designer, is that stochastic innovation involves no implicit “goals” and no guidance toward a particular purpose. The Darwinian paradigm shows how increasing complexity and order can arise from processes that do not involve guidance through intelligence or design. Darwinian evolution is a process of elimination (“evolution-away-from”), rather than a process of design (“evolution-toward”). Stochastic innovation achieves evolution-away-from by search, selection, and lock-in. The present model has the three features of stochastic innovation.
Search. The B agents diffuse randomly through space and find 2s, based on a mutual binding affinity.
Selection. The B agents associate with A agents, if the common resource 2s are not present in the environment, leading to AB complex formation.
Lock-In. If the off-rate for dissociation of AB complexes is slower than the average frequency of resource depletion disasters of the common resource in the environment, then the complexes will be stable beyond the time scale of a single resource disaster.
Here is a possible experimental test of our model. Two enzymes, A and B, would be selected, based on having a common resource (e.g., 2). A's product is known to be B's substrate. The As would be covalently linked to a surface, such as a chromatography stationary phase. A pair of fluorescent probes would be attached: a donor probe is attached to each A molecule and an acceptor probe to each B molecule. This would allow for monitoring the concentration of AB complexes, through fluorescence quenching, for example. Two experiments would be performed. In one experiment, B molecules in a mobile phase would flow across the stationary phase in the absence of added 2s, but in the presence of 1s, the substrate for As, and then the amount of AB complex formation, would be measured. The other experiment would be identical, except that a high concentration of 2s would also be present in the mobile phase. The present model suggests that, if the relative on-rates, off-rates, and diffusion coefficients are roughly as given in SI Table 1, AB complexes should be more concentrated in the first experiment.
Related Modeling Efforts.
The present model differs from others that have been used to explore chemical self-organization. Some agent-based models (10, 11, 30) involve computer-based rules rather than the laws of physical chemistry that are of interest here. Mass-action models of early evolution have been developed (31, 32), but they also are not focused on the microscopic chemical mechanisms. Models of hypercycles (21, 22) pertain to molecular systems that already have a genetic system, whereas our interest here is in molecular systems having no genetic system. Recently, there have been interesting studies of complexity and fragility in biological systems (33, 34), but those treatments presume some preexisting mechanism of stochastic innovation.
Because the work of Turing in the early 1950s (35), much mathematical modeling in chemistry, chemical engineering, and biology has explored pattern formation (36). Pattern formation is often modeled by using lattice models such as the present one, or nonequilibrium coupled spatiotemporal differential equations, sometimes with added stochastic noise terms. We believe that our model, too, could be readily cast in the form of such a coupled set of “reaction–diffusion” equations.
However, the present work differs from those treatments in certain respects. First, ours addresses not just how molecules can move and organize in space, but how chemical reactions can move and organize in space and in so doing become chained together into more complex reactions. Second, pattern formation usually involves some particular nonlinear component term in the model's mathematics, often arising from saturable or cooperative binding, for example (37). Our model, too, involves saturable binding, but the most important nonlinear component of our model is what might be called “reflexive catalysis.” That is, at the same time that our agents catalyze reactions among the resources, our common resources also perform a sort of catalysis in the process of agent–agent association. In addition, our resources can be regarded as “ephemeral catalysts,” catalyzing complex formation only under certain nonequilibrium conditions and losing their catalytic power at equilibrium or when resources are plentiful.
Conclusion
A well known process in chemistry is the binding and association of molecules, driven by thermodynamic forces. Here, we consider whether catalyst molecules might be driven to associate with each other, through typical binding forces, but based on their molecular functions. Functional driving forces are well-known in biology, through the principles of evolution, but are not yet much studied in chemistry. We propose a model for how different Michaelis–Menten enzymes or catalysts might tend to associate, driven by the production or depletion of common resources. The agents do not associate if the common resource is plentiful. We call this the shielding principle. In this way, agents organize adaptively, and complexity can form from simpler systems. In our model, “function dictates structure,” a reversal of the paradigm in which “structure dictates function.”
