Abstract
The interplay between population subdivision and epistasis is investigated by studying the fixation probability of a coadapted haplotype in a subdivided population. Analytical and simulation models are developed to study the evolutionary fate of two conditionally neutral mutations that interact epistatically to enhance fitness. We find that the fixation probability of a coadapted haplotype shows a marked increase when the population is genetically subdivided and subpopulations are loosely connected by migration. Moderate migration and isolation allow the propagation of the mutant alleles across subpopulations, while at the same time preserving the favorable allelic combination established within each subpopulation. Together they create the condition most favorable for the ultimate fixation of the coadapted haplotype. On the basis of the analytical and simulation results, we discuss the fundamental role of population subdivision and restricted gene flow in promoting the evolution of functionally integrated systems, with some implications for the shifting-balance theory of evolution.
GENES are not alone in the genome; instead, they are interconnected in a complex web of gene interaction networks, changing their own function in conjunction with several other genes with which they associate. It is often conceived that many evolved characteristics are the product of complex genetic interactions during ontogeny. Yet we still lack a comprehensive understanding of the factors affecting evolution in multilocus systems. Many of the existing models of adaptive evolution assume, explicitly or implicitly, that evolution proceeds by sequentially accumulating mutational changes at single loci (e.g., Maynard Smith 1970; Gillespie 1983, 1984; Kauffman and Levin 1987; Rice 1990; Hammerstein 1996; Hartl and Taubes 1996, 1998; Orr 1998, 2002; Burch and Chao 1999; Poon and Otto 2000; Griswold and Whitlock 2003; Welch and Waxman 2003; Whitlock et al. 2003; Weinreich et al. 2005). Under this scenario, two loci that could interact epistatically may never be polymorphic simultaneously during evolution, and the fitness effect of each mutation is evaluated in a given, fixed genetic background. Epistatic effects of cosegregating alleles can therefore largely be ignored. On the other hand, when most standing genetic variation within a population is nonadaptive but in fact has a potential to form adaptive gene combinations in a new genetic background, evolutionary changes may be driven by epistatic selection on cosegregating variants, through the simultaneous increase of the interacting alleles that eventually leads to the formation of a coadapted system. These contrasting views on the evolutionary significance of epistasis date back to the conflicting theories of evolution, each founded by Fisher (1930) and Wright (1931, 1932), and in the words of Ewens (1994) represent the true nub of the Wright–Fisher argument (see also Coyne et al. 1997, 2000; Wade and Goodnight 1998; Goodnight and Wade 2000).
Another aspect of evolution emphasized in the Wrightian theory is the genetic structure of geographically subdivided populations and its consequences on adaptive evolution (Wright 1931, 1932). The Fisherian view holds that evolution should proceed in a large, panmictic population such that the effects of genetic subdivision may largely be ignored. In contrast, Wright envisaged that the individuals of a species are distributed into small, partially isolated breeding units, where the effects of random genetic drift and interdemic selection predominate. Evolutionary consequences of population subdivision have also been studied in relation to the fixation probability of a mutant allele under genic selection (Maruyama 1970; Cherry and Wakeley 2003). It has been shown that in the simplest case with conservative migration (i.e., migration that has no directional effects in changing the overall allele frequencies), fixation probabilities are not affected by subdivision (Maruyama 1970; Cherry and Wakeley 2003). Later, more complex (and presumably realistic) models of subdivision that incorporate local extinction and subsequent colonization were developed (Barton 1993; Cherry 2003b, 2004; Roze and Rousset 2003; Whitlock 2003). The effect of dominance has also been included, leading to an alteration in the fixation probability under subdivision, even in the absence of local extinction and recolonization (Slatkin 1981; Cherry 2003a, 2004; Roze and Rousset 2003; Whitlock 2003; Nishino and Tajima 2004).
While these and other studies have seen only a marginal role for population subdivision in driving mutant alleles to fixation in single-locus systems, relatively little has been done formally to elucidate the joint effects of population subdivision and epistasis (Whitlock et al. 1993; Wade and Goodnight 1998; Goodnight 2000; Wade 2002). To investigate the possible role of population subdivision in the evolution of functionally integrated systems, we here extend our preceding formulation for a panmictic population (Takahasi and Tajima 2005) and develop two-locus models for the evolution of a coadapted system in a subdivided population. By focusing on the fixation process of two conditionally neutral mutations that interact epistatically to form a coadapted haplotype, we have previously demonstrated that when both dominance and linkage disequilibrium are absent in a panmictic population, the fixation probability of the coadapted haplotype is simply given by the product of two probabilities: the fixation probability of a selectively neutral mutant, which is 1/(2N) in a population of N diploids, multiplied by the fixation probability of a beneficial allele with a selective advantage s, which is approximately 2s (Haldane 1927). The result holds irrespective of the time T between the two consecutive mutational events, implying that under random mating the epistatic effects of cosegregating variants during evolution do not induce any additional chance of double fixation.
Since we can expect that most natural populations should show some level of subdivision, the following analysis investigates the effect of limited migration by incorporating the finite-island model of population subdivision. The effects of local extinction and recolonization are not considered. Dominant allelic effects in fitness are not included either. Even with these simplifying assumptions, it is demonstrated by stochastic simulations that the fixation probability of a coadapted haplotype critically depends on migration rate, suggesting a role for population subdivision in promoting the evolution of epistatic systems. Analytical theories based on diffusion and birth-and-death models are also developed (in the appendixes), which should approximate high and low migration limits, respectively.
ANALYSIS
The fundamental part of this analysis follows our previous formulation of two-locus evolutionary dynamics in a panmictic population (Takahasi and Tajima 2005). In brief, we focus on the evolutionary fate of two new mutations that jointly form a coadapted haplotype with selective advantage s (> 0), but independently have no fitness consequences, forming selectively neutral intermediate haplotypes (i.e., haplotypes carrying a mutant allele at one of the two loci and an ancestral allele at the other). Since the two loci are equivalent and interchangeable, we may denote the first and second mutations as A1 and B1, respectively, without loss of generality. The corresponding ancestral allele at each locus is indicated by a subscript 0 (A0 and B0). The entire population is genetically subdivided into L subpopulations (or demes) of an equal size N. Hence NT = NL designates the total population size. Specifically, the finite-island model of population subdivision is considered; every generation, a fraction m of individuals in each subpopulation is replaced by immigrants from the common gene pool. Under the island model, the overall allele frequencies in the entire population are unaltered by migration. Generations are discrete, and the life cycle is adult migration, then random mating and reproduction with recombination, followed by local selection on differential viability in the newborns. Mating and epistatic selection occur independently in each subpopulation. When selection is not too strong, as assumed below, the results of the following analysis should not be affected by the order of events in the life cycle.
