Abstract
Understanding the connections among genotype, phenotype, and fitness through evolutionary time is a central goal of evolutionary genetics. Wrinkly spreader (WS) genotypes evolve repeatedly in model Pseudomonas populations and show substantial morphological and fitness differences. Previous work identified genes contributing to the evolutionary success of WS, in particular the di-guanylate cyclase response regulator, WspR. Here we scrutinize the Wsp signal transduction pathway of which WspR is the primary output component. The pathway has the hallmarks of a chemosensory pathway and genetic analyses show that regulation and function of Wsp is analogous to the Che chemotaxis pathway from Escherichia coli. Of significance is the methyltransferase (WspC) and methylesterase (WspF) whose opposing activities form an integral feedback loop that controls the activity of the kinase (WspE). Deductions based on the regulatory model suggested that mutations within wspF were a likely cause of WS. Analyses of independent WS genotypes revealed numerous simple mutations in this single open reading frame. Remarkably, different mutations have different phenotypic and fitness effects. We suggest that the negative feedback loop inherent in Wsp regulation allows the pathway to be tuned by mutation in a rheostat-like manner.
UNDERSTANDING the relationship among DNA sequence, phenotype, and fitness through evolutionary time is a central goal of evolutionary genetics. Theoretical approaches, building on ideas first put forward by Fisher (1930) and Maynard Smith (1970), have led to a definition of the statistical properties expected to characterize the distribution of fitness effects among new mutations exposed to—and fixed by—selection (reviewed in Orr 2005b). However, these models say little about the specific kinds of mutational changes, in terms of the genetic or phenotypic basis of adaptive traits, expected in evolving populations. What is required is an understanding of the relationship (if any) between the underlying genetic architecture of an organism and the ability of the changes at the DNA sequence level to translate through to phenotypically useful solutions (Pepin et al. 2006).
Our work has focused on the ecological and genetic causes of diversity in simple laboratory populations of Pseudomonas fluorescens (strain SBW25). When propagated in spatially structured microcosms, populations of P. fluorescens rapidly diversify, giving rise to a range of niche specialist genotypes (Rainey and Travisano 1998). One common class of niche specialist colonizes the air–liquid interface of static broth microcosms and produces colonies with a characteristic wrinkled morphology on agar plates: these mutants are referred to as “wrinkly spreaders” (WS). WS morphs (of which there are many morphologically distinct types) arise by spontaneous mutation from the ancestral genotype and are substituted in the population because they enjoy a fitness advantage when rare. Their ability to colonize the air–liquid interface stems from the formation of a self-supporting mat that is a product of the combined (cooperative) activities of the individual cells (Rainey and Rainey 2003).
A single WS genotype, the large spreading WS (LSWS), has been the focus of genetic analysis. Suppressor studies have identified numerous genes that contribute either directly or indirectly to mat formation (Spiers et al. 2002; Gehrig 2005), but two loci are of special significance: wss and wrinkly spreader (wsp)—transposon insertions throughout either locus abolish the wrinkled phenotype, niche specialization, and associated traits characteristic of LSWS (Spiers et al. 2002; Goymer et al. 2006).
The wss locus encodes proteins that together produce an acetylated cellulose polymer (Spiers et al. 2002, 2003). In LSWS, the wss-encoded enzymes are constitutively active as a consequence of overproduction of bis-(3′-5-)-cyclic dimeric guanosine monophosphate (c-di-GMP), an allosteric activator of cellulose (Ross et al. 1987; Goymer et al. 2006; Malone et al. 2007). Overproduction of c-di-GMP results in constitutive production of polymer, which, in conjunction with a similarly overproduced proteinaceous adhesin (Spiers et al. 2003), causes daughter cells to remain attached after cell division. The cellulosic polymer thus functions as a cell–cell glue and is the cause of cooperation among cells that form the WS mat (Spiers et al. 2002; Rainey and Rainey 2003).
The proximate cause of excess c-di-GMP in LSWS is wsp, a seven-gene operon that encodes a chemosensory pathway, although, more specifically, the cause is constitutive phosphorylation of the di-guanylate cyclase (DGC) response regulator WspR, the output component of the wsp pathway. When phosphorylated (at Asp67), the C-terminal DGC domain of WspR becomes functionally active and this leads to the joining of two molecules of GTP in a head-to-tail manner, resulting in the formation of c-di-GMP (Malone et al. 2007).
Here we define and genetically characterize the Wsp operon, predict mutational targets, and show that the cause of LSWS is a transversion in wspF. Analysis of wspF nucleotide sequence from another 26 independent WS genotypes showed that 13 harbor simple mutations. Remarkably, different mutations have different phenotypic and fitness effects. We suggest that these different effects are a consequence of the negative feedback loop inherent in Wsp regulation that allows the pathway to realize multiple output states.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and manipulation:
The ancestral (wild-type) “smooth” (SM) strain is P. fluorescens SBW25 and was isolated from the leaf of a sugar beet plant grown at the University Farm, Wytham, Oxford, in 1989 (Rainey and Bailey 1996). The ancestral strain was placed at −80° immediately after isolation to minimize lab adaptation. The niche-specialist wrinkly spreader (LSWS) genotype (PR1200) was derived from the ancestral genotype following 3 days of selection in a spatially structured microcosm (Spiers et al. 2002). PR3522 (LSWSΔpanB) is a pantothenate requiring an auxotroph of LSWS (PR1200) and contains a clean deletion of the entire panB gene (Rainey 1999). The mutation is neutral in a pantothenate replete environment (Spiers et al. 2002). The WS genotypes WSA–WSZ are 26 independent WS genotypes obtained by propagating the ancestral SM genotype in 26 separate microcosms. After 5 days of selection, populations were diluted and plated onto KB agar, and a single WS genotype was selected at random from each microcosm. These genotypes were streaked onto a single colony to check purity and then stored at −80°.
P. fluorescens strains were cultured at 28° in Luria–Bertani (LB) (Miller 1972), Pseudomonas agar F (Difco), or King's medium B (KB) (King et al. 1954). Escherichia coli DH5a, XL1-Blue MRF′ (Stratagene, La Jolla, CA), and S17-1lpir (Simon et al. 1983) were cultured at 37° in LB. Antibiotics were used at the following concentrations: ampicillin 100 μg ml−1, kanamycin 75 μg ml−1 (50 μg ml−1 for E. coli), and tetracycline 25 μg ml−1. Pantothenate was added to media at a final concentration of 0.24% (w/v) where required. CFC (Oxoid) was added to media to select for P. fluorescens strains following conjugation. Congo Red was used at a final concentration of 0.001% (w/v) in KB or LB agar without NaCl (NaCl causes Congo Red to precipitate). Calcofluor [Sigma (St. Louis) Fluorescent Brightener 28] was used in LB at 1–10 μm and cells or the polymer were incubated with the stain for 30 min before examination by fluorescent microscopy.
Plasmid DNA was introduced into E. coli and P. fluorescens by transformation (electroporation) or conjugation using standard procedures. The helper plasmid pRK2013 (Figurski and Helinski 1979) was used to facilitate transfer of plasmids between E. coli and P. fluorescens.
Molecular biology techniques:
Plasmid DNA was isolated from E. coli using QIAprep plasmid kits (QIAGEN, Chatsworth, CA). Standard agarose gel electrophoresis and other recombinant DNA techniques were performed according to Sambrook et al. (1989). DNA fragments were recovered from agarose using a QIAEX II gel extraction kit (QIAGEN). pSK+ (pBluescriptII SK+, Stratagene), pCR2.1 (Invitrogen, San Diego), and pVSP61 (Loper and Lindow 1994) were used for cloning and overexpression of wsp genes. Allelic exchange and the construction of chromosomally integrated ′lacZ operon fusions were performed using the suicide reporter plasmid pUIC3 (Rainey 1999). DNA sequencing was carried out on an ABI310 (Perkin-Elmer, Norwalk, CT) automated sequencer. Oligonucleotide primers used for sequencing, cloning, reverse transcriptase–PCR (RT–PCR), and construction of wspA-F, wspC, wspF, and wspR deletion strains are available upon request and are based upon the available SBW25 genome sequence (Sanger Centre).
