Abstract
Bone formation is carried out by the osteoblast, a mesenchymal cell whose lifespan and activity are regulated by growth factor signaling networks. Growth factors activate phosphatidylinositol 3-kinase (PI3K), which enhances cell survival and antagonizes apoptosis through activation of Akt/PKB. This process is negatively regulated by the Pten phosphatase, which inhibits the activity of PI3K. In this study, we investigated the effects of Akt activation in bone in vivo by conditionally disrupting the Pten gene in osteoblasts by using Cre-mediated recombination. Mice deficient in Pten in osteoblasts were of normal size but demonstrated a dramatic and progressively increasing bone mineral density throughout life. In vitro osteoblasts lacking Pten differentiated more rapidly than controls and exhibited greatly reduced apoptosis in association with markedly increased levels of phosphorylated Akt and activation of signaling pathways downstream of activated Akt. These findings support a critical role for this tumor-suppressor gene in regulating osteoblast lifespan and likely explain the skeletal abnormalities in patients carrying germ-line mutations of PTEN.
Keywords: Akt, bone acquisition, osteoblast survival, high bone mass
The development and maintenance of the mammalian skeleton are controlled by actions of morphogens and growth factors on bone cells. Bone formation is carried out by the osteoblast, a mesenchymal cell whose lifespan and activity are regulated by growth factor signaling networks (1, 2). Skeletal growth factors such as insulin-like growth factor-1 (IGF-1) affect osteoblast proliferation and lifespan by activating anti-apoptotic pathways, increasing cell proliferation, and influencing differentiation (3). A key control point in many anti-apoptotic pathways is the lipid kinase phosphatidylinositol (PI) 3-kinase (PI3K), which is activated in response to various extracellular signals (4, 5). On activation of growth factor receptor tyrosine kinases, a p85 regulatory subunit of PI3K is recruited to phosphorylated tyrosine residues. This action engages and activates a catalytic subunit (p110) and induces phosphorylation of the inositol ring of PI(4)P or PI(4,5)P2 at the D position to generate the second messengers PI(3,4)P2 and PI(3,4,5)P3 (4, 5). A key downstream target of PI3K and PIP3 is the serine–threonine Akt kinase family (also known as protein kinase B). PIP3 generated in the plasma membrane recruits Akt and PI-dependent kinase 1 (PDK1) through an interaction between the PI and the Akt or PDK1 pleckstrin homology (PH) domains. Once recruited to the plasma membrane, Akt is phosphorylated and activated by PDK1. Activated Akt promotes both cell growth and cell survival by regulating numerous downstream pathways including those controlled by GSK3, BAD/Bcl-XL, p53, and TSC2/mTOR (6).
The phosphatase and tensin homologue deleted on human chromosome 10 (PTEN) gene encodes a PIP phosphatase, which negatively regulates PI3K by dephosphorylating PI(3,4,5)P3. PTEN was originally cloned as a tumor suppressor for gliomas (7–9), but is deleted or inactivated in many other tumor types (10). Loss of PTEN function in either embryonic stem cells or human cancer cell lines results in an accumulation of PI(3,4,5)P3 and persistent activation of Akt, leading to increases in cell proliferation, survival, and migration (11–13). In addition, germ-line PTEN mutations are associated with Cowden disease, Bannayan–Zonana syndrome, and Lhermitte–Duclos disease, disorders characterized by hamartomas involving multiple organs (14, 15).
To directly investigate the role of Pten in osteoblasts in vivo, we disrupted the gene encoding Pten by using Cre-mediated gene replacement techniques. Mice carrying an osteoblast-specific deletion of Pten had normal body size but demonstrated progressive increases in bone volume and density throughout life. Osteoblasts from the mutant mice demonstrated a striking decreased susceptibility to apoptosis and accelerated differentiation capacity. These findings provide in vivo evidence that signaling via the PI3K/Pten pathway promotes osteoblast survival and function.
Results
Osteoblast-Specific Disruption of the PtenGene Increases Postnatal Bone Acquisition.
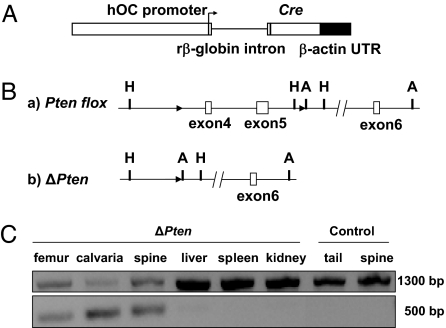
OC-CreTG/+; Ptenflox/flox mice, which were viable and fertile, were crossed with Ptenflox/flox mice to generate litters in which half of the progeny were of the OC-CreTG/+; Ptenflox/flox genotype (referred to as ΔPten), whereas the other half were control littermates (lacking the OC-Cre transgene and, therefore, wild type for Pten gene function). PCR analysis by using DNA templates from tissues of ΔPten offspring confirmed that Cre-mediated recombination occurred exclusively in bone (Fig. 1C).
Fig. 1.
