Abstract
For most common diseases with heritable components, not a single or a few single-nucleotide polymorphisms (SNPs) explain most of the variance for these disorders. Instead, much of the variance may be caused by interactions (epistasis) among multiple SNPs or interactions with environmental conditions. We present a new powerful statistical model for analyzing and interpreting genomic data that influence multifactorial phenotypic traits with a complex and likely polygenic inheritance. The new method is based on Markov chain Monte Carlo (MCMC) and allows for identification of sets of SNPs and environmental factors that when combined increase disease risk or change the distribution of a quantitative trait. Using simulations, we show that the MCMC method can detect disease association when multiple, interacting SNPs are present in the data. When applying the method on real large-scale data from a Danish population-based cohort, multiple interactions are identified that severely affect serum triglyceride levels in the study individuals. The method is designed for quantitative traits but can also be applied on qualitative traits. It is computationally feasible even for a large number of possible interactions and differs fundamentally from most previous approaches by entertaining nonlinear interactions and by directly addressing the multiple-testing problem.
MOST common diseases are multifactorial and influenced by several factors that can be of both genetic and environmental origin (Lander and Schork 1994; Weiss 1994). The more prevalent of these disorders include cancer, diabetes, obesity, hypertension, and premature cardiovascular morbidity and mortality. Numerous genetic variations have been implicated as major pathogenic factors in various Mendelian disorders. However, the success in identifying the genetic factors underlying complex traits has at times been limited (Glazier et al. 2002; Hirschhorn et al. 2002). Many studies have been challenged by a presumed low effect of each genetic variant, small study populations, confounding effects such as population stratification, and, possibly, the use of highly simplistic genetic models.
Even though conditions such as diabetes harbor strong genetic components, not a single or a few single-nucleotide polymorphisms (SNPs) explain most of the genetic variance for these disorders (Risch and Merikangas 1996). It is hypothesized that much of the genetic variation may be caused by the interaction (epistasis) of multiple SNPs and interaction with environmental conditions (Cordell 2002). The disease penetrance associated with each allele is low and the impact of genetic components may vary depending on the genetic and environmental background (Carlson et al. 2004). Great progress has been achieved in the last few years using simple linear models (Cockerham and Zeng 1996; Falconer 1996; Schaid et al. 2002; Baker 2005). However, for large-scale data the methods often cannot detect interactions that are believed to have a substantial impact on the development of complex diseases (Culverhouse et al. 2004), because the models include only linear interactions and because the solutions that have been developed to address the multiple-testing problem lead to a drastic reduction in the statistical power (Cardon and Bell 2001). When modeling all the possible interactions between environmental factors and many SNPs at different loci using classical methods, the number of necessary parameters becomes very large as the number of SNPs increases (Nelson et al. 2001; Culverhouse et al. 2004). This is one of the main reasons why SNP–SNP interactions and SNP–environment interactions are rarely modeled in association studies (Carlborg and Haley 2004) even though these interactions are essential in uncovering the etiological background for complex diseases.
Some of the more successful methods for incorporating higher-order interactions are based on clustering algorithms. The combinatorial partitioning method (Nelson et al. 2001) was one of the first methods to model higher-order interactions without any main effect. This method evaluates all possible partitions and is not computationally feasible for large-scale data. The popular multifactor-dimensionality reduction (MDR) (Ritchie et al. 2001) for binary traits reduces the number of partitions by labeling the possible combinations as either high risk or low risk. The results are then evaluated through cross-validation or permutation testing, which compensates for multiple testing.
Several methods based on regression and classification trees allow for some interactions and are extremely fast (Breiman et al. 1984; Freund and Schapire 1997; Huang et al. 2004). One of these is the random forest method (Breiman 2001). This method uses a bootstrap sample of the data to construct a tree and uses the nonsampled (called “out of bag”) data for cross-validation. Multiple bootstrap samples are used to construct a forest of trees and the significance of each parameter is determined from the out-of-bag samples.
We propose using Markov chain Monte Carlo (MCMC) to overcome some of the problems described. MCMC is a stochastic computational approach that is very useful in Bayesian statistics where it can be used to estimate the posterior distribution of the parameters using Monte Carlo integration. This is achieved by generating samples from an ergodic Markov chain that has the same stationary distribution as the posterior distribution. MCMC provides a convenient method for dealing with a large parameter space when only parts of the posterior distribution are of interest. The new method explores sets of effects (risk sets) that increase the risk, or the phenotypic value, for individuals who fulfill the criterion defined by the sets. A risk set may contain one or more genetic or environmental conditions. The MCMC method then provides a probability that a particular risk set exists, i.e., that the conditions specified by the risk truly cause an increase in the phenotypic value or a higher disease risk. Methods that explore such a large range of models (multiple combinations of effects) often have very little power because they do not efficiently combine the evidence for association from different models. The new Bayesian method addresses this problem by combining information from many different models, for example, by evaluating the effect of all possible interactions when testing the effect of a single SNP. The method is described in materials and methods and the software for the method is called BAMSE and can be found at http://biostat.ku.dk/∼ande/BAMSE
MATERIALS AND METHODS
Risk sets:
In the context of this method, a risk set is a subset of the parameter space that partitions the individuals on the basis of their genotypes and environmental factors. Let Ω denote all possible sets of SNPs and environmental factors (discrete or continuous). Let the jth group of individuals whose genetic and environmental profiles fit a risk set Tj ⊂ Ω denote a potential risk group Gj. Individuals not in any of the risk groups are placed in G0. We set the upper bound, mr, on the number of risk sets so that the number of risk sets nm ∈ {0, 1,…, mr} is finite. An example of a risk set could be {weight > 90 kg, sex is male, SNP3 is a heterozygote} and the individuals who fit this description constitute a risk group. Given a quantitative trait yi for individual i ∈ Gj and assuming that the trait for every individual in a given risk group is independent and normally distributed with equal variance, the observations yi are modeled as 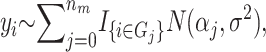 where nm is the number of risk sets, and I is the indicator function. Let
where nm is the number of risk sets, and I is the indicator function. Let 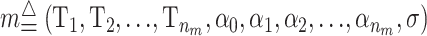 be a state from the space M. If the phenotype is binary (case–control design) the phenotype is modeled using a binomial distribution where α j is the probability for being a case in the jth risk group.
be a state from the space M. If the phenotype is binary (case–control design) the phenotype is modeled using a binomial distribution where α j is the probability for being a case in the jth risk group.