Our model of stochastic innovation for chemical catalysts has three components: (i) a method for sampling a space of possibilities (Bs can search for As or not, depending on the presence of 2s), (ii) selection of a “favorable” outcome (Bs become more productive of 3s when their substrate 2s are depleted, if they associate with As), and (iii) a form of memory that locks in this outcome (the off-rates of Bs from As is slower than the time constant for fluctuations of 2s in the environment. Our process resembles other stochastic innovation processes, such as Darwinian evolution, in that agents compete, cooperate, value consistency, and innovate. The model is experimentally testable. It is a possible model for how metabolic reactions might have become chained together, at least primitively, before the formation of a genetic system. And, it might lead to ways to self-assemble chemical reactions in solution.
Materials and Methods
Fig. 4 shows that B agents can associate with A agents through two routes: (i) slow random migration on the surface, after Bs bind to the lattice, or (ii) rapid diffusion in the bulk, then binding to the lattice. The latter is more targeted, because Bs bind preferentially (through parameter kcoop) where 2s are located, potentially near the As. Hence, the intrinsic rate of AB complexation depends on kon, koff, and Dlattice.
Intermediary 2s can affect AB complexation through two mechanisms. First, AB complexation is promoted if the number of lattice-bound 1s is high and if As are productive (k1–2 is large). The number of 1s on the surface depends on 1s in the bulk (E1) and the adsorption equilibrium constant, kon/koff. The number of surface As is then determined by kcoop and dependent on the concentration of lattice-bound 1s. These bound molecules produce bound 2s at the rate k1–2. However, if catalytic rates are much slower than koff, resources disappear from the surface before they can become concentrated enough to attract agents. Second, AB complexation is slowed by the environmental supply of 2s (E2), which adsorb to the surface randomly and serve as decoys that attract Bs to nonproductive sites (where there are no As).
In order that 2s remain near As long enough to mediate AB complexation, lateral diffusion on the surface is set to be relatively slow, so dissociation of an AB complex on the surface is typically followed by reassociation. In contrast, dissociation of an AB complex in the bulk solution seldom leads to reassociation, because diffusion in the bulk is set to be fast. Therefore, most of the permanent dissociation of AB complexes occurs in the bulk solution. Also, the catalytic rates used in our simulations are large or comparable to the lateral diffusion rates.
Supplementary Material
Acknowledgments
We thank Patsy Babbitt, Don Hilvert, Tack Kuntz, Chris Voigt, John Chodera, and Yigal Nochomovitz for helpful discussions. This work was supported by a National Science Foundation predoctoral fellowship (to J.A.B.) and National Institutes of Health Grant GM34993 and the Sandler Foundation (to K.A.D.).
Abbreviations
- GF
genetics-first
- MF
metabolism-first.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703522104/DC1.