Diffusion approximation:
While the substantial part of this analysis is based on stochastic simulations, I also provide analytical theories based on diffusion and birth-and-death approximations for the limiting cases with high and low migration rates, respectively. The details of the analytical treatments are relegated to the appendixes. I first study the probability of double fixation in a subdivided population using diffusion approximation. Recent applications of the diffusion theory to the related issues for the single-locus situations include Cherry (2003a,b, 2004), Cherry and Wakeley (2003), Roze and Rousset (2003), Whitlock (2003), and Nishino and Tajima (2004, 2005). Gavrilets and Gibson (2002) and Whitlock and Gomulkiewicz (2005) have developed diffusion-based analytical methods to obtain fixation probabilities under spatially heterogeneous selection.
For a panmictic population of N diploids, we have previously shown that when the two loci are in linkage equilibrium and there is no dominance at each locus, the probability for the ultimate fixation of the coadapted haplotype A1B1 is given by
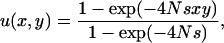 |
(1) |
where x and y denote the initial mutant allele frequencies at the two loci (Takahasi and Tajima 2005, Equation 4); the assumption of linkage equilibrium should be valid as long as recombination is sufficiently frequent and linkage disequilibrium generated by epistatic interaction should soon be removed. On the basis of this result, it is natural to expect that the probability of double fixation in a subdivided population should be approximated by an equivalent formula
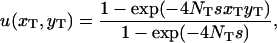 |
(2) |
provided the migration rate among subpopulations is sufficiently high. Here,
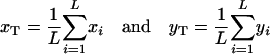 |
are the total frequencies of the mutant alleles A1 and B1 in the entire population, respectively, with xi and yi denoting the frequencies of mutant alleles A1 and B1 in the ith subpopulation (i = 1, 2, 3, … , L). In appendix a, we find that (2) indeed gives the valid fixation probability in a subdivided population if a condition
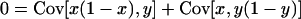 |
(3a) |
or equivalently
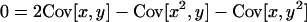 |
(3b) |
is met (see Equation A5 in appendix a). Covariances are here computed across subpopulations, such that
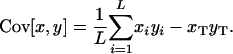 |
(4) |
Mathematically, the condition (3) will always be satisfied if the allele frequencies at the two loci are statistically independent of each other. This situation is more likely to be met when the entire population is genetically homogenized by frequent migration. Later in this section we specify by simulations the range of migration rates where the diffusion approximation may be applicable.
For sufficiently strong selection (4NTs ≫ 1) on new mutations {yT = 1/(2NT) and E[xT] = 1/(2NT), respectively, assuming that the allele A1 has appeared in the population before the introduction of the mutant B1; see Takahasi and Tajima 2005}, the solution (2) reduces to E[u] = 2s/(2NT); here, the expectation is taken over the distribution of xT at the second mutational event.
Birth-and-death approximation:
For the opposite circumstances with extremely low migration, I develop a model based on the birth-and-death approximation (Lande 1979; Slatkin 1981; Takahata 1991; Nishino and Tajima 2004, 2005). In this limit, the migration process is so slow compared to the frequency changes within subpopulations that the fixation or loss of a new allele occurs independently within each subpopulation. As detailed in appendix b, this implies that at any time during evolution each subpopulation may be considered monomorphic for one of the four possible allelic combinations, and the state of the entire population is uniquely determined by the numbers of subpopulations that are fixed for each of the four allelic combinations.
Consider two new mutations A1 and B1, each introduced randomly into a subdivided population. When the entire population is subdivided into L subpopulations of an equal size N, the two new alleles may initially arise in a single subpopulation with probability 1/L. Otherwise they arise in two distinct subpopulations (with probability 1 − 1/L). When the two mutations co-occur in a single subpopulation, the conditional probability of double fixation within the subpopulation is given approximately by 2s/(2N), assuming sufficiently strong selection (4Ns ≫ 1) and linkage equilibrium during the fixation process (Takahasi and Tajima 2005). The assumption of strong selection further entails that once an adaptive gene combination is established in a subpopulation, it is destined to proliferate throughout the entire population. This is because the stochastic loss of the favorable combination from the subpopulation should never happen under this assumption, in spite of the recurrent introduction of ancestral (and deleterious) alleles through migration. Therefore, when the two mutations are initially introduced into a single subpopulation, the contribution to the overall probability of double fixation becomes 2s/(2NL).
When the two mutations arise in distinct subpopulations, each of them must first get fixed independently in its resident subpopulation before they can be exported into other subpopulations through migration. This probability is simply given by {1/(2N)}2, which is the square of the fixation probability of a selectively neutral mutation in a population of N diploids. Summing up the two mutually exclusive possibilities, the expected fixation probability in the low migration limit is expressed as
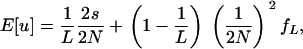 |
(5) |
where fL denotes the conditional probability for the ultimate fixation of the coadapted haplotype, given that the two mutations, initially introduced into distinct subpopulations, are both fixed in the respective subpopulations (appendix b). Note that in (5) the first term in the right-hand side is equivalent to the corresponding fixation probability in a panmictic population of size NL (see Equation 5 in Takahasi and Tajima 2005; see also Equation 2 above). The fixation probability in the low migration limit should therefore be inflated by an amount given by the second term of (5), which should be positive as long as the conditional probability fL is nonzero.
By numerically solving the system of linear equations that describes the transitions between possible states of the population (Equation B1), we obtain the solution for the conditional probability fL, and further by substituting this into (5), we arrive at the numerical solution for the fixation probability in the low migration limit.
Simulations:
For moderate migration, I conduct a set of simulations that follow the forward frequency dynamics of the two-locus system with recombination (at rate c). Initially, the entire population is monomorphic for the ancestral alleles. The two mutations are consecutively introduced, each as a single copy into a randomly chosen subpopulation. The second mutation appears T generations after the introduction of the first allele. No recurrent mutations are assumed afterward. While the waiting time T for the second mutation may in fact be determined by the mutability μ of the B0 allele and the population size NT {such that E[T] = 1/(2NTμ), assuming an exponential distribution of the second mutational event}, we here adopt a simplifying assumption that T is a fixed constant in each set of simulations. The outcome does not differ qualitatively if we assume that the time difference is a random variable. Random genetic drift is implemented in the simulation by the improved version of the pseudosampling (Kimura and Takahata 1983), whereas the expected frequency changes are obtained from the standard two-locus model with selection and recombination (Ewens 2004, Equation 2.94), with additional changes due to migration. Note that in the simulations two-locus genotype frequencies are followed explicitly in each subpopulation, without imposing linkage equilibria as assumed in the diffusion analysis. There is no dominance in fitness at each locus; more specifically, epistatic selection with cis-acting mutations is assumed (see Takahasi and Tajima 2005, Table 2).
Each run of simulations lasts until either of the mutant alleles is lost from the entire population or when both become fixed so that a coadaptation is established. The simulations are repeated until the fixation of the haplotype A1B1 is replicated for 1000 times. The total number of simulation runs, required to produce 1000 fixation events, is recorded to estimate the fixation probability.