Each risk set Tj defines a genetic and environmental profile and all individuals must fit this profile to be part of the associated risk group Gj. The space of the lth component 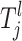 for the jth risk set is defined as Ωl and it is a finite discretization of a compact subspace of R or a finite subset of N, depending on the lth parameter. For example, a SNP parameter has seven states given that individuals who are heterozygous and homozygous for major and minor alleles are all present in the sample (see Equation A4). This corresponds to one state for each possible combination of genotypes. For the continuous-risk parameters, such as environmental factors, a threshold t can be defined that excludes individuals with observations that exceed this threshold. The space is defined as either
for the jth risk set is defined as Ωl and it is a finite discretization of a compact subspace of R or a finite subset of N, depending on the lth parameter. For example, a SNP parameter has seven states given that individuals who are heterozygous and homozygous for major and minor alleles are all present in the sample (see Equation A4). This corresponds to one state for each possible combination of genotypes. For the continuous-risk parameters, such as environmental factors, a threshold t can be defined that excludes individuals with observations that exceed this threshold. The space is defined as either 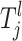 = {a: a > t} or
= {a: a > t} or 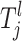 = {a: a < t}. Let the jth risk set Tj be the set of the risk parameters
= {a: a < t}. Let the jth risk set Tj be the set of the risk parameters 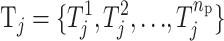 for np parameters.
for np parameters.
In some instances an individual may fit several risk sets. In this case an individual is placed in the risk group with the highest mean αi. Since not all of the observations (SNPs and environmental factors) are necessarily thought to affect the trait in a causative combination, not all of them need to define a risk set. Therefore, only some 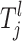 's are needed to restrict the jth risk group and the number of components, henceforth called the active risk components (for risk set j), will therefore be restricted to na ∈ {1, 2,…, ma}, where ma ≤ np. This means that a risk set can be restricted to a maximum of ma active components. If only the kth component is active for the jth risk set then
's are needed to restrict the jth risk group and the number of components, henceforth called the active risk components (for risk set j), will therefore be restricted to na ∈ {1, 2,…, ma}, where ma ≤ np. This means that a risk set can be restricted to a maximum of ma active components. If only the kth component is active for the jth risk set then 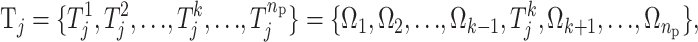 which means that only the kth component restricts the jth risk group.
which means that only the kth component restricts the jth risk group.
Priors:
The means, αj's, for the risk sets are assumed to be normally distributed with the empirical average 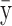 as mean and a variance calculated from the length of the range of the observed values R so that α ∼ N(ξ, κ−1), where κ is a multiple of R−2 and
as mean and a variance calculated from the length of the range of the observed values R so that α ∼ N(ξ, κ−1), where κ is a multiple of R−2 and 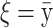 . The prior for κ is the same prior in Richardson and Green (1997).
. The prior for κ is the same prior in Richardson and Green (1997).
For the quantitative traits the variance is chosen to be uniformly distributed on (0, ∞). The priors for the distribution of the active risk components are uniformly chosen. The priors for the number of active risk components na and the number of risk sets nm are chosen to be geometrically distributed, 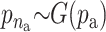 and
and 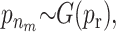 respectively. Both distributions are normalized to sum to one. The priors for components defining the SNP and environmental factors are chosen to be uniformly distributed.
respectively. Both distributions are normalized to sum to one. The priors for components defining the SNP and environmental factors are chosen to be uniformly distributed.
The Markov chain:
The chain is updated using the Metropolis–Hasting algorithm with acceptance probability
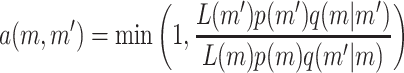 |
(1) |
of jumping from the current state m to a proposed state m′. p(m) is the prior for the mth state, q(m′ | m) is the proposal probability of proposing state m′ given the current state m, and L(m) is the likelihood for state m. The method is implemented so that the parameters are updated one at a time when possible. However, when updating the number of active components or the number of risk sets, updates of several parameters must simultaneously be proposed. A new risk set can be proposed with a random choice of parameters or a risk set can be deleted, which is sometimes called the death/birth move. Details regarding the proposal algorithm are provided in the appendix.
The posterior probability of a certain risk set Tk can be approximated as
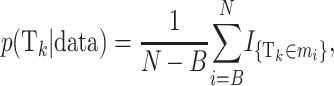 |
where B is the burn-in, N is the number of iterations, and mi is the ith sampled state. Likewise, the posterior probability for the number of risk sets being equal to x is
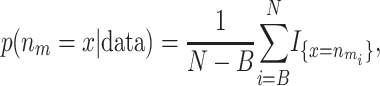 |
where 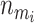 is the ith sampled number of risk sets and I is the indicator function. This probability can be used to evaluate whether there are any factors, genotypical or environmental, that affect the trait.
is the ith sampled number of risk sets and I is the indicator function. This probability can be used to evaluate whether there are any factors, genotypical or environmental, that affect the trait.
Simulations:
To evaluate the sensitivity and selectivity of the method several simulations were performed under different genetic models. Simulations were performed assuming 500 unrelated individuals with 20 uncorrelated SNPs and assuming equal variance for affected and unaffected individuals. The frequency of the minor allele was chosen to be 0.2 for all the SNPs and the mean phenotypical value for unaffected individuals was 100 with a standard deviation of 10. The same variance was chosen for the affected individuals but with varying means. However, in two genetic scenarios linkage disequilibrium (LD) was simulated using coalescent simulations based on the ms program (Hudson 2002), and the frequencies of the SNPs were thus random variables. In these simulations, a region was simulated under a neutral infinite size model assuming a crossover at rate of 1/4N0 for a 50,000-bp-long segment per generation, where N0 is the effective population size. A random selection of 20 SNPs with a minor allele frequency >0.05 was included in the nonepistatic scenario. For the epistatic scenario 20 SNPs were selected from two regions and a SNP with a minor allele frequency between 0.17 and 0.23 from each region was selected as a susceptibility SNP. Throughout, a burn-in of 10,000 iterations and a run time of 100,000 were chosen for the MCMC analysis by examining the convergence of the likelihood score and using the potential scale reduction factor (PSRF), also called the “shrinking factor”(Gelman and Rubin 1992), for the parameters that are always sampled. The shrinking factor was visualized at several intervals of the chains as recommended in Brooks and Gelman (1998). The risk parameters that were frequently, but not always, sampled were transformed into Bernoulli variables (0 for sampled and 1 for nonsampled) and then tested in the same manner as the other parameters.
To evaluate the power of the MCMC-based method compared with that of a linear model and random forests, sensitivity and selectivity were calculated for the three methods under different genetic models. The result is presented as ROC curves for general use of the MCMC method, the random forest, and a one-locus linear model. We evaluate the ability of the methods to detect any effect in the data and to identify specific susceptibility loci.
The sensitivity and selectivity for the linear model is calculated using an F-test comparing the full one-locus model to the null model of no genetic impact (three parameters vs. one parameter). For the MCMC method, the posterior probability of at least one risk set [p(nm > 0 | data)] was used to detect an effect in the data and the posterior probability of SNP i belonging to a risk set divided by the posterior probability for there being at least one risk set [p(Ti ⊂ T | data)/p(nm > 0 | data)] was used for identifying the susceptibility SNPs. The estimated increase in mean squared error was used for identifying SNPs using the random forest method and the estimated explained variance was used to detect an effect in the data. The random forest implementation in the statistical software R version 2.3.1 was used. Five hundred trees for each data set were chosen and seven variables were randomly sampled as candidates at each split. When identifying the SNPs, the susceptibility SNPs act as the true positives and the other SNPs as false positives. For detecting an effect in data, the simulations were compared with another set of simulations without any genetic effect. For the linear model the best (lowest) P-values from the two sets of simulations were compared. The same data sets were used for the three methods.