References
- 1.Mayr E. What Evolution Is. New York: Basic Books; 2001. [Google Scholar]
- 2.Hebb DO. The Organization of Behavior. New York: Wiley; 1949. [Google Scholar]
- 3.Kelso SR, Ganong AH, Brown TH. Proc Natl Acad Sci USA. 1986;83:5326–5330. doi: 10.1073/pnas.83.14.5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goda Y, Davis GW. Neuron. 2003;40:243–264. doi: 10.1016/s0896-6273(03)00608-1. [DOI] [PubMed] [Google Scholar]
- 5.Alberts B, Johnson A, Lewis J, Raff M., Roberts K, Walter P. Mol Biol Cell. New York: Garland Science; 2002. p. 1238. [Google Scholar]
- 6.Ferrara N, Gerber HP, LeCouter J. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 7.Kirschner MW, Gerhardt JC. The Plausibility of Life. New Haven, CT: Yale Press; 2005. pp. 143–176. [Google Scholar]
- 8.Goss S, Aron S, Deneubourg JL, Pasteels JM. Naturwissenschaften. 1989;76:579–581. [Google Scholar]
- 9.Detrain C, Natan C, Deneubourg JL. Naturwissenschaften. 2001;88:171–174. doi: 10.1007/s001140100217. [DOI] [PubMed] [Google Scholar]
- 10.Lenski RE, Ofria C, Pennock RT, Adami C. Nature. 2003;423:139–144. doi: 10.1038/nature01568. [DOI] [PubMed] [Google Scholar]
- 11.Epstein JM, Axtell R. Growing Artificial Societies: Social Science from the Bottom Up. Cambridge, MA: MIT Press; 1996. [Google Scholar]
- 12.Hanczyc MM, Fujikawa SM, Szostak JW. Science. 2003;302:618–622. doi: 10.1126/science.1089904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segre D, Ben-Eli D, Deamer DW, Lancet D. Orig Life Evol Biosph. 2001;31:119–145. doi: 10.1023/a:1006746807104. [DOI] [PubMed] [Google Scholar]
- 14.Wächtershäuser G. Microbiol Rev. 1988;52:452–484. doi: 10.1128/mr.52.4.452-484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Behe M. Darwin's Black Box. New York: Free Press; 1996. [Google Scholar]
- 16.de Duve C. Proc Natl Acad Sci USA. 1987;84:8253–8256. doi: 10.1073/pnas.84.23.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith E, Morowitz HJ. Proc Natl Acad Sci USA. 2004;101:13168–13173. doi: 10.1073/pnas.0404922101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orgel LE. Trends Biochem Sci. 1998;23:491–495. doi: 10.1016/s0968-0004(98)01300-0. [DOI] [PubMed] [Google Scholar]
- 19.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 20.Joyce GF, Orgel LE. In: The RNA World. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1999. pp. 49–77. [Google Scholar]
- 21.Eigen M, Schuster P. J Mol Evol. 1982;19:47–61. doi: 10.1007/BF02100223. [DOI] [PubMed] [Google Scholar]
- 22.Eigen M. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 23.Bartel DP, Szostak JW. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 24.Wright MC, Joyce GF. Science. 1997;276:614–617. doi: 10.1126/science.276.5312.614. [DOI] [PubMed] [Google Scholar]
- 25.Keefe AD, Szostak JW. Nature. 2001;410:715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith E, Morowitz HJ. Proc Natl Acad Sci USA. 2004;101:13168–13173. doi: 10.1073/pnas.0404922101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kauffman SA. J Theor Biol. 1986;119:1–24. doi: 10.1016/s0022-5193(86)80047-9. [DOI] [PubMed] [Google Scholar]
- 28.de Duve C. Blueprint for a Cell. Burlington, NC: Neil Patterson; 1991. pp. 123–170. [Google Scholar]
- 29.Orgel LE. Proc Natl Acad Sci USA. 2000;97:12503–12507. doi: 10.1073/pnas.220406697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Troisi A, Wong V, Ratner MA. Proc Natl Acad Sci USA. 2005;102:255–260. doi: 10.1073/pnas.0408308102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain S, Krishna S. Proc Natl Acad Sci USA. 2001;98:543–547. doi: 10.1073/pnas.021545098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfeiffer T, Soyer OS, Bonhoeffer S. PloS Biol. 2005;3:1269–1275. doi: 10.1371/journal.pbio.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Csete ME, Doyle JC. Science. 2002;295:1664–1669. doi: 10.1126/science.1069981. [DOI] [PubMed] [Google Scholar]
- 34.Carlson JM, Doyle J. Proc Natl Acad Sci USA. 2002;99:2538–2545. doi: 10.1073/pnas.012582499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turing AM. Trans Roy Soc London. 1952;237:37–72. [Google Scholar]
- 36.Murray JD. Mathematical Biology. New York: Springer; 2002. [Google Scholar]
- 37.Strogatz SH. Nonlinear Dynamics and Chaos. Reading, MA: Addison–Wesley; 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.