Construction of deletion mutants, allelic replacements, and lacZ fusions:
Deletion mutants were constructed as described previously (Rainey 1999). First, ∼500 bp of DNA flanking either side of the gene (or genes) to be deleted were spliced together by PCR (Horton et al. 1989). The spliced DNA was cloned into pBluescript, from where it was sequenced to check for errors. The spliced fragment was then excised as a BglII–SpeI fragment and ligated into the suicide vector pUIC-3 (Rainey 1999). To generate wspF allelic replacements, PCR fragments of 3 kb centered on wspF were amplified from chromosomal DNA using wsp-specific primers and cloned into pCR2.1 (Invitrogen). wspF plasmids were reintroduced into P. fluorescens by electroporation and allowed to recombine with the chromosome by homologous recombination. Recombinants from which pCR2.1 had been lost were identified from LB agar plates after a brief period of nonselective growth in KB broth. The change in wspF sequence was confirmed by DNA sequencing. Chromosomal fusions between the last gene of the wsp operon (wspR) and ′lacZ were generated by cloning the entire wspR gene as a BglII–SpeI fragment into pUIC3. The resulting plasmid was introduced into the genome by conjugation, and integration by a single homologous recombination event was ensured by selection for plasmid-encoded tetracycline resistance. Correct placement of the fusion was checked by PCR.
Fitness of genotypes:
Competitive fitness of WS genotypes was determined by direct competition between each WS genotype and PR3522 (a panB deletion derivative of LSWS) and was performed in the presence of the ancestral SM genotype (the presence of the ancestral genotype ensured that the broth phase was colonized and this led to improved reproducibility of fitness measures). All strains were grown overnight in shaken KB broth (supplemented with pantothenate where appropriate) and competitors were introduced into spatially structured KB microcosms (∼105 cells of each competitor) at a ratio of 1:1:1 (SM:LSWS ΔpanB:WSi). Relative fitness was calculated as the ratio of the Malthusian parameters of the two WS genotypes being compared (Lenski et al. 1991) and cell counts were determined after 24 hr of competition. The pantothenate-marked gentoype was distinguished from WSi by plating cells on vitamin-free KB agar supplemented with 4.8 × 10−6% pantothenic acid (on this medium the pantothenate-marked strain is readily distinguished by its greatly reduced size). All cultures were whirli-mixed for 60 sec before dilution plating to maximally disperse clumped cells.
RESULTS
Defining the Wsp chemosensory pathway:
The wsp locus spans ∼8.5 kb and is composed of seven open reading frames designated wspABCDEFR. Analysis of whole-genome sequence data shows that wsp is a highly conserved feature of all Pseudomonas genomes (Stover et al. 2000; Nelson et al. 2002; Feil et al. 2005; Joardar et al. 2005; Paulsen et al. 2005): it is also resident on plasmid pMLb from Mezorhizobium loti (Kaneko et al. 2000). While effects of disrupting Wsp function have been observed in P. aeruginosa (D'Argenio et al. 2002; Hickman et al. 2005) and P. fluorescens (Spiers et al. 2002), the locus has received little direct characterization: in lieu of a genetic model of Wsp regulation, we briefly describe the salient features of each predicted protein (Table 1).
TABLE 1.
Predicted function and domain characteristics of Wsp proteins
| Protein | Sizea | Predicted function | Pfam domain | E-value |
|---|---|---|---|---|
| WspA | 547 | MCP | HAMP | 2 × 10−16 |
| MCP | 8.6 × 10−27 | |||
| WspB | 170 | Scaffold protein | CheW | 7.2 × 10−27 |
| WspC | 419 | Methyltransferase | CheR | 6.7 × 10−33 |
| WspD | 232 | Scaffold protein | CheW | 4.5 × 10−26 |
| WspE | 755 | Histidine kinase/response regulator | Hpt | 2.0 × 10−12 |
| HATPase_c | 9.1 × 10−22 | |||
| CheW module | 3.3 × 10−24 | |||
| Response_reg | 9.1 × 10−33 | |||
| WspF | 336 | Methylesterase | Response_reg | 2.9 × 10−19 |
| CheB_methylest | 2.7 × 10−57 | |||
| WspR | 333 | Di-guanylate cyclase/response regulator | Response_reg | 5.8 × 10−17 |
| GGDEF | 8.5 × 10−68 |
Number of amino acids.
Predicted proteins of the Wsp locus:
WspA (AAO92333) resembles a membrane-associated methyl-accepting chemotaxis protein (MCP). It contains the functional domains of a typical chemotactic transducer (Falke et al. 1997), including an N-terminal (periplasmic) sensory ligand-binding region flanked by two membrane-spanning regions; a C-terminal cytoplasmic domain including the conserved signaling region and methylation sites; a C-terminal domain composed of two subdomains—the conserved HAMP domain involved in regulation of receptor methylation (Chervitz and Falke 1996); and the conserved methyl-accepting domain containing MCP methylation sites (Le Moual and Koshland 1996).
WspB (AAO92334) and WspD (AA092336) are predicted scaffold proteins similar to CheW from E. coli (Levit et al. 2002). WspD has, in addition to a CheW domain, another 60 amino acids at the N terminus.
WspC (AA092335) contains a conserved N-terminal CheR domain and is a probable methyltransferase (Djordjevic and Stock 1997). Unlike CheR, WspC contains an additional 160 residues downstream of the CheR domain, including amino acid sequences that define a tetratrico peptide repeat region. The methyltransferase homolog FrzF of Myxococcus xanthus (McCleary et al. 1990) shows a similar arrangement.
WspE (AA092337) contains regions homologous to both the CheA histidine kinase domain and the CheY response regulator domain. Overall, WspE shows the greatest structural homology to the FrzE hybrid protein of M. xanthus (McCleary and Zusman 1990). The CheA-like domain of WspE comprises three distinct modules: the Hpt module, which contains key residues involved in autophosphorylation of the kinase; the conserved catalytic module responsible for kinase activity; and a module essential for receptor-mediated regulation (which contains the CheW and receptor-binding sites).
WspF (AA092338) has two predicted domains: an N-terminal CheY response regulator domain (Stock et al. 2000) and a CheB methylesterase domain. Overall, it is similar to CheB from E. coli (see supplemental Figure 3 at http://www.genetics.org/supplemental/), which is an esterase that hydrolyzes the methyl esters formed by CheR and restores negatively charged glutamate residues on MCPs (West et al. 1995). The N-terminal response regulator receiver domain contains the major active-site aspartate residues (Asp-10, Asp-11, and Asp-56—the phosphorylation site) (Stock and Surette 1996; Stock et al. 2000). The C-terminal methylesterase domain contains the major methylesterase active sites (Ser-164, His-190, and Asp-286) (West et al. 1995).
WspR (AAL71852) is a DGC response regulator and has been described in detail elsewhere (see Goymer et al. 2006). Like WspE and WspF, it also contains (at its N terminus) a CheY response regulator domain; the C-terminal domain is a DGC whose function is dependent upon phosphorylation of Asp67 (Goymer et al. 2006).
The seven wsp genes form a single transcriptional unit:
The first six genes of the wsp locus are translationally coupled (the stop codon of the preceding gene overlaps the predicted start codon of the following gene), indicating that wspA-F form a single transcriptional unit; however, a 50-nucleotide gap separates the seventh gene wspR from wspF, suggesting the possibility that wspR is a separate transcriptional unit. An RT–PCR strategy was used to test this hypothesis; specifically, mRNA was extracted from the ancestral genotype and cDNA was generated by RT–PCR using primers flanking the 3′-end of wspF and the 5′-start of wspR (see supplemental Figure 1 at http://www.genetics.org/supplemental/ for details). A series of test and control PCR reactions were then performed to determine whether cDNA corresponding to the sequence between wspF and wspR had been generated (as expected if wspF and wspR were transcribed as a single mRNA). A 0.5-kb fragment was obtained using primers that flank the wspF–wspR intergenic region, demonstrating that wspR is part of the same transcriptional unit as wspF. DNA sequence of this fragment confirmed its identity.