Strategy for osteoblast-specific deletion of Pten. (A) Human osteocalcin (hOC) construct used to generate mouse lines expressing a Cre transgene. Cloning of this construct is described in ref. 37. The arrow indicates the transcriptional orientation. (B) Strategy for generation of a loxP-flanked allele of the Pten gene. (a) Pten-floxed allele. Exons 4 and 5 were flanked by two loxP sequences, shown as black arrowheads (A, ApaI; H, HindIII). (b) Cre-mediated deletion produces the ΔPten allele. (C) Allele-specific PCR detection of the ΔPten allele in mutant and control mice. (Upper) PCR fragments generated for the Pten-floxed allele. (Lower) Pten deletion (ΔPten) allele. Deletion of exons 4 and 5 was detected only in skeletal tissues.
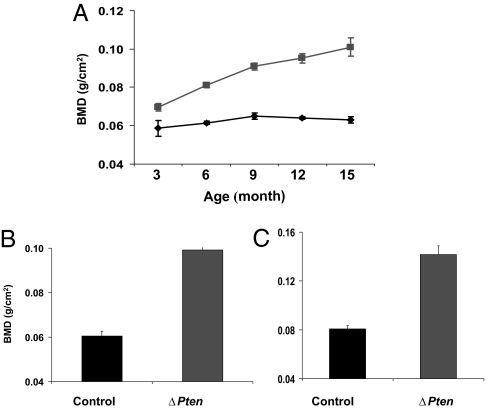
We generated cohorts of these animals and performed a longitudinal analysis of their whole-body bone mineral density (BMD) by using dual-energy x-ray absorptiometry (Fig. 2A). At 3 months of age, mice of both sexes displayed significant increases in BMD relative to their wild-type cohorts (P = 0.002 in both cases). BMD increased in mutant mice relative to controls (P = 0.002) at 6 weeks (data not shown) and increased progressively as the animals aged. By 15 months of age, female ΔPten mice had 71% higher whole-body BMD than controls (P = 2 × 10−6), whereas BMD in males increased by 60% (P = 5 × 10−5). BMD was similarly increased in both axial and peripheral skeletal sites at 12 months (Fig. 2 B and C).
Fig. 2.
Deletion of Pten leads to cumulative increases in BMD. (A) Mice carrying an osteoblast-specific deletion of Pten exhibit progressive increases in BMD with age. ■, ΔPten; ♦, wild-type cohorts (male mice shown). The error bars indicate SEM. At least five mice in each category were examined. The increase in BMD was already statistically significant (P < 0.002) at 3 months of age for both sexes (females not shown), and the differences steadily increased with age. (B and C) BMD was statistically elevated (P < 10−5) for both the lumbar spine (B) and femur (C) in 12-month-old male (shown) and female (data not shown) ΔPten mice relative to control littermates (at least five mice were evaluated for each category).
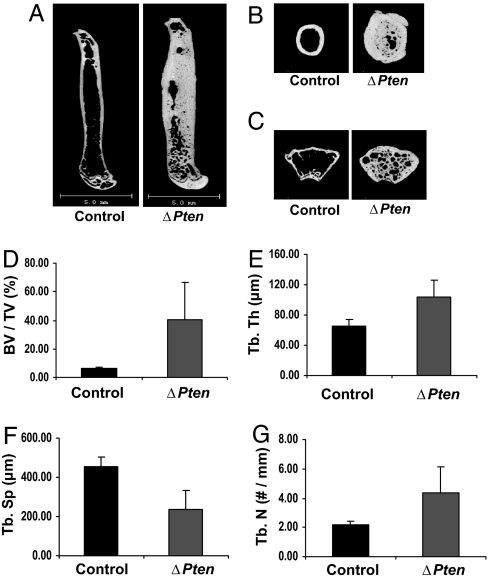
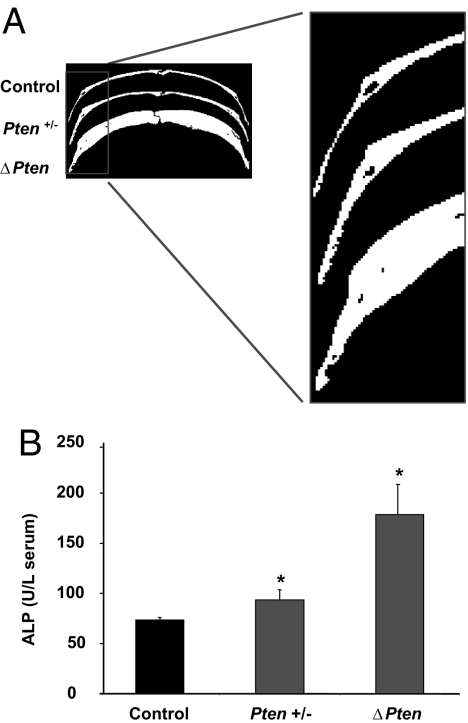
Three-dimensional micro computed tomography (microCT) scanning and histological analysis of mutant long bones showed pronounced increases in both cortical and trabecular bone. Trabeculae from mutant femurs were thicker and less separated than those of controls (Fig. 3). Cortical thickness in the femurs was increased by 43% at 3 months of age (data not shown) and by 250% (256 ± 7 vs. 676 ± 9, mean ± SE) at 12 months of age (Fig. 3). The thickness of calvarial bone was similarly increased in the mutants, suggesting that loss of Pten influences the development of bone formed through intramembranous processes (Fig. 4A). Calvaria from mice heterozygous for the conditional Pten allele (OC-CreTG/+; Ptenflox/+) were slightly but consistently thicker than those of controls (Fig. 4A). In addition, serum alkaline phosphatase levels in the heterozygous mice were intermediate between the wild-type and homozygous mutant animals (Fig. 4B). These results indicate that loss of a single Pten allele results in increased bone mass.