Gene–gene–environment interactions affecting serum triglycerides:
Inter99 is a population-based cohort of 6741 individuals randomly recruited using the central person registry from the western part of Copenhagen County (Glümer et al. 2003). Only individuals with Danish ancestry by self report were included. A second group of individuals consisting of type 2 diabetes patients was recruited at Steno Diabetes Center. An oral glucose tolerance test was used to determine the glucose tolerance status of each individual according to the World Health Organization (1999): normal glucose tolerant (NGT), impaired fasting glycemia (IFG), impaired glucose tolerance (IGT), and type 2 diabetes (T2D). Smoking habits were quantitated on the basis of interviews and questionnaires for the Inter99 cohort.
RESULTS
Simulations:
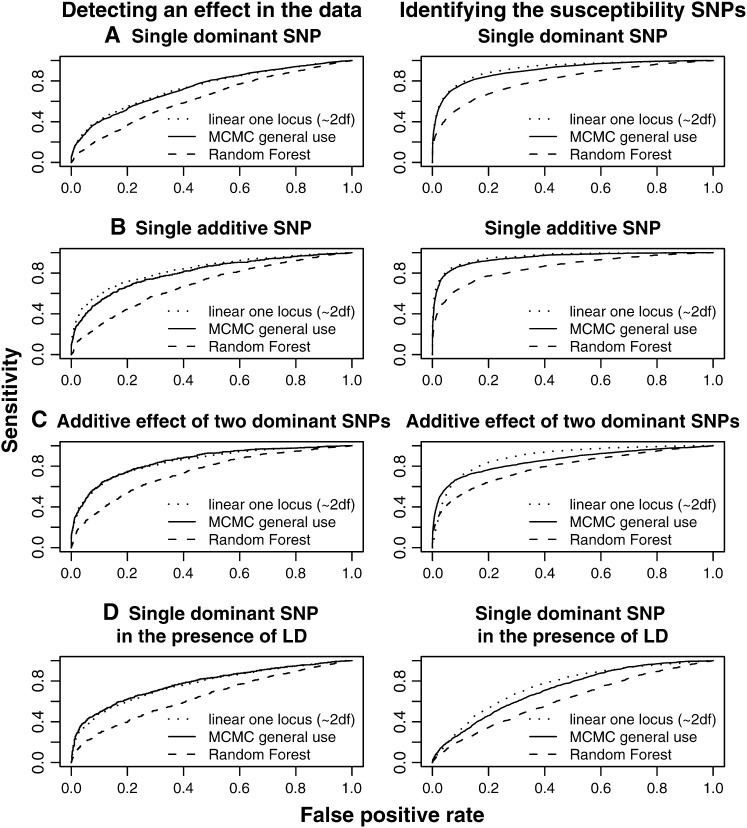
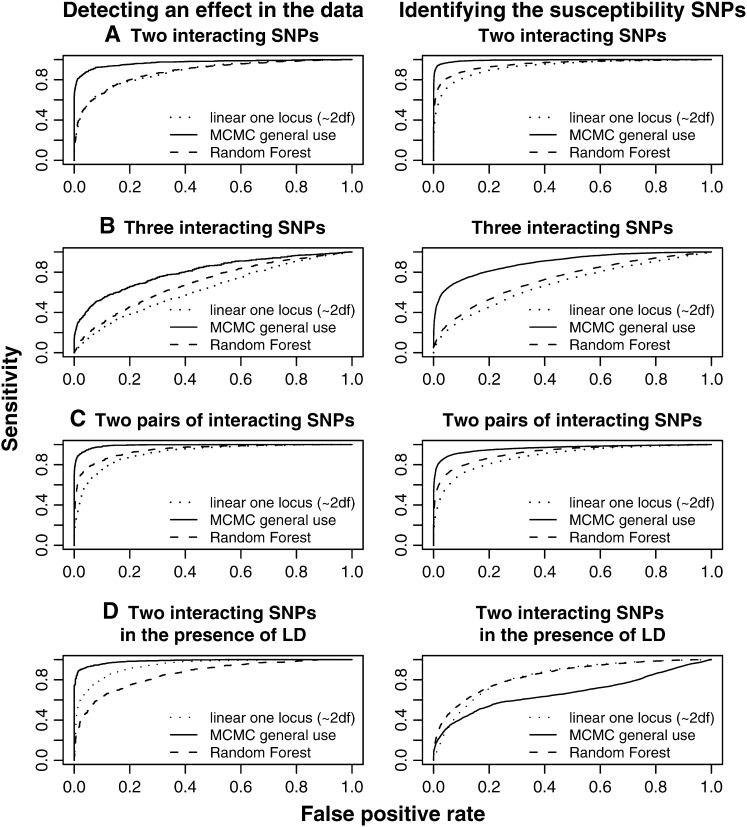
We have performed extensive simulations to evaluate the performance of the new method and compare the results with the performance of the single-locus linear model and the random forest method. Although there are many methods we could also have evaluated we have restricted ourselves to these methods because they can handle quantitative traits and are computationally feasible for the simulated data. The new MCMC method suffers only a slight reduction in power compared to the single-locus linear model assuming a single SNP with a dominant effect or an additive effect (Figure 1). This is true both for the detection of an effect in the data and for the identification of the particular SNP involved. This may be somewhat surprising given that the MCMC method allows for effects of multiple SNPs and interactions among SNPs. In contrast, the random forest method suffers a severe reduction in power compared to the linear model under these parameter settings. Similarly, under additive effects of two SNPs, the linear model and the MCMC method perform almost identically. In contrast, the random forest method has much lower power. In the presence of two interacting SNPs, or two pairs of interacting SNPs, the new MCMC method has a distinct advantage over the two other methods (Figure 2), particularly in detecting an effect. With three interacting SNPs, the effect is even more pronounced. The difference in power is manyfold at a low false-positive rate. However, in the presence of LD the advantage of the MCMC is somewhat reduced, which is due to the varying SNP frequency in the simulations.
Figure 1.—
ROC curves for the three methods in nonepistatic genetic scenarios Each genetic scenario represents 1000 simulations of 500 individuals with 20 SNPs. The prior for the MCMC method is chosen as 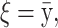
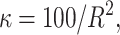 σ ∼ U(0, ∞),
σ ∼ U(0, ∞), 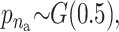 and
and 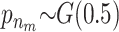 . In all scenarios unaffected individuals have a phenotype drawn from N(100, 100). (A) Affected individuals have at least one minor allele at a specific locus and have a phenotype drawn from N(102.5, 100). (B) Affected individuals have either one or two minor alleles at a specific locus and have a phenotype drawn from N(102.5, 100) or N(105, 100), respectively. (C) Affected individuals have at least one minor allele at one of two specific loci or at both loci and have a phenotype drawn from N(102.5, 100) or N(105, 100), respectively. (D) The same as in A but there is linkage between the loci.