Transcription of wsp is similar in SM and LSWS:
It was not inconceivable that the cause of enhanced WspR activity in LSWS (vs. SM) was a mutation in the wsp promoter that caused enhanced transcription of the operon. A promoterless lacZ fusion was therefore made to wspR (without affecting the wspR open reading frame) and integrated into the genome of both the ancestral SM and derived LSWS genotypes. β-Galactosidase activity was determined in King's medium B after 4, 12, 24, and 48 hr of growth. The wsp operon was transcriptionally active at all stages of growth and, while expression was slightly lower in the ancestral genotype, typically the difference was less than twofold (supplemental Figure 2 at http://www.genetics.org/supplemental/). Such minor differences were not considered significant and the possibility that the cause of LSWS was a mutational change in the promoter region was not further investigated.
The Wsp pathway donates phosphoryl groups to WspR:
Previous analysis of WspR variants [generated by in vitro means (Goymer et al. 2006)] showed (1) that phosphorylation of WspR was necessary for development of the WS phenotype, (2) that in LSWS WspR was constitutively phosphorylated, and (3) that the source of phosphoryl groups in LSWS was “upstream” of WspR. The latter finding stemmed from the analysis of WspR variants with dominant-negative effects, that is, variants that, when overexpressed in LSWS, caused development of the ancestral SM phenotype. Sequence (and genetic) analysis showed that these variants harbored single nonsynonymous amino acid substitutions in the C-terminal DGC domain. The dominant-negative effect was thus attributed to competitive inhibition: variant forms with inactive DGC domains (being in excess) “soaked up” phosphoryl groups that were otherwise destined to activate the DGC activity of the chromosomally encoded WspR. This result indicated that one effect of the mutation(s) responsible for LSWS was overstimulation of a kinase (Goymer et al. 2006). The most likely candidate is WspE, but cross talk among two-component sensing systems meant that kinases beyond the Wsp operon might also be involved.
To test the hypothesis that the Wsp pathway is the source of phosphoryl groups, the first six genes of the wsp operon were deleted from the LSWS genotype. The deletion was constructed in such a way as to ensure minimal disruption to wspR; indeed, the ATG start codon of the first gene of the wsp operon (wspA) became the start codon of wspR so that wspR remained fully under the control of the wsp promoter. The resulting mutant expressed the ancestral smooth morphology, was unable to colonize the air-liquid interface of static broth microcosms, and no longer produced detectable amounts of cellulose as determined by both Congo Red assay and fluorescent microscopy of calcofluor-stained cells. This result confirms Wsp as the source of phosphoryl groups.
Regulation of Wsp:
The discovery that Wsp is the source of phosphoryl groups led to questions regarding Wsp function and regulation. As demonstrated above (Table 1), the Wsp operon bears substantial similarity to the chemotaxis (Che) operon from E. coli: as such, Che provides the basis of a working model.
Che is a complex two-component signal transduction system found in almost all motile bacteria: it has been extensively investigated and reviewed (Stock and Surette 1996; Falke et al. 1997; Stock et al. 2000; Baker et al. 2006). Its primary function is to sense and process environmental stimuli in such a way as to effect change in the direction of rotation of the flagella motor. Central to the pathway are two proteins: CheA (histidine kinase) and CheY (response regulator). CheA catalyzes the transfer of phosphoryl groups from ATP to a conserved histidine residue on the kinase, while CheY catalyzes the transfer of the phosphoryl group from the kinase to a conserved aspartate residue on the response regulator. When phosphorylated, CheY associates with the flagella motor, causing a reversal in the direction of rotation of the flagella bundle. Activity of CheY is also affected by CheZ, a CheY-specific phosphatase (there is no homolog of CheZ in the Wsp pathway). CheA does not act alone, but forms a membrane-associated receptor complex in conjunction with MCPs and the scaffold protein CheW; the receptor complex integrates sensory information to control the level of CheA kinase activity.
Two additional proteins play a critical role in regulating kinase activity: CheR and CheB. Even in an environment devoid of stimulatory ligands, the kinase switches between active and inactive states about once every second due to the opposing activities of CheR and CheB. CheR constantly adds methyl groups to glutamate residues on the cytoplasmic domain of the MCP to the point that the kinase becomes activated; phosphoryl groups pass to CheY (which dissociates from the signaling complex to interact with the flagella motor) and also to CheB. Upon activation, CheB removes methyl groups and thus resets the kinase to an inactive state.
Given that the Wsp operon has proteins with apparent functional equivalence to Che, a model for Wsp function and regulation was developed (Figure 1). Initial attempts to test the model by genetic complementation of a set of defined E. coli che mutants (cheA, cheW, cheR, cheB, and cheY) using predicted homologs from the Wsp pathway (wspE, wspB/D, wspC, wspF, and wspR) met with minimal success (Bantinaki 2002).
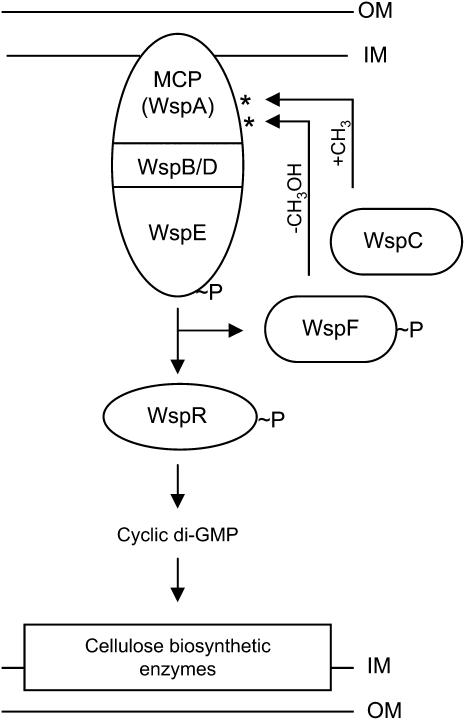
Figure 1.—
Model for the regulation and function of the Wsp pathway based on the Che (chemotaxis) pathway of enteric bacteria. The MCP (WspA), scaffold proteins (WspB and WspD), and kinase (WspE) form a membrane-bound receptor-signaling complex (IM, inner membrane; OM, outer membrane). WspR is a DGC response regulator and the primary output of the pathway. When phosphorylated, WspR catalyzes the synthesis of cyclic-di-GMP, an allosteric activator of cellulose biosynthetic enzymes. The level of kinase activity is controlled by the opposing activities of methyltransferase (WspC) and methylesterase (WspF), which add and remove (respectively) methyl groups from conserved glutamate residues on the signaling domain of the MCP (asterisks). WspC is constitutive, whereas WspF becomes active upon receipt of phosphoryl groups from the kinase. Addition of methyl groups to glutamate residues on the MCP by WspC causes the kinase to autophosphorylate and results in activation of WspF (and WspR); phosphorylated WspF removes methyl groups and thus resets the WspE kinase to an inactive state. As in the Che pathway of E. coli, the model presented here predicts that Wsp continually oscillates between active and inactive states. According to the model, a mutation in WspF that decreases WspF function (or removes functionality altogether) will result in constitutive activation of the kinase, constitutive activation of WspR, overproduction of c-di-GMP, and overproduction of cellulose and other adhesive polymers, resulting in expression of the WS phenotype.
An alternative approach was to test directly the regulatory model by genetic manipulation of the component parts. If the model is correct (and the individual components are functionally equivalent to their Che counterparts), then activity of the output component, WspR, should be dependent upon the activity of the kinase (WspE), which should ultimately be dependent upon the relative activities of the methyltransferase (WspC) and the methylesterase (WspF). It follows, then, that an increase in the level of expression of WspC relative to WspF (but not the reverse) in the ancestral SM genotype should generate the WS colony morphology because an increase in WspC activity relative to WspF will cause methylation of glutamate residues on the MCP to rise to an irreversibly high level, resulting in constitutive activation of the kinase, overactivation of WspR, production of excess c-di-GMP, overactivation of the cellulose biosynthetic enzymes, and a switch in colony morphology from SM to WS (with concomitant effects on cellulose biosynthesis and niche specialization). Conversely, an increase in the level of expression of WspF (relative to WspC) in the derived LSWS genotype should cause development of the ancestral smooth morphology because an increase in WspF activity relative to WspC (but not the reverse) will cause modified glutamate residues on the MCP to be hydrolyzed, leading to a negligible number of modified glutamate residues and thus inactivation of the WspE kinase, inactivation of WspR, no synthesis of c-di-GMP, inactivation of cellulose biosynthetic enzymes, and a switch in colony morphology from LSWS to SM (with concomitant effects on cellulose biosynthesis and niche specialization).