Fig. 3.
Disruption of the Pten gene increases bone volume. MicroCT analysis was performed on femur (A), midshaft femur (B), and distal femur (C) of ΔPten and control mice at 12 months of age. Bar graphs show the trabecular thickness (Tb. th) (P < 0.006) (D), ratio of bone volume to tissue volume (BV/TV) (E), trabecular separation (Tb. Sp) (F), and trabecular number (Tb. N) (G), respectively. Error bars represent SEM.
Fig. 4.
Loss of Pten in osteoblasts is associated with increased calvaria thickness and serum alkaline phosphatase levels. (A) MicroCT analysis was performed on the calvaria of 12-month-old control, Pten+/− (heterozygous for Pten in osteoblasts), and ΔPten mice. (B) Serum alkaline phosphatase (ALP) levels (units/liter) in control, Pten+/−, and ΔPten mice. ∗, P < 0.05. SEM is represented by the error bars.
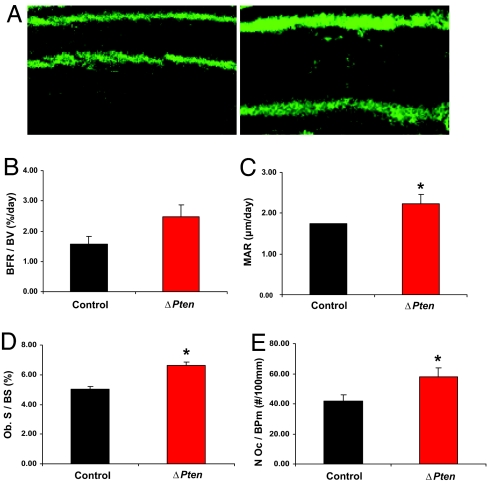
To determine the impact of the Pten mutation on bone-formation rates, dynamic histomorphometric measurements were performed on 6-week-old mice doubly labeled with sequential doses of calcein. Cancellous bone formation and mineral apposition rates were significantly increased in the ΔPten mice compared with controls (Fig. 5). In addition, the number of osteoblasts lining bone surfaces was increased, and serum osteocalcin, a marker of osteoblastic activity, was also elevated, although not to a statistically significant level (data not shown). There was no evidence of defective mineralization; osteoid volume and mineralization lag time were similar to control values (data not shown). Finally, we found no evidence for a defect in bone resorption; osteoclast numbers were increased, suggesting appropriate coupling of bone formation with resorption (Fig. 5E), and circulating osteoprotegerin levels were similar to those measured in control serum (data not shown).
Fig. 5.
Disruption of the Pten gene in osteoblasts increases bone formation rate. Six-week-old male mice were labeled with sequential doses of calcein, and dynamic indices of bone formation were quantitated in epiphyseal trabeculae of the femur of control and ΔPten mice. (A) Representative calcein-labeled sections of distal femur from control (Left) and ΔPten (Right) mice. (B) Bone formation rate/bone volume (BFR/BV). (C) Mineral apposition rate (MAR). (D) Osteoblast surface/bone surface (Ob. S/BS). (E) Osteoclast number/bone perimeter (N Oc/BPm). Black bars, control mice; red bars, ΔPten mice. ∗, P < 0.05. Error bars represent SEM.
Constitutive Activation of Akt in Osteoblasts Deficient in Pten.
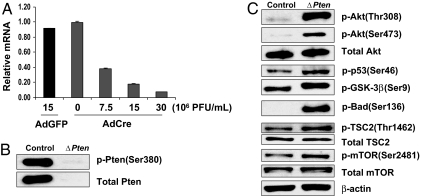
To examine the effect of Pten loss on signaling pathways downstream of Akt, we analyzed the expression of several key Akt targets after Cre-mediated disruption of the Pten gene in calvarial osteoblasts in vitro. Cells derived from mice carrying the floxed alleles were infected with adenovirus expressing the Cre recombinase (Adeno-Cre) or a control adenovirus directing the expression of green fluorescent protein (Adeno-GFP) and then cultured in the presence of β-glycerol phosphate and ascorbate (mineralizing medium). Infection with a virus concentration of 15 × 106 pfu/ml was sufficient to decrease Pten mRNA and protein to 10% of the levels observed in GFP-infected cells (Fig. 6 A and B). As shown in Fig. 6C, osteoblasts deficient in Pten had elevated levels of the phosphorylated forms of Akt, p53, GSK-3β, Bad, TSC-2, and mTOR, consistent with constitutive activation of Akt.
Fig. 6.
Disruption of Pten in osteoblasts in vitro activates the Akt pathway. (A) Establishment of Adeno-Cre viral titer for Pten gene deletion. Primary Ptenflox/floxosteoblasts were infected with the indicated number of plaque-forming units of Adeno-Cre or Adeno-GFP virus. Real-time PCR was used to measure relative Pten gene expression in primary osteoblast cell cultures from Ptenflox/flox calvaria 48 h after infection. (B) Western blot of Pten protein. Pten protein in whole-cell lysates from Ptenflox/flox primary osteoblasts was diminished 48 h after Adeno-Cre infection (ΔPten) compared with Adeno-GFP infection (control). (C) Western blots for Akt and its downstream signaling molecules. Whole-cell lysates from Ptenflox/flox primary osteoblasts were analyzed 48 h after Adeno-Cre (ΔPten) or Adeno-GFP (control) infection. The signaling molecules are phosphorylated (p-) at the residues indicated in parentheses.