. In all scenarios unaffected individuals have a phenotype drawn from N(100, 100). (A) Affected individuals have at least one minor allele at a specific locus and have a phenotype drawn from N(102.5, 100). (B) Affected individuals have either one or two minor alleles at a specific locus and have a phenotype drawn from N(102.5, 100) or N(105, 100), respectively. (C) Affected individuals have at least one minor allele at one of two specific loci or at both loci and have a phenotype drawn from N(102.5, 100) or N(105, 100), respectively. (D) The same as in A but there is linkage between the loci.
Figure 2.—
ROC curves for the three methods in epistatic genetic scenarios Each genetic scenario represents 1000 simulations of 500 individuals with 20 SNPs. The prior for the MCMC method is chosen as 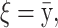
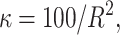 σ ∼ U(0, ∞),
σ ∼ U(0, ∞), 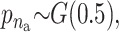 and
and 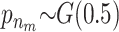 . In all scenarios unaffected individuals have a phenotype drawn from ∼N(100, 100). (A) In this simulation the affected, drawn from N(107.5, 100), are individuals carrying at least one risk allele at two specific loci. (B) In this simulation the affected, drawn from N(107.5, 100), are individuals carrying at least one risk allele at three specific loci. (C) Affected are individuals that have at least one minor allele at one of two specific combinations of two loci. There are two possible risk combinations and four risk loci. (D) The same as in A but there is linkage between the loci.
. In all scenarios unaffected individuals have a phenotype drawn from ∼N(100, 100). (A) In this simulation the affected, drawn from N(107.5, 100), are individuals carrying at least one risk allele at two specific loci. (B) In this simulation the affected, drawn from N(107.5, 100), are individuals carrying at least one risk allele at three specific loci. (C) Affected are individuals that have at least one minor allele at one of two specific combinations of two loci. There are two possible risk combinations and four risk loci. (D) The same as in A but there is linkage between the loci.
In general, the MCMC-based method outperforms the normal linear model when dealing with interactions, especially multiple combinations of interactions, while the linear model in some cases may perform slightly better when dealing with a single susceptibility SNP with additive effects. The random forest method performs better than the linear model at detecting interacting SNPs, but much worse at detecting the effect of a single locus. On the basis of a limited number of simulations the three methods seem fairly robust against phenocopy and small departures from normality (data not shown).
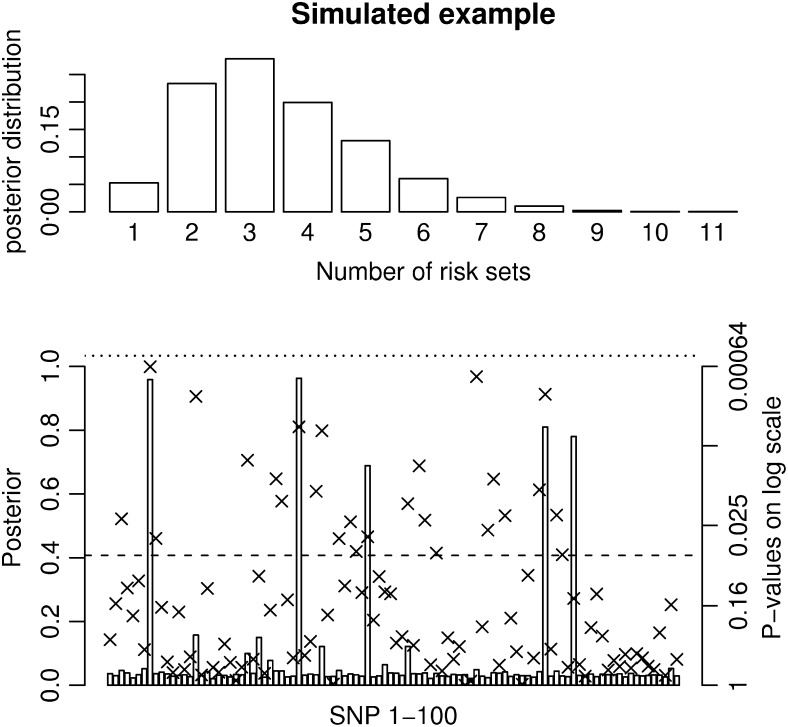
To illustrate the efficacy of the method when the number of individuals is large we simulated 100 SNPs from five regions and 5000 unrelated individuals with effect sizes so that the linear model could not achieve significance after Bonferroni correction. Phenotypes were simulated on the basis of a second- and a third-order interaction, i.e., five susceptibility SNPs. A description of the simulated data and the result from the simulation can be seen in Figure 3 and Table 1. Figure 3 and Table 1 show that the MCMC method provides very strong evidence for association [p(nm > 0) = 1] and that the method easily identifies the five susceptibility SNPs and the two specific combinations. The sampling from the stationary distribution starts within a few thousand iterations, which is <1 min on a standard PC.
Figure 3.—
Results for a simulated scenario with 100 SNPs and 5000 unrelated individuals. Five 500,000-bp-long regions were simulated using the ms program. SNPs with a minor allele frequency of <0.05 and the SNPs in high LD (r2 > 0.95) were removed. Then 20 SNPs were randomly selected from each region and one SNP from each of the five regions with a minor allele frequency between 0.17 and 0.23 was chosen as a susceptibility SNP. Phenotypes were simulated so that the individuals with at least one minor allele at SNP8 and SNP34 had a phenotype drawn from N(103, 100) and individuals with at least one minor allele at SNP46, SNP77, and SNP82 had a phenotype drawn from N(104, 100). Individuals with minor alleles at all five susceptibility SNPs had a phenotype drawn from N(107, 100) and individuals without any of the two combinations had a phenotype drawn from N(100, 100). The prior for the MCMC method is chosen as 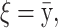
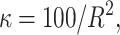 σ ∼ U(0, ∞),
σ ∼ U(0, ∞), 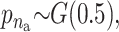 and
and 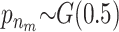 . The posterior distribution for the number of risk sets is shown at the top and the posterior probabilities for a SNP parameter being part of a risk set is shown at the bottom. Also, the P-values for the full single-locus linear model are shown as x's and the dashed and dotted lines denote P-values of 0.05 and 0.0005, respectively. The frequently sampled risk sets can be seen in Table 1.
. The posterior distribution for the number of risk sets is shown at the top and the posterior probabilities for a SNP parameter being part of a risk set is shown at the bottom. Also, the P-values for the full single-locus linear model are shown as x's and the dashed and dotted lines denote P-values of 0.05 and 0.0005, respectively. The frequently sampled risk sets can be seen in Table 1.
TABLE 1.