To test these predictions, wspC and wspF were deleted from both ancestral SM and derived LSWS backgrounds. In addition, both genes were overexpressed (separately) from the constitutive lac promoter of plasmid pVSP61—in both ancestral SM and LSWS backgrounds. The results, shown in Table 2, are fully consistent with predictions of the model: deletion of wspF and overexpression of wspC in the ancestral SM background caused development of the wrinkled colony morphology (no effect on colony morphology occurred when wspC was deleted or wspF was overexpressed). Conversely, the wrinkled morphology of LSWS converted to smooth when wspF was overexpressed and the same conversion was evident upon deletion of wspC (no phenotypic changes were observed when wspF was deleted or when wspC was overexpressed). By way of positive controls, the output component WspR was deleted from SM and derived LSWS and overexpressed in the same two genotypes, and results were consistent with previous findings, namely, that deletion of the wspR in LSWS causes development of SM and that overexpression of wspR in SM causes development of the wrinkled morphology typical of WS (Goymer et al. 2006). The kinase WspE was also cloned and overexpressed on the basis that overexpression might result in an increase in spontaneous autophosphorylation and thus cause SM to convert to WS; however, this did not occur. Interestingly, however, overexpression of wspE in LSWS caused reversion to the SM phenotype. This effect is likely to be attributable to a disruption of the receptor-signaling complex due to an imbalance between the various components.
TABLE 2.
Effect of manipulation of components of the Wsp pathway on colony morphology of ancestral SM and derived LSWS genotypes
| Genotype and colony morphology of initial genotype | Genetic manipulationa | Effect on colony morphology |
|---|---|---|
| SM | ΔwspC | SM |
| SM | ΔwspF | WSb |
| SM | ΔwspR | SM |
| SM | pVSP61-wspC | WSb |
| SM | pVSP61-wspF | SM |
| SM | pVSP61-wspR | WSb |
| SM | pVSP61-wspE | SM |
| LSWS | ΔwspC | SMb |
| LSWS | ΔwspF | WS |
| LSWS | ΔwspR | SMb |
| LSWS | pVSP61-wspC | WS |
| LSWS | pVSP61-wspF | SMb |
| LSWS | pVSP61-wspR | WS |
| LSWS | pVSP61-wspE | SMb |
Manipulation involved either deletion of a gene (e.g., ΔwspC) or overexpression of the gene on plasmid pVSP61 (e.g., pVSP61-wspC).
Conversion of phenotype.
Predicting the mutational cause of LSWS:
The model established for signal transduction through the Wsp pathway allowed for predicting the likely mutational causes of the LSWS genotype. From the very beginnings of the genetic analysis of WS it had been difficult to reconcile the genetic data on the origins of the WS phenotype with the repeatable occurrence of WS mutants in static broth microcosms. The genetic data are consistent with the WS phenotype resulting from a “gain of function” (Spiers et al. 2002), yet the relative ease by which WS mutants arise (estimated at ∼5 × 10−7) suggests that the underlying mutation is a loss of function. The model outlined in Figure 1 (and tested above) indicates that WS genotypes could indeed arise by a loss-of-function mutation: a mutation that decreased or abolished the function of the methylesterase WspF would result in overstimulation of the WspE kinase (due to ever-increasing levels of glutamate modification caused by WspC), with ensuing positive effects on WspR activity, c-di-GMP production, and production of adhesive polymers. Conversely, while mutations causing WS could arise within the methyltransferase WspC—or within WspR itself—that increase activity of either component, these would have to be gain-of-function (or at least increase-in-activity) mutations and thus should be rare. To test these predictions, the nucleotide sequence was obtained from wspF, wspC, and wspR from both SM and the derived LSWS genotype. No change was found in wspC or wspR; however, a single nucleotide change (A901C) (causing a nonsynonymous amino acid change: S301R) was found in wspF of LSWS.
Many independent mutations occur within wspF:
Independent populations evolving in identical environments often show parallel genetic changes and the fact that they do is a strong argument for the changes being adaptive (Cooper et al. 2001, 2003; Zhong et al. 2004; Colosimo et al. 2005; Woods et al. 2006). A collection of 26 independently obtained WS was thus analyzed for changes to the nucleotide sequence of wspF. The results presented in Table 3 show that 13 of the 26 WS genotypes contained simple mutations (transitions, transversions, or short deletions) in wspF; WSN harbored the same genetic change as was found in LSWS (wspF A901C), but all other mutations were unique. A careful analysis of the nucleotide sequence of wspF failed to reveal any evidence of mutable nucleotide tracts, such as repeats or insertion elements. For those WS genotypes that did not contain a mutant wspF allele, the nucleotide sequence was obtained from wspC and wspR, but no changes in these genes were detected.
TABLE 3.
wspF mutations from independent WS genotypes
| WS genotypea | Nucleotide change | Nature of mutation | Amino acid change | Predicted effect on function and groupb |
|---|---|---|---|---|
| LSWS | A901C | Transition | S301R | Reduced activity (2); mutation close to active site |
| WSA | T14G | Transversion | I5S | Reduced activity (2); mutation close to active site |
| WSB | Δ620-674 | Deletion | P206Δ(8)c | No activity (3); C-terminal third of protein lost |
| WSC | G823T | Transversion | G275C | Reduced activity (2); mutation close to active site |
| WSE | G658T | Transversion | V220L | Reduced activity (2) |
| WSF | C821T | Transversion | T274I | Reduced activity (2); mutation close to active site |
| WSG | C556T | Transversion | H186Y | No activity (3); catalytic residue altered |
| WSJ | Δ865-868 | Deletion | R288Δ(3)c | Reduced activity (2); most of protein remains |
| WSL | G482A | Transition | G161D | Reduced activity (2) |
| WSN | A901C | Transition | S301R | Reduced activity (2); same mutation as in LSWS |
| WSO | Δ235-249 | Deletion | V79Δ(6)c | No activity (3); C-terminal two-thirds lost |
| WSU | Δ823-824 | Deletion | T274Δ(13)c | No activity (3); Asp286 active site residue is lost |
| WSW | Δ149 | Deletion | L49Δ(1)c | No activity (3); most of protein lost |
| WSY | Δ166-180 | Deletion (in frame) | Δ(L51-I55) | Reduced activity (2); small in-frame deletion of five residues |
No wspF (wspC or wspR) mutation was detected in WSD, WSH, WSI, WSK, WSM, WSP, WSQ, WSR, WSS, WST, WSV, WSX, and WSZ.
Defects caused by each mutation were scored on the basis of the likely magnitude of the effect (see text): 1, wild-type wspF (13 isolates); 2, reduced activity (9 isolates); 3, no activity (5 isolates).
P206Δ(8) indicates a frameshift; the number of new residues before a stop codon is reached is in parentheses.
Substantial structure–function analyses of the homologous methylesterase CheB (West et al. 1995; Stock and Surette 1996; Djordjevic et al. 1998) have defined domain structure, active sites, and catalytic residues (see above and supplemental Figures 3 and 4 at http://www.genetics.org/supplemental/). Analysis of the particular mutations in light of this knowledge (Table 3) indicates a range of effects from seemingly catastrophic in the case of WSO [WspF V79Δ(6)], which lacks the C-terminal two-thirds of the protein, to seemingly minor in the case of WSE (WspF V220L), which differs from wild type by a single amino acid with similar chemical properties.