Loss of Pten Is Associated with Reduced Osteoblast Apoptosis.
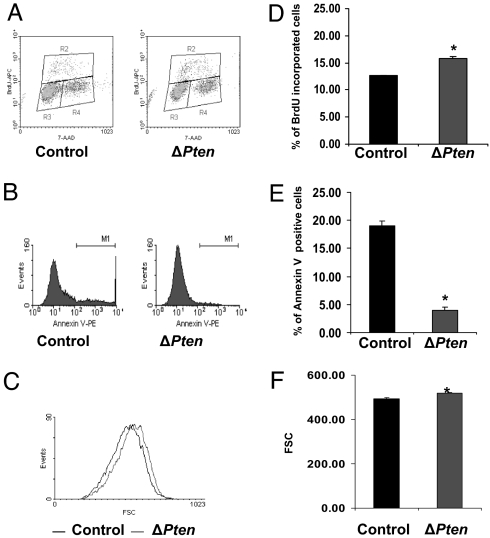
The loss of Pten in osteoblasts and increased signaling through PI3K/Akt pathways would be predicted to increase cell survival. Direct measurements of apoptosis in osteoblasts in vivo are problematic owing to the low frequency of apoptosis in this cell type (16). We therefore determined the effect of Pten deletion in vitro by measuring BrdU incorporation and annexin V staining in Pten-deficient and control primary osteoblasts by using flow cytometry. Osteoblasts deficient in Pten proliferated marginally faster than controls when grown in 10% serum (Fig. 7 A and D). By contrast, the rate of apoptosis induced by culturing in reduced serum was markedly decreased in the mutant osteoblasts relative to control cells (Fig. 7 B and E). These results are compatible with the finding of increased osteoblast numbers in the Pten mutant mice and suggest that deficiency for Pten increases osteoblast proliferation and reduces apoptosis. Pten-deficient osteoblasts also exhibited a significant increase in cell size (Fig. 7 C and F), consistent with the effects of activation of the Akt and mTOR pathways (17, 18).
Fig. 7.
Loss of Pten in osteoblasts in vitro reduces apoptosis. (A and D) BrdU incorporation. Primary osteoblasts were infected with Adeno-Cre or Adeno-GFP virus at a multiplicity of infection (moi) of 100 for 48 h. Cells were then cultured in αMEM with 1% serum for 24 h and then αMEM with 10% serum for an additional 12 h before harvesting cells for BrdU and 7-aminoactinomycin C (7-AAD) staining. BrdU was added to the medium 1 h before harvesting cells. (A) Density plot of the cell population in G0/G1 (region R3), G2/M (region R4), and S phase (region R2). (D) Percentage of BrdU incorporation (mean ± SD) in ΔPten (with Adeno-Cre) and control (with Adeno-GFP) osteoblasts (n = 3, P = 0.000315). (B and E) Annexin staining. Primary osteoblasts were infected with Adeno-Cre or Adeno-GFP virus at a moi of 100 for 48 h. Cells were then exposed to αMEM with 1% serum for 24 h and serum-free αMEM for an additional 12 h to induce apoptosis before harvesting cells for annexin V staining. Samples were analyzed by FACSCalibur immediately after staining. (B) Representative histograms of annexin V-positive cells (M1). PE, phycoerythrin. (E) Percentage of annexin V-positive cells in ΔPten (with Adeno-Cre) and control (with Adeno-GFP) osteoblasts (n = 3, P = 1.21 × 10−5). (C) Pten deletion increases forward scatter (FSC) of osteoblast cells. Pten-deficient cells are represented by the trace shifted to the right. (F) Mean FSC of control and ΔPten osteoblasts. ∗, Significantly different from control at P < 0.05.
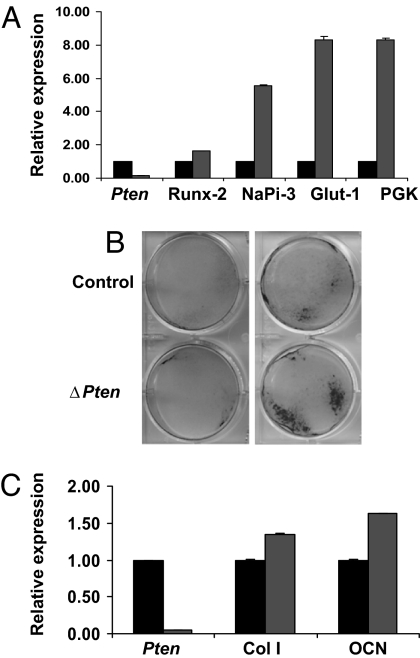
To determine the effect of loss of Pten on osteoblast differentiation, cells were differentiated in the presence of β-glycerol phosphate and ascorbate. Osteoblasts deficient in Pten showed increased expression of several genes, including Runx-2, NaPi3, Glut-1, and PGK, which in our hands are expressed early in the differentiation of primary mouse osteoblasts (Fig. 8A). At later times, disruption of Pten was associated with increased alkaline phosphatase expression (Fig. 8B) and augmented expression of gene markers for the differentiated osteoblast, including collagen I and osteocalcin (Fig. 8C). Overall, these results are compatible with the increased numbers and activity of osteoblasts seen in mice lacking Pten in vivo.