Frequently sampled risk sets from a simulation of 100 SNPs and 5000 individuals
| Posterior probability |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk set | SNP8 | SNP16 | SNP27 | SNP34 | SNP46 | SNP53 | SNP77 | SNP82 | N | Mean | General | Specific |
| 1 | HE/HO | HE/HO | 447–587 | 103.1 (102.3–103.9) | 0.97 | 0.72 | ||||||
| 2 | HE/HO | HE/HO | HE/HO | 317–380 | 103.7 (102.8–104.6) | 0.69 | 0.47 | |||||
| 3 | HE/HO | HE/HO | HE/HO | 196–267 | 104.2 (103.0–105.4) | 0.09 | 0.05 | |||||
| 4 | HE | 376–2053 | 100.6 (100.1–101.3) | 0.14 | 0.06 | |||||||
| 5 | HE/HO | 166–872 | 101.1 (100.2–102.1) | 0.08 | 0.04 | |||||||
N, the number of individuals in the risk group; HE, heterozygote; HO, homozygote; mean, the posterior estimate of the mean phenotype of the risk group. Results are shown for the simulated scenario described in Figure 3. Posterior probabilities were calculated as the probability of a parameter combination being part of a risk set (general) and the probability of a combination being part of a risk set excluding combinations where other parameters are also included in the risk set (specific). Large differences between the general and specific posterior probability mean that other parameters are important for this group of risk sets. For illustration of risk set grouping all combinations with a general posterior probability of >0.05 are included. Because of a low number of homozygotes some of the risk sets are very similar and are manually grouped together so that the SNP parameters (HE or HO) and (HE) were grouped together. Although this method performs no formal test for interaction, interactions are suggested when some parameters are sampled only in combination and rarely alone. The number of individuals in a risk set and the mean for each risk set are given as 90% credibility intervals. The nonsusceptibility SNPs SNP16, SNP27, and SNP53 are in LD with the true susceptibility SNPs SNP8, SNP34, and SNP46 respectably (D′ > 0.98, r2 < 0.7).
Gene–gene–environment interactions affecting serum triglycerides:
To illustrate the method we applied it to the data described in materials and methods for a −1131 T > C polymorphism in the APOA5 gene and a −250 G > A, an IVS1 + 49 C > T, and a Ser215Asn polymorphism in the LIPC gene. APOA5 and LIPC are two of several genes where common alleles, many in high linkage disequilibrium, have shown a strong association with serum lipids such as triglycerides and cholesterol (Kao et al. 2003; Lai et al. 2003; Klos et al. 2005; Oliva et al. 2005). It has been hypothesized that APOA5 interacts with lipoprotein lipases that hydrolyze the apolipoproteins. Furthermore, it has been shown that APOA5 binds to the lipases (Merkel et al. 2005). APOA5 has been shown to be expressed only in the liver (Pennacchio et al. 2001), where the hepatic lipase encoded by LIPC is also found. Hepatic lipase hydrolyzes triglycerides and has been shown to enhance the uptake of lipoproteins (Thuren 2000). The effect of the −1131 T > C polymorphism in the APOA5 promoter has shown a consistent association with serum triglycerides in various studies in different ethnic groups. Recently, a small association study (Jiang et al. 2005) indicated that plasma glucose levels may interact with an APOA5 polymorphism, giving higher plasma triglyceride levels among type 2 diabetic patients, but failed to show any interactions among nondiabetic subjects. Association studies of the APOA5 −1131 T > C variant in the Inter99 cohort have shown that the effect on serum triglyceride of the deleterious allele is modulated by other factors that affect serum triglyceride levels (G. Andersen, T. Sparsø, A. Albrechtsen, S. Castella, C. Glümer, K. Borch-Johnsen, T. Jørgensen, R. Nielsen, T. Hansen and O. Pedersen unpublished results). These factors include glucose tolerance status, gender, and smoking habits. The interactions were found using a linear model with two-way interaction terms. Using a linear model it was not possible to include all possible interaction terms among all factors. No interaction was found with the three SNPs in LIPC and the APOA5 variant using a linear model. However, since the effect of the APOA5 polymorphism is strongly modulated through glucose tolerance status, gender, and smoking habits it is possible that an association with serum triglycerides can be observed only through higher-order interactions. Therefore, the APOA5 variant, the three LIPC variants, sex, smoking habits, and glucose tolerance status were included in the MCMC method. The adjustment factors age and body mass index (BMI) were also included, assuming a linear relationship. The glucose tolerance status and the smoking habits each have four categories (NGT, IFG, IGT, and T2D and never smoked, used to smoke, occasional smoker, daily smoker, respectively). Both environmental factors were assumed to be discrete ordinal variables.
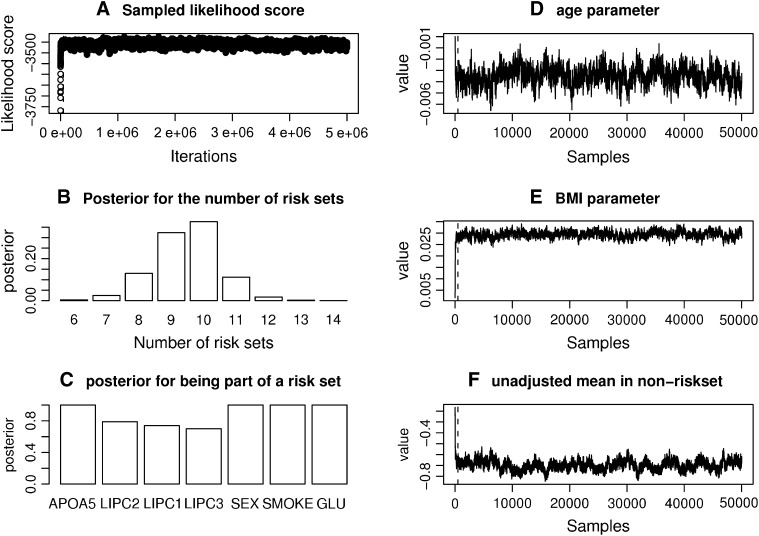
We applied the MCMC method two times on 5300 individuals without missing data from the Inter99 cohort using 5,000,000 iterations in each run. The two chains gave similar results and the parameters sampled in each iteration can be seen in Figure 4. Convergence diagnostics were performed using the method of Gelman and Rubin (1992) and the multivariate shrinking factor was <1.01. The serum triglyceride levels were logarithmically transformed and two extreme outliers were excluded (6–8 SD from the mean after transformation). The method sampled 6–14 risk sets (see Figure 4B), where 10 was the most frequent sample. All the factors used in the method were frequently sampled (see Figure 4C). Not surprisingly, the environmental factors were sampled in each iteration.