Allelic replacements confirm the adaptive significance of wspF mutations:
While parallel genetic changes in wspF, combined with a clear understanding of the physiological effects of defects in methylesterase function, provide a persuasive case for the mutations in wspF being causal, we sought direct evidence by recreating three different wspF mutations (LSWS wspF A901C, WSA wspF T14G, and WSG wspF C556G) in the ancestral genotype.
Replacement of the wild-type allele in the ancestral SM genotype by each variant wspF allele in independent allelic replacement experiments resulted in genotypes with WS phenotypes. In each instance, the recreated WS genotype bore the subtle, but nonetheless distinct, phenotypic characteristics unique to the WS genotype from which the mutant allele was obtained. In addition, detailed proteomic analysis reported elsewhere shows that 52 proteins differentially expressed between the ancestral and derived LSWS genotypes showed consistent changes in the recreated wspF A901C genotype (Knight et al. 2006). To further examine the evolutionary significance of the wspF A901C mutation (in LSWS), this mutant allele was replaced with the wild-type sequence (wspF C901A): in all phenotypic respects this genotype manifested the ancestral SM type.
While analysis of the phenotypic effects of different wspF alleles has made clear the connection between the nucleotide sequence of wspF and phenotype, the relationship between the wspF allele and fitness effects required investigation. To this end, the fitness of each recreated WS mutant was compared (in triplicate assay) to the fitness of the original WS genotype (LSWS, WSA, WSG) from which the wspF allele was originally obtained (in each case fitness was determined relative to LSWS panB and in the presence of the SM ancestor; see materials and methods). In each instance the fitness of the original WS genotype and the recreated WS genotype was not significantly different [F1,17 = 0.100, P = 0.760]. Taken together, these data provide strong evidence that the mutations identified in wspF are solely responsible for both the phenotypic and fitness effects of these WS genotypes.
Different mutations in wspF have a range of fitness effects:
The class of adaptive mutant referred to as WS encompasses an extensive range of diversity at both phenotypic and fitness levels. Just how a single genome can give rise to such a broad repertoire of diversity is an enigma. A clue was provided in a previous study (Goymer et al. 2006): variation in both form and fitness of WS morphs could be generated by alterations in the level of activity of the DGC regulator WspR.
With knowledge of the mutational origins of 14 independent WS—and being in possession of a further 13 WS genotypes with no mutational change in wspF—we sought understanding of the extent to which DNA sequence variation in wspF corresponds to variation in competitive fitness by determining the fitness of the independently derived wrinkly spreader genotypes in competition against the LSWS containing a neutral genetic marker (ΔpanB) in the presence of the SM ancestor.
Substantial quantitative genetic variation in fitness was observed among the entire set of independently derived wrinkly spreader genotypes (Figure 2) and this was shown to be highly significant by ANOVA [F26,79 = 52.81, P < 0.0001]. A significant difference in fitness was also evident among only those WS genotypes with mutations in wspF (i.e., genotypes with no wspF mutation were removed from the analysis) [F13,42 = 60.65, P < 0.0001]. Further analysis revealed that neither knowledge of the different kinds of mutations within wspF (deletion, transition, or transversion) nor the predicted activity of the wspF protein (Table 3) were capable of explaining significant amounts of genetic variation in fitness [type of mutation (deletion, transition, transversion): F2,11 = 0.14, P = 0.87; predicted effect on protein activity (reduced or inactive; Table 3): F1,12 = 1.44, P = 0.25 (nested ANOVA with genotypes, nested within the main effect of mutation type or predicted function, used as the error term)]. These data show that subtle, and usually small, changes in DNA sequence within the coding region of a single gene produce quantitative variation at the level of fitness that cannot be predicted on the basis of the particular kind of mutation nor the predicted activity of the resulting protein.
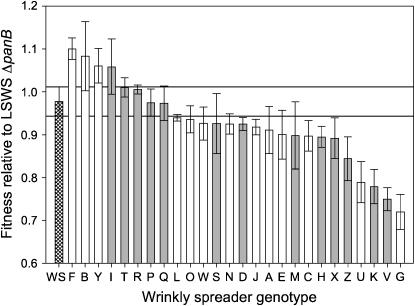
Figure 2.—
Fitness of the 26 independently isolated wrinkly spreader mutants relative to ΔpanB-marked LSWS (hatched bar). Fitness was measured in static broth microcosms over the course of 24 hr. A fitness of 1.0 means that the fitness is identical to LSWS ΔpanB. Open bars represent strains with mutations within wspF; shaded bars represent mutants that are wild type for wspF. Error bars are means and 95% confidence intervals from four replicate assays. In a separate experiment, the fitness of a defined wspF null mutant (SM ΔwspF) was determined relative to LSWS ΔpanB: the fitness of SM ΔwspF was indistinguishable to WSG (see text).
Given the lack of correspondence between the wspF mutation and ensuing fitness effects, it was of interest to determine the fitness consequences of a clearly defined wspF null mutation. To this end, the fitness of three independently generated SM ΔwspF genotypes (which are phenotypically WS; see above) was determined relative to the panB-marked LSWS genotype (in triplicate assay). In the same experiment, the fitness of independent WS genotypes WSG and WSB was also measured (as representative of WS genotypes with low and high fitness, respectively). Genotypes WSG and WSB showed qualitatively similar fitness to those reported in Figure 2 and ANOVA revealed a significant difference among means [F4,10 = 11.87, P = 0.0008]. Tukey's honestly significantly different (HSD) test showed that the fitness of WSG was significantly less than the fitness of WSB (P < 0.05), but indistinguishable from the three SM ΔwspF genotypes (α = 0.05). This indicates that a true wspF null mutation produces a WS genotype with a fitness (relative to LSWS) that is at the low end of the spectrum of WS fitness measurements.
DISCUSSION
Two decades ago one of us became fascinated with the adaptive radiation that occurs each time Pseudomonas populations are propagated in spatially structured microcosms. Examination of an agar plate displaying the net outcome of such a radiation reveals an abundance of ecologically significant diversity, elegant in form and with different types seemingly fitting into a multiplicity of niches (see Rainey and Travisano 1998). Noting both the pace of diversification (a mere 50 generations) and the degree of fit between different types and their environment, it is tempting to suggest that the radiation is genetically programmed. While analysis is yet to extend beyond the WS class of variant, the evidence presented here indicates that the diverse array of WS types arises as a consequence of nothing more than simple, random (spontaneous) mutation aided by intense diversifying selection.
While the mutations are simple, their effects are both phenotypically and ecologically profound. A functionally compromised WspF methylesterase results in constitutive activation of the WspR DGC (Goymer et al. 2006), overproduction of c-di-GMP (Malone et al. 2007), and enhanced production of adhesive substances (Spiers et al. 2002, 2003; Spiers and Rainey 2005), most importantly, of a partially acetylated cellulose polymer (Spiers et al. 2002). The net effect is to cause daughter cells to remain attached after cell division. The cellulosic polymer thus functions as a cell–cell glue and is the cause of cooperation among cells that form the WS mat (Rainey and Rainey 2003). Given appropriate ecological conditions, WS genotypes follow distinctively new evolutionary trajectories compared to the ancestral type. The potential for the evolution of further complexity at the group level is of considerable interest and possibility.
Predicting adaptive evolution:
Predicting the course of evolutionary change is the holy grail of evolutionists (Orr 2005a). On the basis of a variety of different experimental and theoretical approaches, accurate predictions have been made in a small number of instances. For example, a phylogenetic analysis of nucleotide polymorphisms within the influenza hemagglutinin gene enabled accurate predictions of the mutant lineage most likely to persist in the following year (Bush et al. 1999). Similarly, using a biochemical model of lactose flux developed for E. coli populations inhabiting lactose-limited chemostats, Dean (1989) and colleagues (Dean et al. 1986; Dykhuizen et al. 1987) predicted the target of selection—the lactose permease (Zhang and Ferenci 1999). More recently, mutational trajectories leading to enhanced levels of antibiotic detoxifying enzymes have enabled detailed predictions as to the future evolution of antibiotic resistance (Barlow and Hall 2003; Hall and Barlow 2004; Weinreich et al. 2006).