Fig. 8.
Loss of Pten accelerates osteoblast differentiation in vitro. (A and C) Effect of Pten deletion on osteoblast gene expression. Primary osteoblasts were infected with Adeno-Cre or Adeno-GFP virus at a moi of 100 for 48 h (A) or 21 days (C). Relative expression of the indicated mRNAs in osteoblasts expressing (black bars) or lacking (gray bars) Pten was determined by real-time PCR. Col I, collagen I; OCN, osteocalcin. (B) Effect of Pten deletion on alkaline phosphatase expression. Primary osteoblasts were infected with Adeno-Cre or Adeno-GFP virus at a moi of 100 and then grown in medium containing ascorbate and β-glycerol phosphate. Monolayers were stained for alkaline phosphatase at 10 (Left) and 21 days (Right) of culture. (C) Expression of Collagen I (Col I) and osteocalcin (OCN) was also enhanced in the absence of Pten.
Discussion
In this study, we investigated the role of the Pten phosphatase in bone by using Cre-mediated recombination strategies to conditionally disrupt this gene in osteoblasts. We show that loss of Pten profoundly increases bone mass at all skeletal sites by increasing the number of osteoblasts, most likely through retarding apoptosis.
The role of PTEN as a tumor suppressor is illustrated by the fact that it is frequently mutated or lost in human cancers (19–21). Homozygosity for the null mutation (Pten−/−) in mice results in early embryonic lethality, indicating that the function of Pten is critical for normal development (22–24). Pten heterozygous mice develop a spectrum of neoplasias in organs such as the breast, thyroid, endometrium, and prostate, which closely resembles the situation in humans heterozygous for a PTEN mutation (22, 25–27). The bone mass in such individuals has not been reported.
Growth factors such as insulin and IGF-1 signal through the PI3K pathway, which promotes proliferation and inhibits apoptosis via activation of Akt/PKB in a variety of cell types (28). Previous studies also support a role for this pathway in bone progenitor cell differentiation. For example, in mesenchymal stromal cells, blockade of insulin/IGF-1-activated PI3K/AKT pathways decreased bone morphogenetic protein-induced alkaline phosphatase and osteopontin expression in human mesenchymal stem cells (6). In addition, enhanced osteoblastic differentiation by forced expression of the transcription factor Runx-2 in C3H/10T1/2 mouse embryo fibroblasts was blocked by treatment with IGF-1 antibody, inhibiting Akt signaling with LY294002, or through adenoviral introduction of dominant-negative AKT. The current observations provide in vivo validation of the importance of Pten in regulating PI3K signaling in osteoblasts and suggest that sustained activation of the PI3K pathway in mature osteoblasts lacking Pten enables these cells to extend their lifespan.
The precise downstream targets of PI3K/Akt in osteoblasts are unknown. In a number of tissues, the biological response to the activation of the PI3K/Akt pathway appears to manifest in a tissue-specific fashion. For example, Pten deficiency in the prostate (29, 30) leads to the activation of FOXO3a and p70S6K pathways and enhanced proliferation, whereas in the bladder, which is less sensitive to tumor formation, Pten deficiency leads to decreased levels of p27, increased levels of p21, and decreased proliferation rates (31). Previous studies using genetic methods similar to those used here have shown that selective deletion of Pten in the liver (32) and skin (33) resulted in increased organ size. Surprisingly, however, disruption of Pten in adipocytes (34, 35) and muscle (36) increased sensitivity to insulin, but did not change the number of fat or muscle cells or alter the overall size of these organs. In the case of the adipocyte, a reduction in the level of resistin, a secretory protein that plays a key role in the development of insulin resistance, may mediate the enhanced sensitivity to insulin (36). Thus, it appears that signals generated by activation of the PI3K pathway have an impact on different functional gene pathways even among cells of the mesenchymal lineage.
In conclusion, we demonstrate that loss of Pten function in murine osteoblasts positively influences postnatal bone acquisition through constitutive activation of the PI3K pathway. We attribute this anabolic effect to a decreased rate of apoptosis, which would prolong the time each osteoblast spends in synthesizing and mineralizing bone matrix.
Materials and Methods
Generation of OC-CreTG/+; Ptenflox/flox Mice.
Mice lacking Pten in osteoblasts were generated by crossing OC-Cre mice (37, 38) with homozygous conditional mutants carrying modified Pten alleles (with loxP sites flanking exons 4 and 5) (39) to generate OC-CreTG/+; Ptenflox/flox progeny and control littermates (lacking the OC-Cre gene; Fig. 1). Mice homozygous for floxed Pten alleles were crossed to animals homozygous for the osteocalcin-Cre transgene to generate OC-CreTG/+; Ptenflox/+ mice. These mice were then crossed with Ptenflox/flox mice to generate litters that contained approximately ¼ OC-CreTG/+; Ptenflox/flox mice, which were then used for subsequent crosses. The specificity and efficiency of recombination at several loci were reported previously by using these animals (37). All experiments performed were in compliance with the guiding principles of the Guide for the Care and Use of Laboratory Animals (ref. 40, available at www.nap.edu/books/0309053773/html) and approved before use by the Institutional Animal Care and Use Committee of the Van Andel Research Institute.
Genotype Analysis.