Figure 4.—
Result for the MCMC analysis of SNPs and environmental factors affecting triglyceride. A total of 5300 individuals with three SNPs and three environmental factors were tested against fasting serum triglycerides. The triglyceride levels were logarithmically transformed before testing. 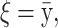
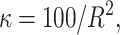 σ ∼ U(0, ∞), nm ∼ G(0.5), and na ∼ G(0.5) but <5. The total run time was 5,000,000 with a thinning factor of 100. (A) The sampled likelihood score before removing a 50,000-iteration-long burn-in. (B) The posterior distribution of the number of risk sets. Only nonempty risk sets were counted. (C) The posterior for a parameter being part of a risk set. LIPC1 is the LIPC IVS1 + 49 C > T SNP, LIPC2 is the LIPC Ser215Asn SNP, and LIPC3 is the −514 T > C variant. GLU is the glucose tolerance status. (D and E) The values of the adjustment factors age and BMI. (F) The unadjusted mean for the individuals not placed in a risk set. The burn-in is shown as a dashed line.
σ ∼ U(0, ∞), nm ∼ G(0.5), and na ∼ G(0.5) but <5. The total run time was 5,000,000 with a thinning factor of 100. (A) The sampled likelihood score before removing a 50,000-iteration-long burn-in. (B) The posterior distribution of the number of risk sets. Only nonempty risk sets were counted. (C) The posterior for a parameter being part of a risk set. LIPC1 is the LIPC IVS1 + 49 C > T SNP, LIPC2 is the LIPC Ser215Asn SNP, and LIPC3 is the −514 T > C variant. GLU is the glucose tolerance status. (D and E) The values of the adjustment factors age and BMI. (F) The unadjusted mean for the individuals not placed in a risk set. The burn-in is shown as a dashed line.
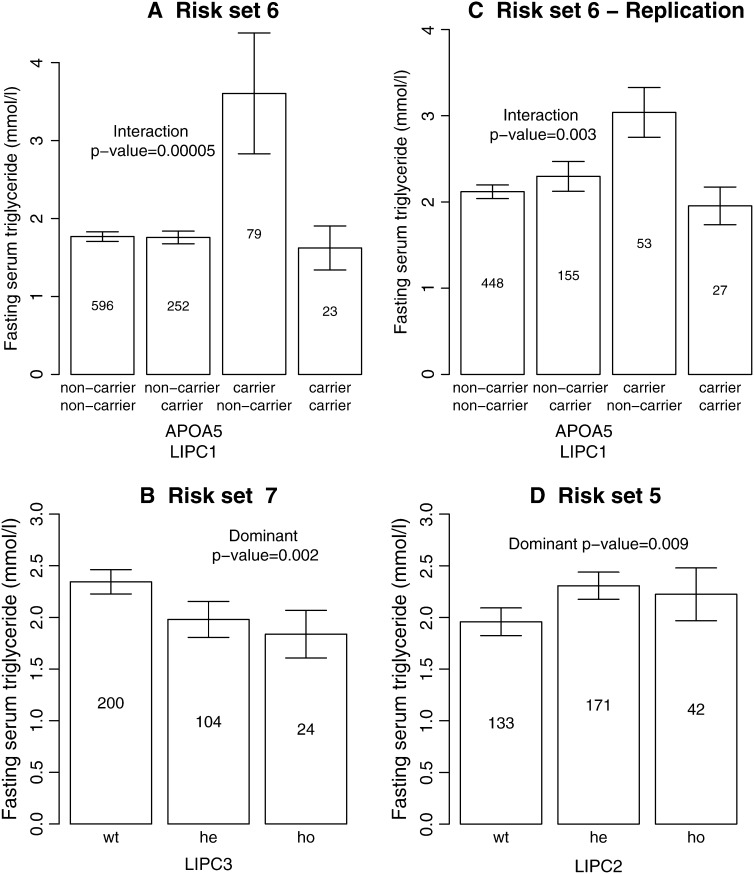
The final results can be seen in Table 2. Even though there was no main effect of the three LIPC variants, they all show an association with serum triglyceride levels in combination with both genetic and environmental factors. Using the same criteria for the risk sets as seen in Table 2, plots for the genotypes and mean serum triglyceride levels when LIPC −250 G > A, LIPC IVS1 + 49 C > T, or LIPC Ser215Asn were part of the criteria are shown in Figure 5. The presence of several factors in a risk set does not necessarily imply an interaction but could also be additive effects between the factors. One of the risk sets that indicates a strong epistatic effect is the risk set consisting of smoking nonnormal glucose tolerance status (IFG, IGT, T2D) individuals with a combination of APOA5 and LIPC IVS1 + 49 C/T alleles (see Figure 5A). The (unadjusted) P-values using a linear model for these stratifications of the data are also shown for illustration purposes. For risk set 6 we tested for epistasis between these SNPs for the smoking IFG, IGT, and type 2 diabetics using a linear model with covariates for being carriers of the minor alleles. To verify this, using standard methods, we followed up this finding by also performing the test for interactions on another cohort of 1008 IFG, IGT, and type 2 diabetic individuals consisting mostly of type 2 diabetics. A total of 683 of the individuals were smokers. The P-value of replication was 0.003 and it was the same allele combination that gave heightened serum triglyceride levels. It should be noted that most of the individuals in the follow-up group are under treatment, which may potentially influence the results.
TABLE 2.
Results of the MCMC method for gene–gene–environment interactions affecting serum triglycerides
| Posterior probability |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk set | APOA5a | LIPC2 | LIPC1b | LIPC3b | SEX | SMOKE | GLU | N | Mean | General | Specific |
| 1 | HE/HO | HO | Y | IFG/IGT/T2D | 5–9 | 2.73 (2.13–3.47) | 0.24 | 0.24 | |||
| 2 | M | IFG/IGT/T2D | 65–630 | 1.5 (1.41–1.78) | 0.86 | 0.69 | |||||
| 3 | HE/HO | WT/HO | T2D | 16–22 | 3.03 (2.62–3.51) | 0.16 | 0.13 | ||||
| 4 | HE/HO | T2D | 18–26 | 2.85 (2.47–3.3) | 0.68 | 0.58 | |||||
| 5 | HE/HO | M | Y | IGT/T2D | 91–181 | 2 (1.82–2.16) | 0.52 | 0.52 | |||
| 6 | HE/HO | WT/HO | Y | IFG/IGT/T2D | 35–74 | 2.07 (1.81–2.38) | 0.45 | 0.45 | |||
| 7 | WT/HO | M | Y | IGT/T2D | 103–210 | 2 (1.82–2.19) | 0.50 | 0.50 | |||
| 8 | HE/HO | M | 195–291 | 1.4 (1.31–1.5) | 0.98 | 0.78 | |||||
| 9 | M | Y | 266–1096 | 1.2 (1.16–1.32) | 0.98 | 0.72 | |||||
| 10 | HE/HO | IFG/IGT/T2D | 6–108 | 2.08 (1.7–2.9) | 0.15 | 0.15 | |||||
| 11 | IFG/IGT/T2D | 278–905 | 1.28 (1.2–1.44) | 0.99 | 0.96 | ||||||
| 12 | Y | T2D | 12–119 | 1.79 (1.6–2.19) | 0.20 | 0.16 | |||||
| 13 | Y | 106–1067 | 1.08 (1.01–1.54) | 1.00 | 0.91 | ||||||
| 14 | IGT/T2D | 48–178 | 1.34 (1.21–1.94) | 0.66 | 0.54 | ||||||
| 15 | M | 93–1016 | 1.1 (0.99–1.49) | 0.66 | 0.54 | ||||||
N, the number of individuals in the risk group; GLU, glucose tolerance status; WT, wild type; HE, heterozygote; HO, homozygote; M, man; Y, yes; IFG, impaired fasting glycemia; IGT, impaired glucose tolerance; T2D, screen-detected type 2 diabetes patients; Mean, the posterior estimate of the mean phenotype of the risk group. Posterior probabilities of the risk sets affecting serum triglyceride levels are shown. For explanation of the posterior probabilities see Table 1.