Interaction between epistatic selection and migration:
To see how limited gene flow interacts with epistatic selection acting independently within each subpopulation, we first focus on two new mutations that arise simultaneously (T = 0 in the present notation) and investigate how subsequent migration should facilitate or inhibit the formation of coadaptation. In so doing, we consider two situations separately, where the two mutations together arise in a single subpopulation and where they arise separately in two distinct subpopulations, respectively. The probability for the ultimate fixation of the coadapted haplotype would be much different depending on the initial distribution of the new mutations.
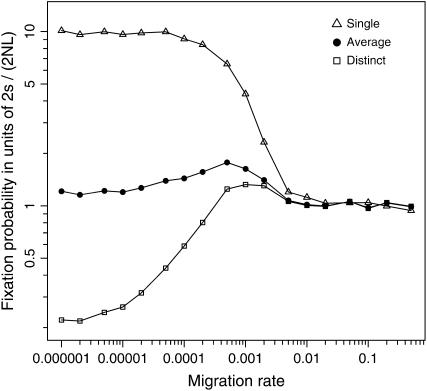
When the two mutations initially co-occur in a single subpopulation, Figure 1 shows that with restricted migration the fixation probability may be increased by a factor of L (= 10 in Figure 1), roughly ranging from 2s/(2NL) (= 10−6) for m ≥ 0.005 to 2s/(2N) (= 10−5) for m ≤ 0.0002. Such dependence on migration rate is expected because limited gene flow will keep the two mutations clumped together in the subpopulation, and the evolutionary increase of the coadapted haplotype will be effectively promoted by epistatic selection, which favors the combination of two alleles that initially cosegregate at low frequencies in the subpopulation. Figure 2A shows that when m ≤ 0.0002 the correlation between allele frequencies at the two loci, computed across subpopulations (as in Equation 4), is kept high for a relatively long period after the introduction, suggesting that the two mutations share a greater chance to cosegregate within a same subpopulation during the initial stages of evolution. As the migration rate becomes larger, the correlation across subpopulations decays at a faster rate. This implies that too much migration would rapidly reallocate the two mutations into distinct localities, thereby limiting the role of epistatic selection in promoting the joint increase of mutant alleles within subpopulations. The conditional probability of double fixation should therefore increase monotonically with decreasing rates of migration.
Figure 1.—
Fixation probabilities of two new mutations that arise simultaneously (T = 0). Simulation results are illustrated for two mutations that co-occur initially in a single subpopulation (open triangles) and for mutations that arise in two distinct subpopulations (open squares). Noting that the former situation should occur with probability 1/L and the latter with probability (1 − 1/L), average (unconditional) values (solid circles) are obtained by appropriately taking the weighted sum of the two conditional probabilities. Parameters are N = 1000, L = 10 (NT = 10,000), s = 0.01, and c = 0.5.
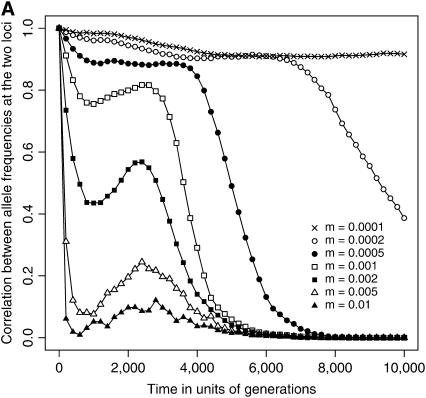
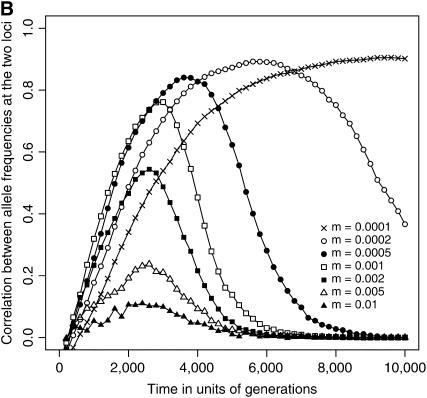
Figure 2.—
Temporal dynamics of the correlation between allele frequencies at the two loci across subpopulations. Simulation results, conditional on the eventual fixation of the coadapted haplotype, are illustrated for 0.0001 ≤ m ≤ 0.01. (A) Two new mutations initially co-occur in a single subpopulation. (B) Mutations are initially introduced into two distinct subpopulations. The abscissa designates the time (in generations) since the introduction of the two mutations, and the ordinate gives the correlation coefficient, averaged over the 1000 replicate runs that resulted in double fixation. As in Figure 1, parameters are N = 1000, L = 10 (NT = 10,000), s = 0.01, c = 0.5, and T = 0.
When the mutations arise separately in two distinct subpopulations, the formation of an advantageous haplotype should be impeded by restricted migration, because the two mutations would have only a smaller chance of being combined together in a single subpopulation. Figure 2B shows that roughly when m ≤ 0.0002, the correlation between allele frequencies at the two loci is established only after a substantial time has elapsed since the introduction of the two mutations. This result indicates that the two mutations should be less likely to cosegregate in the same subpopulation initially when they are still at low frequencies. The potential role of epistatic selection in driving the evolution of coadaptation would therefore be much constrained. In contrast, when the migration rate is slightly higher (m = 0.0005 or 0.001), a significant correlation is established rather rapidly (Figure 2B), and consequently the scaled conditional probability exceeds unity (Figure 1). This implies that in certain cases limited gene flow may promote the evolution of coadaptation even when the interacting alleles initially arise in distinct localities. (It should be emphasized here that in Figure 2 correlations were computed over a subset of simulation runs that eventually resulted in the joint fixation of the two new mutations.)
Taken together, the unconditional probability for the ultimate fixation of the coadapted haplotype should be most facilitated by an intermediate rate of migration, roughly when m = 0.0005 with the parameter values studied here (Figure 1). Simulations also show that when m < 0.01 correlations between x and y(1 − y) [or equivalently between x(1 − x) and y, since we here assume T = 0 so that the two loci are indistinguishable] deviate significantly from zero (result not presented), suggesting that the condition (3) required for the diffusion approximation is satisfied only when the entire population is genetically homogenized by frequent migration, as expected reasonably.