DNA was prepared from tail biopsies by using an AutoGenprep 960 automated DNA isolation system (AutoGenM, Holliston, MA). PCR-based strategies were then used to genotype these mice (details available on request). The Cre transgene was detected by PCR (1 min at 94°C, 1 min at 53°C, and 1 min at 72°C, for 30 cycles) using the primers 5′-CAAATAGCCCTGGCAGATTC-3′ (forward) and 5′-TGATACAAGGGACATCTTCC-3′ (reverse) to generate a 260-bp product corresponding to a portion of the OC promoter and the rabbit β-globin intron. The status of the Pten locus was determined by PCR with the primers 5′-GTCACCAGGATGCTTCTGAC-3′ (forward) and 5′-GAAACGGCCTTAACGACGTAG-3′ (reverse).
Demineralized Bone Histology.
Tissue samples were fixed in formalin overnight, decalcified in Immunocal decalcifying agent (Decal, Baltimore, MD) overnight, and then dehydrated through a graded alcohol series in a Ventana Renaissance processor (Ventana Medical Systems, Tucson, AZ). Tissues were paraffin-embedded, and 5-μm sections were adhered to glass slides. Slides were deparaffinized for immunohistochemistry.
Mineralized Bone Histology.
Femurs were fixed in ethanol at room temperature, dehydrated, and embedded in methyl methacrylate. Three-micrometer sections were cut with a Microm microtome (Thornwood, NY) and stained with modified Mason–Goldner trichrome stain. The number of osteoblasts and osteoclasts per bone perimeter was measured at standardized sites under the growth plate by using a semiautomatic method (Osteoplan II; Kontron, Munich, Germany) at a magnification of ×200. These parameters comply with guidelines of the nomenclature committee of the American Society of Bone and Mineral Research (38).
Dual Energy X-Ray Absorptiometry.
Mice were anesthetized through inhalation of 2% isoflurane with oxygen (1.0 liter/min) before and during the procedure (≤5 min). The mice were placed on a specimen tray in a Lunar PIXImus II bone densitometer (GE Healthcare, Piscataway, NJ) for analysis. BMD was calculated by the PIXImus software based on the active bone area in the subcranial region within the total body image, specifically in the femur and spine. Calibrations were performed with a phantom by using known values, and quality assurance measurements were performed daily with this same phantom.
MicroCT.
High-resolution images of the femur were acquired by using a desktop microtomographic imaging system (MicroCT40; Scanco Medical, Basserdorf, Switzerland). The femur was scanned at 45 keV with an isotropic voxel size of 6 μm, and the resulting 2-dimensional cross-sectional images are shown in gray scale. Scanning was started in the mid-epiphysis and extended proximally for ≈3.6 mm (600 microCT slices).
Osteoblast Isolation and Culture.
Osteoblasts were isolated from calvaria of newborn Ptenflox/flox mice by serial digestion in αMEM (Mediatech, Herndon, VA) containing 10% BSA, 25 mM Hepes at pH 7.4, 0.2 mg/ml collagenase type I (Worthington, Lakewood, NJ), 0.7 mg/ml collagenase type 2 (Worthington), and 1 mM CaCl2. Calvaria were digested for 15 min at 37°C with constant agitation. The digestion solution was collected, washed with fresh medium, and digested an additional five times. Digestions 3–6 (containing the osteoblasts) were centrifuged, washed with αMEM containing 10% FBS and 1% penicillin/streptomycin, and cultured for 48 h at 37°C.
Adenovirus Infection.
Monolayer osteoblasts were infected with control adenovirus (Adeno-GFP) or Cre-recombinase virus (Adeno-Cre; Microbix Biosystems, Toronto, ON, Canada) at a moi of 100 for most experiments. Osteoblasts were harvested after 48 h. mRNA was extracted from osteoblasts for Pten mRNA quantitation by real-time PCR.
Osteoblast Proliferation.
After exposure to adenovirus, osteoblasts were plated on 100-mm plates (1.0 × 106 per plate) and cultured in αMEM with 1% FBS for 48 h and αMEM with 10% serum for 12 h before harvesting cells for BrdU and 7-AAD staining. BrdU was added to the medium 1 h before harvesting cells. After staining with anti-BrdU-APC and 7-AAD (BD PharMingen, San Diego, CA), cells were analyzed by FACSCalibur (Becton Dickinson, Franklin Lakes, CA), and 20,000 events were collected. Results were analyzed with WinMDI version 2.8 (The Scripps Research Institute, La Jolla, CA).
Osteoblast Apoptosis.
Osteoblasts were plated on 100-mm plates (1.0 × 106 per plate) and cultured in αMEM with 1% FBS for 48 h and serum-free αMEM for 12 h before harvesting cells for annexin V-PE (BD PharMingen) staining. After staining with annexin V-PE, cells were analyzed immediately by FACSCalibur (Becton Dickinson), and 20,000 events were collected.
Protein Extraction and Immunoblots.
Proteins were extracted from cultured osteoblasts by using M-PER (Pierce, Rockford, IL) with protease inhibitors (Pierce). The extracts were separated on SDS/10% polyacrylamide gels and transferred to nitrocellulose membranes. After probing with primary antibodies (Cell Signaling Technologies, Danvers, MA), the membranes were incubated with horseradish peroxidase-linked secondary anti-rabbit antibodies (Cell Signaling Technologies), and bound antibodies were visualized using the Supersignal West Femto Maximum Sensitivity Substrate (Pierce).