The SNP parameters (HE or HO) and (HE) were grouped together because they are never sampled in the same iteration (very few homozygotes).
The SNP parameters (WT or HO) and (WT) were likewise grouped together. The mean number of individuals not in a risk set is 1236. All combinations with an average general posterior probability of >0.1 are included.
Figure 5.—
Bar plots for some of the risk sets with high posterior probability. The mean serum triglyceride levels, with standard error bars, distributed on the genotypes are shown. The cohort is stratified according to the environmental factors for the risk sets in Table 2. Due to individuals belonging to two or more risk groups, the number of individuals in the bar plot might differ from the risk sets in Table 2. The numbers in the bars represent the number of individuals with this genotype.
DISCUSSION
The MCMC approach appears to have overcome many of the major difficulties in modeling higher-order interactions in large-scale population-based data. For many SNPs no method can explore all of the possible interactions. However, the MCMC method uses the marginal effects for a low-order interaction to find the higher-order interactions. For example, if a fifth-order interaction exists in the data, then the phenotypical mean of the group of individuals with three of the five factors will probably be different from that for the rest of the individuals. The Markov chain will then spend more time in this state and by exploring the “local” area will find the fifth-order interaction (see appendix for updates of the Markov chain).
A related fully Bayesian MCMC method, called BAMA, has recently been developed (Kilpikari and Sillanpaa 2003). However, there are some important differences between our method and BAMA. The BAMA method assumes no interactions among loci, that all alleles have different effects, and that the effects from each locus are additive. Another related method is the Monte Carlo version of logic regression (Kooperberg and Ruczinski 2005). This method uses Boolean expressions to model covariates in a linear model and thus allows for higher-order interactions. Monte Carlo logic regression uses maximum-likelihood estimates for the coefficients in the model. The current MCMC method models all multiple combinations of SNPs and does not assume any linear relationship between the effects of any combination of SNPs and environmental factors. However, our method does allow adjustment factors to be included in the model if a linear relationship is assumed. We chose to compare our method with random forest because it does not make any linear assumptions.
Computational speed:
The speed of the algorithm is highly dependent on the number of individuals and the number of sampled risk sets. For the simulated data, where the number of sampled risk sets is rather low and the number of individuals is few (500), application of the method takes <1 min, while under the larger simulation condition (5000 individuals, 100 SNPs) each replicate takes a few minutes. When the method was applied on the Inter99 sample, the MCMC method found multiple risk sets defined by up to four factors, which could not be accomplished using most other methods. For example, modeling all possible interactions using a standard linear model would entail including 2592 parameters in the model.
Power:
The main advantage of the MCMC method is that it allows multiple effects simultaneously, thereby reducing the variance and allowing information to be pooled among effects. This gives the method more power than the linear models in most genetic scenarios when dealing with multiple susceptibility SNPs. Simulations showed that the MCMC can have more than twice the power of the linear model to detect two two-way interactions when the average number of false positives is low (<1). When there are interactions present in the data, then many combinations of SNPs will, to some extent, be associated with the trait even without any main effect. However, when compensating for multiple testing only information from the most strongly associated combination is used. The MCMC method combines information from many SNPs and risk sets in the calculation of the posterior probability and, thus, if many combinations are associated, the MCMC method compensates for multiple testing at a much lower cost.
While no method can entirely circumvent the problem of “the curse of dimensionality” (Bellman 1961), our simulations show that the new method is feasible for at least 100 SNPs. Although it may not be worthwhile to entertain the possibility of three-way or four-way interactions in data sets with thousands of SNPs (there are 1011 possible three-way interactions for 10,000 SNPs), the method can still be efficiently applied to such large data sets if the state space is constrained to exclude high-order interactions.
The new method is too slow for modeling, for example, 500,000 SNPs even without interactions. Nonetheless, while we see our method as most suitable for candidate gene studies, we also note that application in large-scale genomewide studies is possible. For these studies, we recommend using the method by applying it independently to different genic regions, in addition to using standard methods. If multiple SNPs within the same genic region have an effect, such an application of the method should greatly increase the mapping power compared to methods that analyze each SNP separately.
Priors and assumptions:
The MCMC method seems to perform well under a range of different genetic models and does not assume the same genetic model for the different risk sets. The factors used in the method can be either biallelic SNPs or environmental risk factors that can be continuous or discrete ordinal. All environmental factors are treated as binary traits, but the threshold that divides the data need not be defined in advance. The environmental factor can be included in two ways, either as the measured values or transformed to the sorting order of the values. By using the latter, the prior for the number of individuals included or excluded from a risk set is then uniformly distributed where, if the actual values were used, the prior for the threshold would be uniformly distributed in the range of the values of the environmental factor. The priors for the mean of each risk set (η, κ) can have a great effect on the posterior distribution of the number of risk sets (see, for example, Richardson and Green 1997 with discussion). If the mean is chosen as the midrange of the trait instead of the empirical average, the MCMC method is likely to predict a higher number of risk sets. This, however, is highly dependent on κ (the prior variance for the means) or the multiple of κ, where a small multiple gives a rather flat prior distribution.
Caveats:
Label switching, where different risk sets partition the data in the same way, is a potential problem in this MCMC method. Defining the risk sets to have a higher mean than the set of individuals not in a risk set ∀i: αi > α0 largely eliminates this problem. However, a label-switching problem can still remain. For example, a risk group might contain all or none of the individuals, which means the data are partitioned in the same way as if there were no risk sets. This problem can be addressed in the calculation of posterior probabilities by appropriate editing of the MCMC output data, and it is, in any case, partially alleviated by assigning low prior probability to states with a high number of risk sets. In our implementation of the method, the number of nonempty risk sets is calculated.
It is important to note that the posterior probabilities estimated using the MCMC method do not have a frequentist interpretation. For example, in repeated simulations without a genetic effect, a posterior probability of >0.4, for there being at least one risk set, was virtually never observed. Practitioners desiring to report a frequentist P-value can do so by applying the method in combination with a standard permutation procedure. For small data sets, similar to the data sets simulated in this article, such permutations are easily completed on a single stand-alone computer. Permutation testing on large and complex data sets would require access to a cluster of processors.