Time difference between two mutational events:
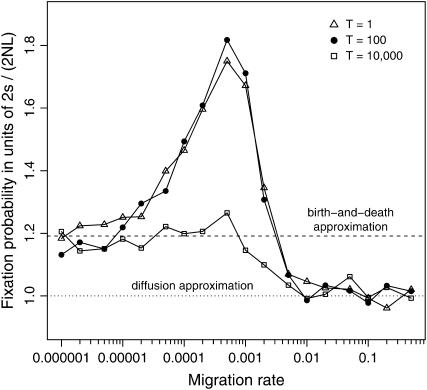
As we have just seen for two mutations arising simultaneously (T = 0), simulations show that when the time interval between the two mutational events is relatively short (T = 1 or 100) there is an intermediate rate of migration (hereafter denoted mmax) that maximizes the fixation probability of the coadapted haplotype (Figure 3). This dependence of fixation probability on migration rate becomes less evident when the second mutation appears long after the first mutational event (T = 10,000). With a sufficiently long time interval T, the first mutation is more likely to be fixed or lost by the time of the second mutational event, and in the limiting case of T → ∞ the probability for the ultimate fixation of the coadapted haplotype eventually converges to
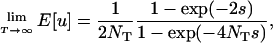 |
(6) |
which is a product of the two fixation probabilities, one for a neutral mutation and the other for a beneficial allele with selective advantage s, both in a subdivided population of NT diploids (Maruyama 1970). The expected probability (6) is independent of the migration rate m and mathematically equivalent to the corresponding probability in a single panmictic population of size NT (Takahasi and Tajima 2005), suggesting that the effect of population subdivision may be negligible when the time interval T is too long. Put another way, the facilitation of double fixation by subdivision should be expected only when the two mutations appear sufficiently close in time during evolution (e.g., T = 1 or 100 in Figure 3).
Figure 3.—
Effects of time difference between two mutational events on the fixation probability of the coadapted haplotype A1B1 in a subdivided population. Simulation results are illustrated for T = 1, 100, and 10,000. Other parameters are N = 1000, L = 10 (NT = 10,000), s = 0.01, and c = 0.5. Note that when T = 1 and 100, the results practically overlap with each other, whereas for T = 10,000, the effect of migration is less significant. Theoretical expectations derived from the diffusion and birth-and-death models are also illustrated; the scaled probabilities are 1.000 (dotted line, diffusion approximation) and 1.191 (dashed line, birth-and-death approximation), respectively.
Figure 3 also indicates that in any case the analytical solution (2), which predicts that E[u] ≈ 2s/(2NL), provides accurate estimates only for relatively high migration rates (roughly when m ≥ 0.01 or equivalently Nm ≥ 10, with the parameter values studied here). Hence, as expected, the diffusion model (A3) in appendix a should be valid when the entire population is effectively panmictic. On the opposite extreme, in the low migration limit, the simulations show that the fixation probability is more or less increased by an amount corresponding to the second term in the right-hand side of (5).
Subpopulation number and size:
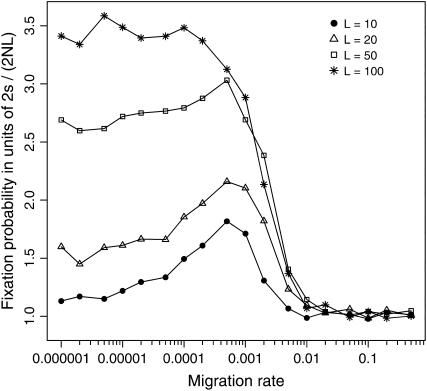
When the second mutation is introduced shortly after the first mutational event, the simulations show that the effect of population subdivision in promoting the fixation of the coadapted haplotype becomes more profound as the number of subpopulations L becomes larger and the entire population gets more fragmented. This may be observed either when the total population size NT is kept constant (so that the subpopulation size is altered along with the subpopulation number; Figure 4) or when the subpopulation size N is constant (so that the total population size is altered along with the subpopulation number; Table 1). In the former case, the subpopulation size is smaller in a more fragmented population (with larger L), implying that the relative strength of random genetic drift becomes more prevailing. In the latter case, as the subpopulation number L gets larger, the total population size also gets larger and accordingly the (absolute) fixation probability becomes smaller (Table 1). Still, the effect of subdivision is more pronounced for larger L; by contrasting the scaled fixation probabilities in units of 2s/(2NL), we can see that the fixation probability shows an almost threefold increase when L = 50, but the increment is only 1.7-fold when L = 10 (Table 1).
Figure 4.—
Effects of subpopulation size and number on the fixation probability of the coadapted haplotype A1B1 in a subdivided population. Simulation results are illustrated for subdivided populations with L = 10, 20, 50, and 100. In each case, the total population size NT is kept constant (at 10,000) so that the subpopulation size N (= NT/L) is altered accordingly. Other parameters are s = 0.01, c = 0.5, and T = 100. Note that for L ≤ 50 the optimal migration rate mmax is almost invariant despite the changes in the subpopulation size N.
TABLE 1.
Effects of subpopulation number on the fixation probability of the coadapted haplotype
| L = 10: |
L = 20
|
L = 50
|
|||
|---|---|---|---|---|---|
| Migration rate | Absolute (× 10−6) | Absolute (× 10−6) | Scaled | Absolute (× 10−6) | Scaled |
| Low migration limit | 1.191 | 0.675 | 1.350 | 0.337 | 1.686 |
| 0.000001 | 1.131 | 0.642 | 1.283 | 0.313 | 1.566 |
| 0.00001 | 1.219 | 0.677 | 1.354 | 0.345 | 1.723 |
| 0.0001 | 1.493 | 0.866 | 1.732 | 0.444 | 2.218 |
| 0.001 | 1.711 | 0.971 | 1.942 | 0.562 | 2.808 |
| 0.01 | 0.986 | 0.529 | 1.059 | 0.214 | 1.068 |
| 0.1 | 0.978 | 0.483 | 0.966 | 0.203 | 1.014 |
| High migration limit | 1.000 | 0.500 | 1.000 | 0.200 | 1.000 |
Fixation probabilities (× 106) of the coadapted haplotype A1B1 in a subdivided population (with L = 10, 20, and 50) are listed. In each set of simulations, the subpopulation size N is kept constant (at 1000) so that the total population size NT (= NL) is altered accordingly. For L = 20 and 50, scaled probabilities in units of 2s/(2NL) are also provided. (For L = 10, the scaled probabilities coincide with the absolute values × 106.) Throughout, s = 0.01, c = 0.5, and T = 100. Also listed are theoretical expectations derived from the diffusion and birth-and-death models, which should approximate high and low migration limits, respectively (see appendixes a and b).
Table 1 also lists the expectations derived by the analytical methods based on diffusion and birth-and-death approximations (appendixes a and b). Again, the diffusion approximation is accurate roughly when m = 0.01 (Nm = 10) or higher. On the other extreme, when the migration rate is substantially low (∼1 × 10−6), it appears that the fixation probabilities converge to (or become somewhat smaller than) the estimates obtained from the birth-and-death model.