Knowing that complete elimination of the methylesterase activity of WspF was sufficient to generate the WS phenotype, combined with the fact that most mutations are deleterious (Kimura 1968), led to the prediction that the evolutionary cause of LSWS was a mutation in wspF. This proved to be correct. While gain-of-function mutations in other components of the Wsp pathway might reasonably cause WS—e.g., a mutation in WspC that leads to enhanced methyltransferase activity or a mutation in WspR that relieves N-terminal inhibition of the DGC domain (Goymer et al. 2006)—such mutations are expected to be rare, relative to the wspF loss-of-function mutations. Indeed, in this study, of those additional components of the Wsp pathway that were sequenced (in those WS genotypes that harbored no mutation in wspF), no mutations were detected.
The genetics of phenotypic innovation:
While there is interest in predicting the future course of evolution, attention has also been devoted to understanding the nature of mutations responsible for major fitness effects, for example, those responsible for developmental innovations among multicellular organisms (Raff 1996; Carroll 2005). There is general acceptance of the conjecture that major phenotypic innovations are likely to arise from changes in gene regulation and particularly from changes in transcriptional control (King and Wilson 1975). Indeed, the literature of evolutionary developmental biology is replete with examples that show that changes in transcriptional regulation, particularly of cis regulatory sequences (Stern 2000), are a significant source of evolutionary novelty (Doebley et al. 1997; de Rosa et al. 1999; Simpson 2002; Levine and Tjian 2003; Wray 2003; Shapiro et al. 2004).
The genetic changes responsible for the evolution of WS share two significant similarities with these “major mutations.” First, the phenotypic effects have important ecological implications and are macro-evolutionary in terms of effects (although wrought by the simplest of possible mutations). Second, wspF mutations change the level of activity of an existing pathway and are thus regulatory in nature. However, there is an important difference: the mutations in wspF exert their primary effect at a post-translational level. That their effect manifests at this level shows that changes at “alternate regulatory levels” can be evolutionarily significant (Alonso and Wilkins 2005).
Arguably, there is a third similarity: the net effect of the wspF mutations is the generation of a phenotype that is not achievable by the ancestral genotype. The mutation does more than simply result in constitutive expression of a trait that the ancestral type manifests given an appropriately intense stimulatory signal. The firmness of this statement is based on an understanding of Wsp regulation (and particularly of the integral control system composed of WspF and WspC) and is discussed more fully below, but this work adds to the growing body of evidence that shows that the range of phenotypic possibilities arising from relatively simple alterations in pathway control can be difficult to predict and highly innovative.
Genetic architecture of Wsp:
Central to the problem of explaining variation is an understanding of the relationship between the underlying genetic architecture of an organism and the ability of changes at the DNA sequence level to translate through to phenotypically useful solutions. Among the more significant challenges posed by the P. fluorescens radiation is an explanation for the array of phenotypic diversity evident within the WS class of mutants. The discovery of multiple independent mutations within wspF with a range of different fitness effects suggests that the pathway (and the structural components that it regulates) can accommodate the production of heritable phenotypic variation. Received wisdom says that nonsynonymous mutations in a single gene ought to have a limited spectrum of effects: most are expected to abolish function and thus different mutations should generate phenotypically equivalent outcomes. Two interrelated factors may be significant in terms of an explanation for the system's capacity to generate phenotypic novelty: first, the connectivity of the Wsp signaling pathway and, second, the negative regulatory function of WspF.
The Che pathway of E. coli is the most thoroughly studied of any signal transduction cascade: not only is the function of the individual components well understood, but also analysis of the system as a whole has revealed important principles of network design. In particular, the Che pathway represents “a minimal topology providing high robustness to physiological perturbation” (Kollmann et al. 2005, p. 507)—a design principle that appears to be widespread among characterized Che pathways from a range of organisms (Szurmant and Ordal 2004), including pathways in eukaryotes under identical control (Raser and O'Shea 2004). Central to this robustness is integral feedback control, control that is defined by the combined activities of methyltransferase (CheR/WspC) and methylesterase (CheB/WspF). Together, these two components ensure that the difference between the actual output and the steady-state conditions are fed back into the system (Yi et al. 2000).
While Wsp does not function to control chemotaxis, the functional equivalence among Che and Wsp components, and particularly the experimentally demonstrated integral feedback loop, indicates that Wsp conforms to the same robust design principles. Attention has been drawn to a fundamental tension between a robust design and evolvability (the capacity to generate heritable phenotypic variation); the seemingly problematic nature of this tension has led to the development of complex arguments and theoretically tortuous solutions (e.g., Wagner 2005a,b). We see no a priori reason for such a tension.
In the case of Wsp, the integral feedback loop is critically important to the pathway's capacity to accommodate the generation of phenotypic novelty. Drawing upon knowledge of Che function, the activity state of Wsp (even in the absence of a stimulatory ligand) is expected to fluctuate rapidly between active and inactive states. Even when maximally stimulated, the pathway will never deliver a constant output signal because the opposing activities of WspF (CheB) and WspC (CheR) constantly reset the signaling status of the kinase. Given the critical control exerted by WspF-WspC, it is not unreasonable to expect function of this feedback loop to be especially sensitive to genetic changes that affect the capacity of either component to balance the activity of the other. Moreover, any such disturbance, even a very minor one, will destroy the capacity of the pathway to fluctuate between activity states, producing instead a steady-state output. The precise strength of the output signal will depend on the degree to which WspF function is reduced and will be maximal when WspF function is completely eliminated. Put another way, the connectivity of the pathway is such that the output signal from the pathway can be converted from an oscillatory to a steady-state signal and the precise level can be tuned by mutation in a rheostat-like manner (Figure 3). While this model requires experimental validation, it would appear that Wsp is robust to physiological change, and yet fragile to genetic change, while at the same time possessing a topology and connectivity to other cellular components that leads to this genetic fragility manifesting as heritable phenotypic variation.
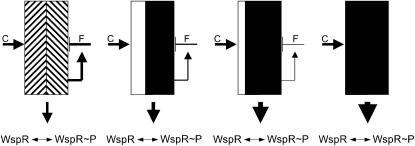
Figure 3.—
A model to account for the effect of wspF mutations on the signaling status of the Wsp pathway. The Wsp signaling machinery (WspA, WspB, WspD, WspE) is shown as a box with the letters C and F indicating the proteins WspC (methyltransferase) and WspF (methylesterase), respectively. The links indicate activations (arrows) or repressions (bar ends). In the absence of a stimulatory ligand, the signaling machinery oscillates between an active (solid) and inactive (open) state, depending on the combined (and opposing) effects of WspC and WspF, which cause the cell to achieve a particular balance between phosphorylated and nonphosphorylated WspR. (Left) The signaling state of the wild-type Wsp pathway that spends time in both active (upward diagonal lines) and inactive (downward diagonal lines) states. Mutations in wspF that reduce protein function, even just slightly, destroy the capacity of the pathway to fluctuate between activity states, producing instead a steady-state output (a constant level of phosphorylated WspR). The precise strength of the output signal depends on the degree to which WspF function is reduced and is maximal when WspF function is completely eliminated (right). Different mutations are predicted to tune the pathway to different output levels (middle panels).
Phenotype–genotype map:
Our model predicts a correlation between the fitness effects of wspF mutations and the extent to which the mutations compromise the function of the enzyme. The fact that the fitness of the wspF deletion strain is at the lower end of the spectrum of fitness effects indicates that the relationship between mutational effect and fitness is an inverse one. While this relationship holds for some WS genotypes, it by no means holds for all. For example, the fitness of WSG and WSU [WspF H186Y and WspF T274Δ(13), respectively] was among the lowest of all WS genotypes and is consistent with the fact that these mutations abolished predicted active sites and therefore should abolish function. However, WSO [V79Δ(6)] carries a deletion that causes a frameshift such that two-thirds of the protein is expected to be absent and yet this genotype has an intermediate fitness. A number of possible explanations can be offered. First, having performed allelic exchange analyses of only three WS genotypes, it is not possible to rule out the possibility that some WS genotypes harbor additional mutations (although unlikely, given that selection was no longer than ∼50 generations). Further, accurate predictions of the effects of various mutations are almost impossible to achieve and the pleiotropic effects can be complex and far reaching (Knight et al. 2006). For a start, WspF has two functional domains [in E. coli, the C-terminal domain of CheB functions constitutively in the absence of the input domain (Lupas and Stock 1989)]; mutations could therefore have effects ranging from impacts on the flow of phosphoryl groups to activity of the methylesterase domain. Even in the case of frameshift mutations, it is not possible to rule out polymerase slippage, which might be sufficiently significant to ensure that at least some functionally active protein is produced. In addition, there might be subtle transcriptional effects; indeed, transcriptional effects on wss [due to changes in the phosphorylation status of WspR (Spiers et al. 2002)] could further complicate understanding. Perhaps more than anything else, this result serves to show how little we do know about the relationship among genotype, phenotype, and fitness.