Alkaline Phosphatase Staining.
Alkaline phosphatase activity was measured in cell layers as previously described by using a p-nitrophenyl phosphate substrate and an incubation temperature of 37°C (41). Two days after adenovirus treatment, osteoblasts were replated in six-well plates with 1.0 × 105 cells per well. Cells were cultured in αMEM with 10% FBS (GIBCO, Carlsbad, CA) and supplemented with l-ascorbic acid (50 μg/ml; Sigma, St. Louis, MO) and β-glycerol phosphate (10 mM; Sigma) for 21 days. Media were changed every other day.
Real-Time PCR.
Total RNA was extracted from cells by using the TRIzol (Life Technologies, Carlsbad, CA) extraction protocol. First-strand cDNA was synthesized by using the Invitrogen kit. Real-time PCR was performed at 57°C for 30 cycles in the Opticon Continuous Fluorescent Detector by using IQ SYBR Green supermix (Bio-Rad, Hercules, CA). Samples were run in triplicate and the results were normalized to β-actin expression. Primer sequences used were as follows: Pten, F5′-CATAACCCACCACAGCTAG-3′ and R5′-GCAGACCACAAACTGAGG-3′; Runx-2, F5′-CCAAATTTGCCTAACCAGAATG-3′ and R5′-GAGGCTGTGGTTTCAAAGCAC-3′; NaPi-3, F5′-CACCCATATGGCTTCTGCTT-3′ and R5′-CAGGAATTCATAGCCCAGGA-3′; Glut-1, F5′-CAGTTTCGAGAAGAACATGAG-3′ and R5′-GC-GGAATTCAATGCTGATGAT-3′; PGK, F5′-GGAAGCG-GGTCGTGATGA-3′ and R5′-GCCTTGATCCTTTGGTTGTTTT-3′; β-actin, F5′-CTGAACCCTAAGGCCAACCGTG-3′ and R5′-GGCATACAGGGACAGCACAGCC-3′.
Statistical Analysis.
Results were expressed as mean ± SD or SEM as indicated. All statistical tests were two sided. An assigned significance level of 0.05 was used. Comparability of the two groups of data was assessed by using Student's t test.
Acknowledgments
We thank Tak Mak (University of Toronto, Toronto, ON, Canada) for providing Pten-flox mice, Pam Swiatek and the Laboratory of Germline Modification core facility for Autogen DNA processing, Bryn Eagleson and the VARI Vivarium core facility for expert animal husbandry, and Cynthia Knight and David Nadziejka for assistance in preparing the manuscript. This work was supported by National Institutes of Health Grants AR49410, AR52746, and P30 AR046031 and a Merit Review Grant from the Department of Veterans Affairs (to T.L.C.). Additional support was provided by the Van Andel Institute. We acknowledge the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for supporting these cores (Michigan Animals Models Consortium Grant 085P1000815).
Abbreviations
- 7-AAD
7-aminoactinomycin D
- BMD
bone mineral density
- IGF-1
insulin-like growth factor-1
- microCT
micro computed tomography
- moi
multiplicity of infection
- PI
phosphatidylinositol
- PDK1
PI-dependent kinase 1
- PI3K
PI 3-kinase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Mundy GR, Boyce B, Hughes D, Wright K, Bonewald L, Dallas S, Harris S, Ghosh-Choudhury N, Chen D, Dunstan C. Bone. 1995;17:71S–75S. doi: 10.1016/8756-3282(95)00182-d. [DOI] [PubMed] [Google Scholar]
- 2.Karsenty G. Matrix Biol 2000. 2000;19:85–89. doi: 10.1016/s0945-053x(00)00053-6. [DOI] [PubMed] [Google Scholar]
- 3.Nakae J, Kido Y, Accili D. Endocr Rev. 2001;22:818–835. doi: 10.1210/edrv.22.6.0452. [DOI] [PubMed] [Google Scholar]
- 4.Cantley LC. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 5.Cantley LC, Neel BG. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Downward J. Semin Cell Biol. 2004;15:177–182. doi: 10.1016/j.semcdb.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Li DM, Sun H. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 8.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R., et al. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 9.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, et al. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 10.Simpson L, Parsons R. Exp Cell Res. 2001;264:29–41. doi: 10.1006/excr.2000.5130. [DOI] [PubMed] [Google Scholar]
- 11.Liliental J, Moon SY, Lesche R, Mamillapalli R, Li D, Zheng Y, Sun H, Wu H. Curr Biol. 2000;10:401–404. doi: 10.1016/s0960-9822(00)00417-6. [DOI] [PubMed] [Google Scholar]
- 12.Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, Ruland J, Penninger JM, Siderovski DP, Mak TW. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 13.Sun H, Lesche R, Li DM, Liliental J, Zhang H, Gao J, Gavrilova N, Mueller B, Liu X, Wu H. Proc Natl Acad Sci USA. 1999;96:6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pilarski R, Eng C. J Med Genet. 2004;41:323–326. doi: 10.1136/jmg.2004.018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thiffault I, Schwartz CE, Der Kaloustian V, Foulkes WD. Am J Med Genet A. 2004;130:123–127. doi: 10.1002/ajmg.a.30335. [DOI] [PubMed] [Google Scholar]