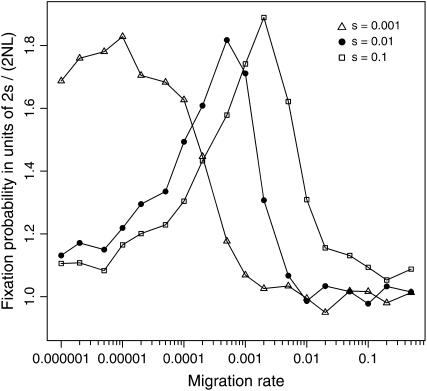
Additional simulations show that mmax is primarily affected by the selection intensity s and not by other factors such as the subpopulation size N (see Figures 4 and 5). Previously, we have demonstrated that there is an optimal rate of recombination, given approximately by c = s/2, that most facilitates the evolution of coadaptation in a panmictic population (Takahasi and Tajima 2005). While simulations suggest that mmax also changes monotonically with s, it appears that the relationship is rather complex.
Figure 5.—
Effects of selection on the fixation probability of the coadapted haplotype A1B1 in a subdivided population. Simulation results are illustrated for s = 0.001, 0.01, and 0.1. Other parameters are N = 1000, L = 10 (NT = 10,000), c = 0.5, and T = 100. Note that the optimal migration rate mmax becomes greater with stronger selection (larger s).
Linkage:
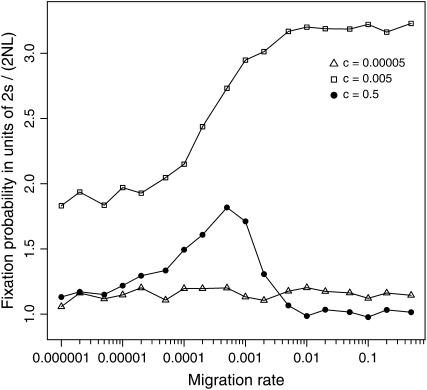
Whereas the primary focus of this study is centered on two unlinked loci, effects of linkage can be investigated by setting c < 0.5 in the simulations. Under random mating, the probability of double fixation is most facilitated by an intermediate rate of recombination, because too much recombination breaks up the selectively favorable combination of alleles in the coupling phase, whereas two mutations in repulsion are hardly combined together on a single chromosome when the recombination rate is too low (Takahasi and Tajima 2005). In accord with the panmictic model, it is found that for a given rate of migration the fixation of the coadapted haplotype is most facilitated by an intermediate rate of recombination (c = 0.005 in Figure 6), although the effect of linkage becomes less significant as the migration rate becomes smaller. This is because the two mutations are less likely to cosegregate in the same subpopulation when the migration rate is too low (as noted above), while recombination should affect the probability of double fixation only when the two loci are simultaneously polymorphic (Takahasi and Tajima 2005). In the extreme case of complete linkage (c = 0), the present model reduces essentially to a single-locus system. Therefore, the observed interplay between population subdivision and epistasis should be absent in asexual lineages.
Figure 6.—
Effects of linkage on the fixation probability of the coadapted haplotype A1B1 in a subdivided population. Simulation results are illustrated for c = 0.00005, 0.005, and 0.5 (i.e., free recombination). Other parameters are N = 1000, L = 10 (NT = 10,000), s = 0.01, and T = 100. The results recapture the preceding observation that the fixation probability is maximized by an intermediate degree of linkage, roughly when c = s/2 (Takahasi and Tajima 2005), although the effect of linkage becomes less pronounced as the migration rate becomes smaller.
Weak selection:
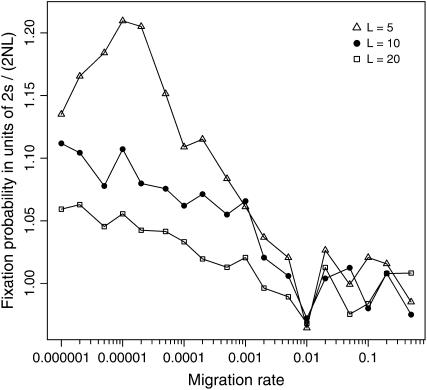
We have so far concentrated on situations where epistatic selection is relatively strong and efficiently promotes the joint increase of the two mutations in each subpopulation; specifically, we have assumed 4Ns ≥ 4 (equality holds when L = 100 in Figure 4 or when s = 0.001 in Figure 5). When this condition is violated, simulations show that in contrast to the situations under strong selection the effect of population subdivision may be less pronounced in a more fragmented population. As shown in Figure 7, when the total population size is kept constant at NT = 2000 and s = 0.001 (hence 4NTs = 8), the probability of double fixation is increased only slightly in a population fragmented into L = 20 subpopulations (hence 4Ns = 0.4), whereas in a less fragmented population with L = 5 (4Ns = 1.6) or 10 (4Ns = 0.8), the effect of subdivision is more evident; still, the increment is only up to a factor of ∼1.2 at most (when L = 5). These simulation results also suggest that the primary role of restricted gene flow is to colocalize simultaneously segregating mutations that interact epistatically, thereby creating the condition favorable for the effective operation of epistatic selection within subpopulations. When 4Ns < 1 and the advantageous haplotype is only weakly selected in small subpopulations, random genetic drift may override the effect of epistatic selection in driving the evolution of coadaptation within each subpopulation, even if selection could be effective at the entire population level (4NTs > 1).
Figure 7.—
Effects of population subdivision on the fixation probability of the coadapted haplotype A1B1 under weak epistatic selection (s = 0.001). Simulation results are illustrated for subdivided populations with L = 5, 10, and 20. In each case, the total population size NT is kept constant (at 2000) so that the subpopulation size N (= NT/L) is altered accordingly. Other parameters are c = 0.5 and T = 1.
DISCUSSION
This study illustrates the importance of population subdivision in promoting the evolution of epistatic systems. Simulation analyses show that when the migration rate is moderate, analytical theories (based on diffusion and birth-and-death approximations, respectively) fail to provide accurate estimates for the probability of double fixation. The diffusion model is applicable only when the migration rate is relatively high and the entire population is effectively panmictic. On the opposite extreme, in the low migration limit, the fixation probability is more or less increased by an amount corresponding to the second term in the right-hand side of (5). For moderate migration, where both approximations fail, simulations show that the chance of double fixation may be exaggerated even more substantially.
When the habitat range of an organism is fragmented into local subpopulations, the two mutations may initially arise in distinct localities. If migration among subpopulations is severely constrained (as in the low migration limit), it is unlikely that the two mutations should ever have a chance to see each other in a single locality before getting fixed (or lost) within a subpopulation. The fixation of a mutant allele within each subpopulation should then be driven completely by random genetic drift, and there is not much room left for epistatic selection to promote the joint increase of the two mutations. When the two mutations are initially introduced into a single subpopulation, limited migration will keep the two mutations together in the subpopulation, and the evolutionary increase of the coadapted haplotype will be effectively promoted by epistatic selection. However, too much migration would rapidly disperse the two mutations away from each other, thereby reducing the effect of epistatic selection in the formation of a favorable allelic combination within each subpopulation. Moderate migration and isolation allow the propagation of the mutant alleles across subpopulations, while at the same time preserving the favorable allelic combination established within each subpopulation. Together they create the condition most favorable for the ultimate fixation of the coadapted haplotype. Moreover, the facilitation of double fixation in a subdivided population should be expected only when the time difference between the two mutational events is sufficiently short (see Figure 3), suggesting the vital role played by epistatic selection during the initial stages of evolution, when both mutations are still segregating at low frequencies.