The microbiological significance of Wsp:
The biological function and ecological significance of Wsp in the ancestral genotype remains a mystery. While this work shows that Wsp regulates the production of acetylated cellulose polymer, all our insights stem from the analysis of variants in which ordinary functioning of the Wsp pathway has been disrupted. Given our understanding of the regulation of the Wsp pathway—complete with integral feedback control—it is safe to assume that the WS phenotype is never achieved by the ancestral genotype, no matter how activated the pathway. The WS phenotype is an altogether novel state achievable only when the Wsp regulation is compromised by mutations that inactivate the integral feedback control. Our hunch is that Wsp, in response to signals unknown, controls the activity of adhesive components in such a way as to effect a kind of colony-based mobility. However, even here we cannot be sure, given that conclusions are largely based on the analysis of constitutive mutants. For example, the primary function of WspR may not be to activate the cellulose synthase enzymes: such activation in WS may be an indirect consequence of a cellular abundance of c-di-GMP. Indeed, the recent discovery of alternate mutation routes to WS—both involving constitutive activation of di-guanylate cyclases that also cause overproduction of the acetylated cellulose polymer—lends weight to this possibility (M. J. McDonald, S. M. Gehrig and P. B. Rainey, unpublished results).
Conclusion:
As others and we have previously commented, adaptive evolution, even in supposedly simple experimental microbial populations, is extraordinarily complex. The fitness effects of the simplest possible mutations can be profound and the capacity of “simple” genomes to generate phenotypic novelty can be quite beyond expectation. Unraveling this complexity is a necessary part of any attempt to shed mechanistic light on the evolutionary process and is necessary to ground theory that ties together the connections among genotype, phenotype, and fitness through evolutionary time. Indeed, the more we discover, the greater the parallels between the kinds of genetic changes and their phenotypic effects observed here and the kinds of events thought to have been significant for the evolution of complex metazoan body plans. This appears to be so, even though our analysis at this stage has been confined to genotypes accessible through minimal (single-step) mutational events from the ancestral type.
Finally, with more than passing interest, we note the recent detection of spontaneous wspF mutations in P. aeruginosa populations colonizing the airways of cystic fibrosis patients (Smith et al. 2006). Given that nonpolar transposon insertions in wspF of P. aeruginosa PAO1 generate WS (D'Argenio et al. 2002)—albeit WS that are not cellulose based—it appears that wrinkly spreaders have an evolutionary significance beyond the bottle.
Acknowledgments
We thank Bertus Beaumont and Dominik Refardt for comments on the manuscript. We thank the Biotechnology and Biological Sciences Research Council and the Natural Environmental Research Council (UK) for financial support. R.K. thanks the Natural Sciences and Engineering Research Council of Canada and St. Hugh's College, Oxford.
References
- Alonso, C. R., and A. S. Wilkins, 2005. Opinion: the molecular elements that underlie developmental evolution. Nat. Rev. Genet. 6: 709–715. [DOI] [PubMed] [Google Scholar]
- Baker, M. D., P. M. Wolanin and J. B. Stock, 2006. Signal transduction in bacterial chemotaxis. BioEssays 28: 9–22. [DOI] [PubMed] [Google Scholar]
- Bantinaki, E., 2002. Characterization of a novel chemosensory pathway underlying adaptive evolution in experimental population of Pseudomonas fluorescens SBW25. Ph.D. Thesis, University of Oxford, Oxford.
- Barlow, M., and B. G. Hall, 2003. Experimental prediction of the natural evolution of antibiotic resistance. Genetics 163: 1237–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, R. M., C. A. Bender, K. Subbarao, N. J. Cox and W. M. Fitch, 1999. Predicting the evolution of human influenza A. Science 286: 1921–1925. [DOI] [PubMed] [Google Scholar]
- Carroll, S. B., 2005. Endless Forms Most Beautiful: The New Science of Evo Devo and the Making of the Animal Kingdom. W. W. Norton, New York.
- Chervitz, S. A., and J. J. Falke, 1996. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc. Natl. Acad. Sci. USA 93: 2545–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo, P. F., K. E. Hosemann, S. Balabhadra, G. Villarreal, M. Dickson et al., 2005. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science 307: 1928–1933. [DOI] [PubMed] [Google Scholar]
- Cooper, T. F., D. E. Rozen and R. E. Lenski, 2003. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl. Acad. Sci. USA 100: 1072–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, V. S., D. Schneider, M. Blot and R. E. Lenski, 2001. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. J. Bacteriol. 183: 2834–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Argenio, D. A., M. W. Calfee, P. B. Rainey and E. C. Pesci, 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184: 6481–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rosa, R., J. K. Grenier, T. Andreeva, C. E. Cook, A. Adoutte et al., 1999. Hox genes in brachiopods and priapulids and protostome evolution. Nature 399: 772–776. [DOI] [PubMed] [Google Scholar]
- Dean, A. M., 1989. Selection and neutrality in lactose operons of Escherichia coli. Genetics 123: 441–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, A. M., D. E. Dykhuizen and D. L. Hartl, 1986. Fitness as a function of beta-galactosidase activity in Escherichia coli. Genet. Res. 48: 1–8. [DOI] [PubMed] [Google Scholar]
- Djordjevic, S., and A. M. Stock, 1997. Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure 5: 545–558. [DOI] [PubMed] [Google Scholar]
- Djordjevic, S., P. N. Goudreau, Q. Xu, A. M. Stock and A. H. West, 1998. Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc. Natl. Acad. Sci. USA 95: 1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., A. Stec and L. Hubbard, 1997. The evolution of apical dominance in maize. Nature 386: 485–488. [DOI] [PubMed] [Google Scholar]
- Dykhuizen, D. E., A. M. Dean and D. L. Hartl, 1987. Metabolic flux and fitness. Genetics 115: 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke, J. J., R. B. Bass, S. L. Butler, S. A. Chervitz and M. A. Danielson, 1997. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 13: 457–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil, H., W. S. Feil, P. Chain, F. Larimer, G. DiBartolo et al., 2005. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 102: 11064–11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski, D. H., and D. R. Helinski, 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function supplied in trans. Proc. Natl. Acad. Sci. USA 76: 1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, R. A., 1930. The Genetical Theory of Natural Selection. Oxford University Press, Oxford.
- Gehrig, S. M., 2005. Adaptation of Pseudomonas fluorescens SBW25 to the air-liquid interface: a study in evolutionary genetics. Ph.D. Thesis, University of Oxford, Oxford.