- 16.Xing L, Boyce BF. Biochem Biophys Res Commun. 2005;328:709–720. doi: 10.1016/j.bbrc.2004.11.072. [DOI] [PubMed] [Google Scholar]
- 17.Backman S, Stambolic V, Mak T. Curr Opin Neurobiol. 2002;12:516–522. doi: 10.1016/s0959-4388(02)00354-9. [DOI] [PubMed] [Google Scholar]
- 18.Plas DR, Thompson CB. Oncogene. 2005;24:7435–7442. doi: 10.1038/sj.onc.1209097. [DOI] [PubMed] [Google Scholar]
- 19.Bose S, Chandran S, Mirocha JM, Bose N. Mod Pathol. 2006;19:238–245. doi: 10.1038/modpathol.3800525. [DOI] [PubMed] [Google Scholar]
- 20.Yan X, Fraser M, Qiu Q, Tsang BK. Gynecol Oncol. 2006;102:348–355. doi: 10.1016/j.ygyno.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 21.Davies MA, Kim SJ, Parikh NU, Dong Z, Bucana CD, Gallick GE. Clin Cancer Res. 2002;8:1904–1914. [PubMed] [Google Scholar]
- 22.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki H, Freije D, Nusskern DR, Okami K, Cairns P, Sidransky D, Isaacs WB, Bova GS. Cancer Res. 1998;58:204–209. [PubMed] [Google Scholar]
- 24.Suzuki A, Tsutomi Y, Akahane K, Araki T, Miura M. Oncogene. 1998;17:931–939. doi: 10.1038/sj.onc.1202021. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki A, de la Pompa JL, Stambolic V, Elia AJ, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W, et al. Curr Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- 26.Stambolic V, Tsao MS, MacPherson D, Suzuki A, Chapman WB, Mak TW. Cancer Res. 2000;60:3605–3611. [PubMed] [Google Scholar]
- 27.Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, Yamada KM, Cordon-Cardo C, Catoretti G, Fisher PE, Parsons R. Proc Natl Acad Sci USA. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine AJ, Feng Z, Mak TW, You H, Jin S. Genes Dev. 2006;20:267–275. doi: 10.1101/gad.1363206. [DOI] [PubMed] [Google Scholar]
- 29.Grunwald V, DeGraffenried L, Russel D, Friedrichs WE, Ray RB, Hidalgo M. Cancer Res. 2002;62:6141–6145. [PubMed] [Google Scholar]
- 30.Trotman LC, Alimonti A, Scaglioni PP, Koutcher JA, Cordon-Cardo C, Pandolfi PP. Nature. 2006;441:523–527. doi: 10.1038/nature04809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fradet Y, Lacombe L. Curr Opin Urol. 2000;10:441–445. doi: 10.1097/00042307-200009000-00013. [DOI] [PubMed] [Google Scholar]
- 32.Stiles B, Wang Y, Stahl A, Bassilian S, Lee WP, Kim YJ, Sherwin R, Devaskar S, Lesche R, Magnuson MA, Wu H. Proc Natl Acad Sci USA. 2004;101:2082–2087. doi: 10.1073/pnas.0308617100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Backman SA, Ghazarian D, So K, Sanchez O, Wagner KU, Hennighausen L, Suzuki A, Tsao MS, Chapman WB, Stambolic V, Mak TW. Proc Natl Acad Sci USA. 2004;101:1725–1730. doi: 10.1073/pnas.0308217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakashima N, Sharma PM, Imamura T, Bookstein R, Olefsky JM. J Biol Chem. 2000;275:12889–12895. doi: 10.1074/jbc.275.17.12889. [DOI] [PubMed] [Google Scholar]
- 35.Tang X, Powelka AM, Soriano NA, Czech MP, Guilherme A. J Biol Chem. 2005;280:22523–22529. doi: 10.1074/jbc.M501949200. [DOI] [PubMed] [Google Scholar]
- 36.Wijesekara N, Konrad D, Eweida M, Jefferies C, Liadis N, Giacca A, Crackower M, Suzuki A, Mak TW, Kahn CR, et al. Mol Cell Biol. 2005;25:1135–1145. doi: 10.1128/MCB.25.3.1135-1145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang M, Xuan S, Bouxsein ML, von Stechow D, Akeno N, Faugere MC, Malluche H, Zhao G, Rosen CJ, Efstratiadis A, Clemens TL. J Biol Chem. 2002;277:44005–44012. doi: 10.1074/jbc.M208265200. [DOI] [PubMed] [Google Scholar]
- 38.Holmen SL, Zylstra CR, Mukherjee A, Sigler RE, Faugere MC, Bouxsein ML, Deng L, Clemens TL, Williams BO. J Biol Chem. 2005;280:21162–21168. doi: 10.1074/jbc.M501900200. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki A, Yamaguchi MT, Ohteki T, Sasaki T, Kaisho T, Kimura Y, Yoshida R, Wakeham A, Higuchi T, Fukumoto M, et al. Immunity. 2001;14:523–534. doi: 10.1016/s1074-7613(01)00134-0. [DOI] [PubMed] [Google Scholar]
- Institute of Laboratory Animal Resources. Guide for the Care and Use of Laboratory Animals. Washington, DC: Natl Acad Press; 1996. [Google Scholar]
- 41.Manolagas SC, Burton DW, Deftos LJ. J Biol Chem. 1981;256:7115–7117. [PubMed] [Google Scholar]