Our findings may seem to render support for the shifting-balance theory of evolution (Wright 1931, 1932; reviewed in Wright 1980), which advocates the primacy of epistasis and random genetic drift as the causes of evolution in small, partially isolated populations. Although the present study also focuses on the interplay between population subdivision and epistasis, there is an important difference between the Wrightian theory and the present theories. According to the conventional views, the Wrightian shifting-balance process proceeds through the rugged fitness landscape with peaks and valleys generated by epistatic interactions among alleles at multiple loci (Whitlock et al. 1995). The fitness valleys represent the class of unfavorable genotypes that may be formed during evolution. In contrast, the present formulation does not include unfit intermediate haplotypes; instead, it involves haplotypes connected by a neutral network, where evolution may proceed without crossing the intervening fitness valleys (Gavrilets 2004; Wagner 2005).
The role of conditionally neutral mutations in adaptation has recently renewed interest in the theories of evolution in multilocus systems, where, for example, biological systems achieve functional robustness by buffering genetic perturbations (e.g., van Nimwegen et al. 1999; Rutherford 2000; Wilke 2001; Gibson and Dworkin 2004; Hermisson and Wagner 2004; Wagner 2005). Classically, the adaptive significance of neutral variation has been noted in the neutral theory of evolution, when Kimura (1983) introduced the Dykhuizen–Hartl effect (Dykhuizen and Hartl 1980) to describe the potential role of neutral mutations in creating the foundation of adaptation (see also Hartl et al. 1985; Kimura 1986; Zhang et al. 1998).
The stochastic dynamics of the shifting-balance process have also been studied by two-locus epistatic selection models, where mutations are individually deleterious but become neutral (or may even be better fit) when combined with the compensatory allele at the other locus (e.g., Kimura 1990, 1991; Phillips 1996; but see also Stephan 1996). In a subdivided population, mutant alleles would individually be kept at low frequencies in each subpopulation if sufficiently strong selection acts against the intermediate haplotypes. Consequently, it is highly unlikely that the two compensatory mutations, each formerly in a distinct locality, should be brought together into a single subpopulation through migration (Phillips 1996). Instead, each subpopulation has to wait for the production of a new mutation that compensates the deleterious effect of the preexisting allele, without the aid of migration. Therefore, the evolutionary trajectories follow very different paths under selection models with or without the detrimental fitness effects on the intermediates, and the dependence of fixation probability on migration rate as demonstrated in the present study would not be expected when multiple fitness peaks are separated by valleys of low fitness. The conditional neutrality thus plays a decisive part in the present theory of evolution in a subdivided population.
It has been known that locally restricted interactions affect the global foundation of interactive systems in various ways (e.g., Eshel and Cavalli-Sforza 1982; Wade and Goodnight 1998; Thompson 1999; Chave et al. 2002; Briggs and Hoopes 2004). The present study also finds a primary role for subpopulation structure in creating the favorable condition for the evolution of coadaptation. As the genomewide polymorphism data accumulate, it is now possible to draw a detailed inference on the genetic structure and history of natural populations (Hey and Machado 2003; Wakeley 2004). Together with our current knowledge on the genetic architecture of phenotypic diversity, insights into the historical demography will cast a new light on the role of population subdivision and migration in determining the course of evolution in multilocus systems.
Acknowledgments
I thank Jo Nishino and Fumio Tajima for comments and discussion. This research was supported by grants from the Japan Ministry of Education, Culture, Sports, Science, and Technology and the National Institute of Genetics Cooperative Research Programs.
APPENDIX A: DIFFUSION MODEL
Here and in appendix b, I develop analytical theories based on diffusion and birth-and-death approximations, each of which should apply to the limiting cases with high and low migration rates, respectively.
In the diffusion limit, the expected frequency changes within each subpopulation are given respectively by
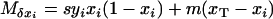 |
(A1a) |
and
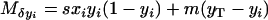 |
(A1b) |
(see Equations 1 in Takahasi and Tajima 2005), assuming the finite-island model of population subdivision and linkage equilibrium within each subpopulation (i.e., Pi − xiyi = 0 for all i, where Pi designates the frequency of haplotype A1B1 in the ith subpopulation). The expected variances of the changes in mutant frequencies are obtained from the ordinary binomial model of random genetic drift,
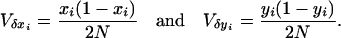 |
(A2) |
Using these notations, fixation probability u(x1, … , xL, y1, … , yL) satisfies the equation
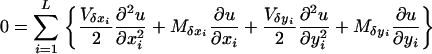 |
(A3) |
Here we want to know under what conditions the partial differential equation (A3) has a solution (2), which is mathematically equivalent to the panmictic solution (1). Given that the probability u is defined by (2), we have
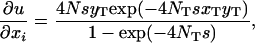 |
(A4a) |
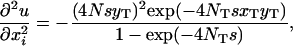 |
(A4b) |
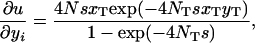 |
(A4c) |
and
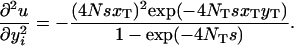 |
(A4d) |
Substituting (A1), (A2), and (A4) into the right-hand side of (A3), we arrive at the desired condition,
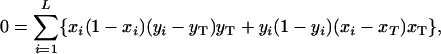 |
(A5) |
which may be expressed as an equality (Equation 3) by using covariances across subpopulations (as defined in Equation 4).