- Goymer, P., S. G. Kahn, J. G. Malone, S. M. Gehrig, A. J. Spiers et al., 2006. Adaptive divergence in experimental populations of Pseudomonas fluorescens. II. Role of the GGDEF regulator, WspR, in evolution and development of the wrinkly spreader phenotype. Genetics 173: 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, B. G., and M. Barlow, 2004. Evolution of the serine beta-lactamases: past, present and future. Drug Resist. Updat. 7: 111–123. [DOI] [PubMed] [Google Scholar]
- Hickman, J. W., D. F. Tifrea and C. S. Harwood, 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 102: 14422–14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen and L. R. Pease, 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77: 61–68. [DOI] [PubMed] [Google Scholar]
- Joardar, V., M. Lindeberg, R. W. Jackson, J. Selengut, R. Dodson et al., 2005. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 187: 6488–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato et al., 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7: 331–338. [DOI] [PubMed] [Google Scholar]
- Kimura, M., 1968. Evolutionary rate at the molecular level. Nature 217: 624–626. [DOI] [PubMed] [Google Scholar]
- King, E. O., M. K. Ward and D. C. Raney, 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44: 301–307. [PubMed] [Google Scholar]
- King, M. C., and A. C. Wilson, 1975. Evolution at two levels in humans and chimpanzees. Science 188: 107–116. [DOI] [PubMed] [Google Scholar]
- Knight, C. G., N. Zitzmann, S. Prabhakar, R. Antrobus, R. Dwek et al., 2006. Unravelling adaptive evolution: how a single point mutation affects the protein co-regulation network. Nat. Genet. 38: 1015–1022. [DOI] [PubMed] [Google Scholar]
- Kollmann, M., L. Lovdok, K. Bartholome, J. Timmer and V. Sourjik, 2005. Design principles of a bacterial signalling network. Nature 438: 504–507. [DOI] [PubMed] [Google Scholar]
- Le Moual, H., and D. E. J. Koshland, 1996. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J. Mol. Biol. 261: 568–585. [DOI] [PubMed] [Google Scholar]
- Lenski, R. E., M. R. Rose, S. C. Simpson and S. C. Tadler, 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am. Nat. 138: 1315–1341. [Google Scholar]
- Levine, M., and R. Tjian, 2003. Transcription regulation and animal diversity. Nature 424: 147–151. [DOI] [PubMed] [Google Scholar]
- Levit, M. N., T. W. Grebe and J. B. Stock, 2002. Organization of the receptor-kinase signaling array that regulates Escherichia coli chemotaxis. J. Biol. Chem. 277: 36748–36754. [DOI] [PubMed] [Google Scholar]
- Loper, J. E., and S. E. Lindow, 1994. A biological sensor for iron available to bacteria in their habitats on plant-surfaces. Appl. Environ. Microbiol. 60: 1934–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas, A., and J. Stock, 1989. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J. Biol. Chem. 264: 17337–17342. [PubMed] [Google Scholar]
- Malone, J. G., R. Williams, M. Christen, U. Jenal, A. J. Spiers et al., 2007. The structure function relationship of WspR: a Pseudomonas fluorescens response-regulator with a GGDEF output domain. Microbiology 153: 980–994. [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J., 1970. Natural selection and the concept of a protein space. Nature 225: 935–949. [DOI] [PubMed] [Google Scholar]
- McCleary, W. R., and D. R. Zusman, 1990. FrzE of Myxococcus xanthus is homologous to both CheA and CheY of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 87: 5898–5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleary, W. R., M. J. McBride and D. R. Zusman, 1990. Developmental sensory transduction in Myxococcus xanthus involves methylation and demethylation of FrzCD. J. Bacteriol. 172: 4877–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J., 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert et al., 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4: 799–808. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 2005. a The genetic theory of adaptation: a brief history. Nat. Rev. Genet. 6: 119–127. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 2005. b Theories of adaptation: what they do and don't say. Genetica 123: 3–13. [DOI] [PubMed] [Google Scholar]
- Paulsen, I. T., C. M. Press, J. Ravel, D. Y. Kobayashi, G. S. Myers et al., 2005. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 23: 873–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin, K. M., M. A. Samuel and H. A. Wichman, 2006. Variable pleiotropic effects from mutations at the same locus hamper prediction of fitness from a fitness component. Genetics 172: 2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff, R. A., 1996. The Shape of Life: Genes, Development, and the Evolution of Animal Form. University of Chicago Press, Chicago.
- Rainey, P. B., 1999. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ. Microbiol. 1: 243–257. [DOI] [PubMed] [Google Scholar]
- Rainey, P. B., and M. J. Bailey, 1996. Physical and genetic map of the Pseudomonas fluorescens SBW25 chromosome. Mol. Microbiol. 19: 521–533. [DOI] [PubMed] [Google Scholar]
- Rainey, P. B., and K. Rainey, 2003. Evolution of cooperation and conflict in experimental bacterial populations. Nature 425: 72–74. [DOI] [PubMed] [Google Scholar]
- Rainey, P. B., and M. Travisano, 1998. Adaptive radiation in a heterogeneous environment. Nature 394: 69–72. [DOI] [PubMed] [Google Scholar]
- Raser, J. M., and E. K. O'Shea, 2004. Control of stochasticity in eukaryotic gene expression. Science 304: 1811–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Ohana et al., 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325: 279–281. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shapiro, M. D., M. E. Marks, C. L. Peichel, B. K. Blackman, K. S. Nereng et al., 2004. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428: 717–723. [DOI] [PubMed] [Google Scholar]
- Simon, R., U. Priefer and A. Puhler, 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1: 784–791. [Google Scholar]
- Simpson, P., 2002. Evolution of development in closely related species of flies and worms. Nat. Rev. Genet. 3: 907–917. [DOI] [PubMed] [Google Scholar]
- Smith, E. E., D. G. Buckley, Z. Wu, C. Saenphimmachak, L. R. Hoffman et al., 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 103: 8487–8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers, A. J., and P. B. Rainey, 2005. The Pseudomonas fluorescens SBW25 wrinkly spreader biofilm requires attachment factor, cellulose fibre and LPS interactions to maintain strength and integrity. Microbiology 151: 2829–2839. [DOI] [PubMed] [Google Scholar]
- Spiers, A. J., S. G. Kahn, J. Bohannon, M. Travisano and P. B. Rainey, 2002. Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics 161: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers, A. J., J. Bohannon, S. M. Gehrig and P. B. Rainey, 2003. Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 50: 15–27. [DOI] [PubMed] [Google Scholar]
- Stern, D., 2000. Evolutionary developmental biology and the problem of variation. Evolution 54: 1079–1091. [DOI] [PubMed] [Google Scholar]
- Stock, A. M., V. L. Robinson and P. N. Goudreau, 2000. Two-component signal transduction. Annu. Rev. Biochem. 69: 183–215. [DOI] [PubMed] [Google Scholar]
- Stock, J. B., and M. G. Surette, 1996. Chemotaxis, pp. 123–145 in Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, edited by F. Neidhardt. American Society for Microbiology, Washington, DC.
- Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener et al., 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406: 959–964. [DOI] [PubMed] [Google Scholar]
- Szurmant, H., and G. W. Ordal, 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68: 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, A., 2005. a Robustness and Evolvability in Living Systems. Princeton University Press, Princeton, NJ.
- Wagner, A., 2005. b Robustness, evolvability, and neutrality. FEBS Lett. 579: 1772–1778. [DOI] [PubMed] [Google Scholar]
- Weinreich, D. M., N. F. Delaney, M. A. Depristo and D. L. Hartl, 2006. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312: 111–114. [DOI] [PubMed] [Google Scholar]
- West, A. H., E. Martinez-Hackert and A. M. Stock, 1995. Crystal structure of the catalytic domain of the chemotaxis receptor methylesterase, CheB. J. Mol. Biol. 250: 276–290. [DOI] [PubMed] [Google Scholar]
- Woods, R., D. Schneider, C. L. Winkworth, M. A. Riley and R. E. Lenski, 2006. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc. Natl. Acad. Sci. USA 103: 9107–9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray, G. A., 2003. Transcriptional regulation and the evolution of development. Int. J. Dev. Biol. 47: 675–684. [PubMed] [Google Scholar]
- Yi, T. M., Y. Huang, M. I. Simon and J. Doyle, 2000. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc. Natl. Acad. Sci. USA 97: 4649–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, E., and T. Ferenci, 1999. OmpF changes and the complexity of Escherichia coli adaptation to prolonged lactose limitation. FEMS Microbiol. Lett. 176: 395–401. [DOI] [PubMed] [Google Scholar]
- Zhong, S., A. Khodursky, D. E. Dykhuizen and A. M. Dean, 2004. Evolutionary genomics of ecological specialization. Proc. Natl. Acad. Sci. USA 101: 11719–11724. [DOI] [PMC free article] [PubMed] [Google Scholar]