APPENDIX B: BIRTH-AND-DEATH MODEL
For the low migration limit, I study the probability of fixation based on the birth-and-death approximation. In this limit, the fixation of an allele within a subpopulation occurs almost instantaneously in comparison to its transmission across subpopulations. This entails that the two loci should both be monomorphic within each subpopulation; i.e., either (x, y) = (0, 0), (0, 1), (1, 0) or (1, 1) holds, where x and y here denote the mutant frequencies in a subpopulation. Denoting the numbers of subpopulations with (x, y) = (0, 1), (1, 0), and (1, 1) by n01, n10, and n11, respectively, the state of the entire population S(n01, n10, n11) can be uniquely determined by these three variables. (The summation n01 + n10 + n11 does not exceed the total subpopulation number L.) Noting that the fixation takes place independently within each subpopulation in the low migration limit, and further assuming that a subpopulation receives immigrants from a randomly chosen subpopulation during a migration event, the ultimate fixation probability f(n01, n10, n11), conditional on the initial state S(n01, n10, n11), satisfies the relation
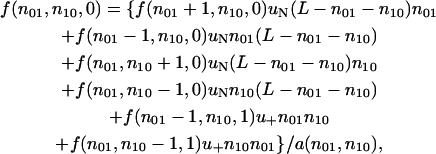 |
(B1) |
where a(n01, n10) = 2{uN(n01 + n10)(L − n01 − n10) + u+n01n10} is the standardizing variable, uN designates the probability that a subpopulation, which receives selectively neutral immigrants during a migration event, becomes fixed for the incoming haplotype, while u+ denotes the corresponding probability for a subpopulation that receives a beneficial haplotype with selective advantage s. In the low migration limit, the two probabilities should be proportional to 1/(2N) and 2s, respectively (for a more thorough discussion, see Nishino and Tajima 2004). The boundary conditions are f(0, n10, 0) = f(n01, 0, 0) = 0, f(n01, n10, 1) = 1, and f(n01, L − n01, 0) = 1 for n01 ≠ 0, L. Note also that fL = f(1, 1, 0). The conditional probability fL can be obtained by numerically solving a sparse system of (L − 1) (L − 2)/2 linear equations for (L − 1) (L − 2)/2 unknowns.
On deriving the relation (B1), it is assumed that if a subpopulation with (x, y) = (0, 1) receives immigrants with haplotype A1B0 from another subpopulation with (x, y) = (1, 0), the possible outcome is either (x′, y′) = (1, 1) (fixation of the incoming allele A1, with probability u+) or (x′, y′) = (0, 1) (no change, with probability 1 − u+), where the primes denote the frequencies after the migration event. An equivalent argument applies when a subpopulation with (x, y) = (1, 0) receives immigrants with haplotype A0B1. This simplifying assumption, which further entails the boundary condition f(n01, L − n01, 0) = 1 (for n01 ≠ 0, L), neglects the possibilities of (x′, y′) = (0, 0) (loss of the preexisting allele B1) and (x′, y′) = (1, 0) (fixation of A1 accompanied by loss of B1) and should be valid only when the selection is sufficiently strong (4Ns ≫ 1). Therefore, when the entire population is fragmented into many small subpopulations such that the required condition 4Ns ≫ 1 does not hold, the present formulation based on (B1) should overestimate the probability of double fixation.
References
- Barton, N. H., 1993. The probability of fixation of a favoured allele in a subdivided population. Genet. Res. 62: 149–157. [Google Scholar]
- Briggs, C. J., and M. F. Hoopes, 2004. Stabilizing effects in spatial parasitoid-host and predator-prey models: a review. Theor. Popul. Biol. 65: 299–315. [DOI] [PubMed] [Google Scholar]
- Burch, C. L., and L. Chao, 1999. Evolution by small steps and rugged landscapes in the RNA virus φ6. Genetics 151: 921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chave, J., H. C. Muller-Landau and S. A. Levin, 2002. Comparing classical community models: theoretical consequences for patterns of diversity. Am. Nat. 159: 1–23. [DOI] [PubMed] [Google Scholar]
- Cherry, J. L., 2003. a Selection in a subdivided population with dominance or local frequency dependence. Genetics 163: 1511–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, J. L., 2003. b Selection in a subdivided population with local extinction and recolonization. Genetics 164: 789–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, J. L., 2004. Selection, subdivision and extinction and recolonization. Genetics 166: 1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry, J. L., and J. Wakeley, 2003. A diffusion approximation for selection and drift in a subdivided population. Genetics 163: 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., N. H. Barton and M. Turelli, 1997. Perspective: a critique of Sewall Wright's shifting balance theory of evolution. Evolution 51: 645–671. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., N. H. Barton and M. Turelli, 2000. Is Wright's shifting balance process important in evolution? Evolution 54: 306–317. [DOI] [PubMed] [Google Scholar]
- Dykhuizen, D., and D. L. Hartl, 1980. Selective neutrality of 6PGD allozymes in E. coli and the effects of genetic background. Genetics 96: 801–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel, I., and L. L. Cavalli-Sforza, 1982. Assortment of encounters and evolution of cooperativeness. Proc. Natl. Acad. Sci. USA 79: 1331–1335 (corrigendum: Proc. Natl. Acad. Sci. USA 80: 1135). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens, W. J., 1994. The changing role of population genetics theory, pp. 186–197 in Frontiers in Mathematical Biology, edited by S. A. Levin. Springer, Berlin.
- Ewens, W. J., 2004. Mathematical Population Genetics I: Theoretical Introduction, Ed. 2. Springer, New York.
- Fisher, R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford.
- Gavrilets, S., 2004. Fitness Landscapes and the Origin of Species. Princeton University Press, Princeton, NJ.
- Gavrilets, S., and N. Gibson, 2002. Fixation probabilities in a spatially heterogeneous environment. Popul. Ecol. 44: 51–58. [Google Scholar]
- Gibson, G., and I. Dworkin, 2004. Uncovering cryptic genetic variation. Nat. Rev. Genet. 5: 681–690. [DOI] [PubMed] [Google Scholar]
- Gillespie, J. H., 1983. A simple stochastic gene substitution process. Theor. Popul. Biol. 23: 202–215. [DOI] [PubMed] [Google Scholar]
- Gillespie, J. H., 1984. Molecular evolution over the mutational landscape. Evolution 38: 1116–1129. [DOI] [PubMed] [Google Scholar]
- Goodnight, C. J., 2000. Modeling gene interactions in structured populations, pp. 129–145 in Epistasis and the Evolutionary Process, edited by J. B. Wolf, E. D. Brodie III and M. J. Wade. Oxford University Press, New York.
- Goodnight, C. J., and M. J. Wade, 2000. The ongoing synthesis: a reply to Coyne, Barton and Turelli. Evolution 54: 317–324. [DOI] [PubMed] [Google Scholar]
- Griswold, C. K., and M. C. Whitlock, 2003. The genetics of adaptation: the roles of pleiotropy, stabilizing selection and drift in shaping the distribution of bidirectional fixed mutational effects. Genetics 165: 2181–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1927. A mathematical theory of natural and artificial selection. V. Selection and mutation. Proc. Camb. Philos. Soc. 23: 838–844. [Google Scholar]
- Hammerstein, P., 1996. Darwinian adaptation, population genetics and the streetcar theory of evolution. J. Math. Biol. 34: 511–532. [DOI] [PubMed] [Google Scholar]
- Hartl, D. L., and C. H. Taubes, 1996. Compensatory nearly neutral mutations: selection without adaptation. J. Theor. Biol. 182: 303–309. [DOI] [PubMed] [Google Scholar]
- Hartl, D. L., and C. H. Taubes, 1998. Towards a theory of evolutionary adaptation. Genetica 102/103: 525–533. [PubMed] [Google Scholar]
- Hartl, D. L., D. E. Dykhuizen and A. M. Dean, 1985. Limits of adaptation: the evolution of selective neutrality. Genetics 111: 655–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson, J., and G. P. Wagner, 2004. The population genetic theory of hidden variation and genetic robustness. Genetics 168: 2271–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey, J., and C. A. Machado, 2003. The study of structured populations—new hope for a difficult and divided science. Nat. Rev. Genet. 4: 535–543. [DOI] [PubMed] [Google Scholar]
- Kauffman, S., and S. Levin, 1987. Towards a general theory of adaptive walks on rugged landscapes. J. Theor. Biol. 128: 11–45. [DOI] [PubMed] [Google Scholar]
- Kimura, M., 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, UK.
- Kimura, M., 1986. DNA and the neutral theory. Philos. Trans. R. Soc. Lond. Ser. B 312: 343–354. [DOI] [PubMed] [Google Scholar]
- Kimura, M., 1990. Some models of neutral evolution, compensatory evolution, and the shifting balance process. Theor. Popul. Biol. 37: 150–158. [DOI] [PubMed] [Google Scholar]
- Kimura, M., 1991. Recent development of the neutral theory viewed from the Wrightian tradition of theoretical population genetics. Proc. Natl. Acad. Sci. USA 88: 5969–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., and N. Takahata, 1983. Selective constraint in protein polymorphism: study of the selectively neutral mutation model by using an improved pseudosampling method. Proc. Natl. Acad. Sci. USA 80: 1048–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande, R., 1979. Effective deme sizes during long-term evolution estimated from rates of chromosome rearrangement. Evolution 33: 234–251. [DOI] [PubMed] [Google Scholar]
- Maruyama, T., 1970. On the fixation probability of mutant genes in a subdivided population. Genet. Res. 15: 221–225. [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J., 1970. Natural selection and the concept of a protein space. Nature 225: 563–564. [DOI] [PubMed] [Google Scholar]
- Nishino, J., and F. Tajima, 2004. Effect of dominance on heterozygosity and the fixation probability in a subdivided population. Genes Genet. Syst. 79: 41–48. [DOI] [PubMed] [Google Scholar]
- Nishino, J., and F. Tajima, 2005. Effect of population structure on the amount of polymorphism and the fixation probability under overdominant selection. Genes Genet. Syst. 80: 287–295. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 1998. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution 52: 935–949. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 2002. The population genetics of adaptation: the adaptation of DNA sequences. Evolution 56: 1317–1330. [DOI] [PubMed] [Google Scholar]
- Phillips, P. C., 1996. Waiting time for a compensatory mutation: phase zero of the shifting-balance process. Genet. Res. 67: 271–283. [DOI] [PubMed] [Google Scholar]
- Poon, A., and S. P. Otto, 2000. Compensating for our load of mutations: freezing the meltdown of small populations. Evolution 54: 1467–1479. [DOI] [PubMed] [Google Scholar]
- Rice, S. H., 1990. A geometric model for the evolution of development. J. Theor. Biol. 143: 319–342. [Google Scholar]
- Roze, D., and F. Rousset, 2003. Selection and drift in subdivided populations: a straightforward method for deriving diffusion approximations and applications involving dominance, selfing and local extinctions. Genetics 165: 2153–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford, S. L., 2000. From genotype to phenotype: buffering mechanisms and the storage of genetic information. BioEssays 22: 1095–1105. [DOI] [PubMed] [Google Scholar]
- Slatkin, M., 1981. Fixation probabilities and fixation times in a subdivided population. Evolution 35: 477–478. [DOI] [PubMed] [Google Scholar]
- Stephan, W., 1996. The rate of compensatory evolution. Genetics 144: 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi, K. R., and F. Tajima, 2005. Evolution of coadaptation in a two-locus epistatic system. Evolution 59: 2324–2332. [PubMed] [Google Scholar]
- Takahata, N., 1991. Genealogy of neutral genes and spreading of selected mutations in a geographically structured population. Genetics 129: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. N., 1999. Specific hypotheses on the geographic mosaic of coevolution. Am. Nat. 153: S1–S14. [Google Scholar]
- van Nimwegen, E., J. P. Crutchfield and M. Huynen, 1999. Neutral evolution of mutational robustness. Proc. Natl. Acad. Sci. USA 96: 9716–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, M. J., 2002. A gene's eye view of epistasis, selection and speciation. J. Evol. Biol. 15: 337–346. [Google Scholar]
- Wade, M. J., and C. J. Goodnight, 1998. Perspective: the theories of Fisher and Wright in the context of metapopulations: when nature does many small experiments. Evolution 52: 1537–1553. [DOI] [PubMed] [Google Scholar]
- Wagner, A., 2005. Robustness and Evolvability in Living Systems. Princeton University Press, Princeton, NJ.
- Wakeley, J., 2004. Metapopulation models for historical inference. Mol. Ecol. 13: 865–875. [DOI] [PubMed] [Google Scholar]
- Weinreich, D. M., R. A. Watson and L. Chao, 2005. Perspective: sign epistasis and genetic constraint on evolutionary trajectories. Evolution 59: 1165–1174. [PubMed] [Google Scholar]
- Welch, J. J., and D. Waxman, 2003. Modularity and the cost of complexity. Evolution 57: 1723–1734. [DOI] [PubMed] [Google Scholar]
- Whitlock, M. C., 2003. Fixation probability and time in subdivided populations. Genetics 164: 767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock, M. C., and R. Gomulkiewicz, 2005. Probability of fixation in a heterogeneous environment. Genetics 171: 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock, M. C., P. C. Phillips and M. J. Wade, 1993. Gene interaction affects the additive genetic variance in subdivided populations with migration and extinction. Evolution 47: 1758–1769. [DOI] [PubMed] [Google Scholar]
- Whitlock, M. C., P. C. Phillips, F. B.-G. Moore and S. J. Tonsor, 1995. Multiple fitness peaks and epistasis. Annu. Rev. Ecol. Syst. 26: 601–629. [Google Scholar]
- Whitlock, M. C., C. K. Griswold and A. D. Peters, 2003. Compensating for the meltdown: the critical effective size of a population with deleterious and compensatory mutations. Ann. Zool. Fenn. 40: 169–183. [Google Scholar]
- Wilke, C. O., 2001. Adaptive evolution on neutral networks. Bull. Math. Biol. 63: 715–730. [DOI] [PubMed] [Google Scholar]
- Wright, S., 1931. Evolution in Mendelian populations. Genetics 16: 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., 1932. The roles of mutation, inbreeding, crossbreeding, and selection in evolution. Proc. 6th Int. Cong. Genet. 1: 356–366. [Google Scholar]
- Wright, S., 1980. Genic and organismic selection. Evolution 34: 825–843. [DOI] [PubMed] [Google Scholar]
- Zhang, J., H. F. Rosenberg and M. Nei, 1998. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc. Natl. Acad. Sci. USA 95: 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]