The MCMC method can detect a range of different genetic effects, whether they be main effects, epistatic, gene–environment, or a mixture of them. In most genetic scenarios with multiple causative factors, the method has equal or more power to detect an effect and identify the causal combination of genetic factors than the more conventional linear model. The method can model higher-order interactions and find significant reproducible combinations of both genetic and environmental factors that influence serum triglyceride levels.
Acknowledgments
We thank Charlotte Glümer, Knut Borch-Johnsen, and Torben Jørgensen for generously supplying the Inter99 data. This work was supported by the Danish research counsel and by National Institutes of Health grants NIGMS R01HG003229, U01HL084706.
APPENDIX
Pseudocode:
The algorithm can be written in pseudocode as
runtime=N
while (runtime – –){
update SNP parameters
update environmental parameters
update a mean for a random risk set
update adjustment factors
update the number of active risk components
update the positions of the active risk component
update the missing genotypes
update a mean for a random risk set
update the number of risk sets using delete or create
mean_update=5
while(mean_update – –){
update of variance
update a mean for a random risk set
}
sample the parameters
}
where N is the number of iterations. In each update a proposal update is either accepted or rejected.
Update of means:
Since a reasonable choice for a mean should be no higher than the maximum phenotypic value maxp and no lower than the minimum phenotypic value minp, the parameter space for a mean is defined as [minp, maxp]. The proposed mean 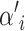 of risk set i is sampled either from N(ξ, κ−1) or from U(minp, maxp). The latter is used when the acceptance rate of the updates falls below a specified threshold. This may typically happen when the Markov chain visits states with many risk sets.
of risk set i is sampled either from N(ξ, κ−1) or from U(minp, maxp). The latter is used when the acceptance rate of the updates falls below a specified threshold. This may typically happen when the Markov chain visits states with many risk sets.
The acceptance probability for this update is
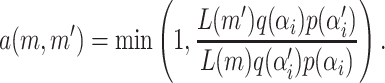 |
(A1) |
To avoid some of the problems with label switching the risk sets 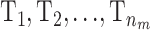 can be chosen to have a higher mean than T0. When updating the mean for one of the risk sets it is restricted to [α0, maxp] and T0 is restricted to
can be chosen to have a higher mean than T0. When updating the mean for one of the risk sets it is restricted to [α0, maxp] and T0 is restricted to 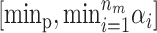 .
.
Update of the variance:
The variance is updated by simulating a uniform U(σ − w, σ + w) random variable, where w is some specified constant. If the proposed value, σ′, is outside (0, ∞), i.e., less than zero, it is mirrored back into the space
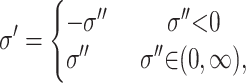 |
(A2) |
where σ′′ is the unmirrored proposal variance sampled from U(σ − w, σ + w). This ensures that q(m | m′) = q(m′ | m) and reversibility.
The acceptance probability for this update is
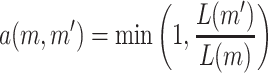 |
(A3) |
because the prior densities are uniformly distributed.
Update of adjustment factors:
This update is performed similarly to the update of the variance, but without any restrictions in R.
Update of risk parameters:
There are seven different possible partitions, excluding the empty space, of the discrete space of the SNP risk parameters, when there are three possible one-locus genotypes,
 |
(A4) |
where partition 7 allows all genotypes. Updates are proposed by sampling from their uniform prior density U{1, 2, 3,…, 6} and the acceptance probability is the same as (A3). The updates for being active are proposed separately.
The environmental parameters allow the individuals to enter risk sets if they have environmental values that are below or higher than a given threshold 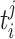 . The ith environmental parameter takes values in the interval between the observed minimum and maximum environmental values. Again the proposals are sampled from their uniform prior density and the acceptance probability becomes (A3).
. The ith environmental parameter takes values in the interval between the observed minimum and maximum environmental values. Again the proposals are sampled from their uniform prior density and the acceptance probability becomes (A3).
Updating the number of active risk components:
In the presence of many environmental and genetic factors, a random risk set is likely to result in an empty risk group. Therefore, it will in most cases lead to significant savings of computational time to define an upper limit for the number of components that are active in a risk set. The maximum number of active components in a risk set, ma, can be defined by the user.
The probability of deactivating a given active risk component for risk set i when there currently are 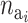 active components is defined as 1/
active components is defined as 1/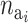 and the probability of activating a given inactive risk component is
and the probability of activating a given inactive risk component is 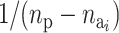 . The acceptance probability for deactivating a risk component becomes
. The acceptance probability for deactivating a risk component becomes
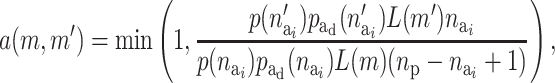 |
(A5) |
where p(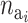 ) is the prior for the number of active components in the ith risk set and
) is the prior for the number of active components in the ith risk set and 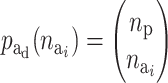 is the prior for the distribution of the active components.
is the prior for the distribution of the active components.
The acceptance probability for activating a risk component is
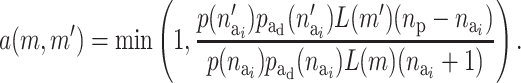 |
(A6) |
Updating the active risk component:
Risk sets are also updated by simultaneously proposing a deactivation of one component and an activation of another component. The components chosen to be activated or deactivated are chosen with equal probability. Because the priors are also uniformly distributed, the resulting acceptance probability is given by (A3).
Updating the number of risk sets:
To allow different combinations of genotypic and phenotypic factors to have different effects, we allow multiple risk sets with different means. Using reversible jumps (Green 1995), the Markov chain can jump between parameter spaces of different dimensionality. To ensure reversibility, a random component c is used so that the mapping from m to m′ is one-to-one. The mapping, when creating a new risk set, is defined as
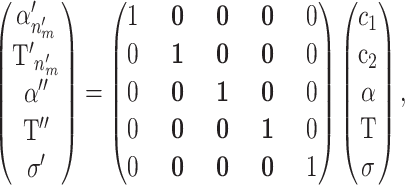 |
(A7) |
where 1 is the identity matrix, 0 is a zero matrix, 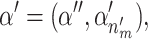 and
and 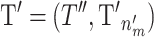 . The Jacobian for this mapping is the identity matrix and thus does not need to be included in the acceptance probability.
. The Jacobian for this mapping is the identity matrix and thus does not need to be included in the acceptance probability.
The number of active risk components in the new risk set 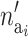 is uniformly proposed from {1, 2,…, ma}, and the positions of each active risk component are also chosen with equal probability. When proposing a new risk set the mean of the new risk set, the SNPs, and the environmental factors are sampled from their prior density. The acceptance probability then reduces to
is uniformly proposed from {1, 2,…, ma}, and the positions of each active risk component are also chosen with equal probability. When proposing a new risk set the mean of the new risk set, the SNPs, and the environmental factors are sampled from their prior density. The acceptance probability then reduces to
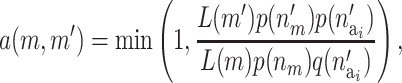 |
(A8) |
where 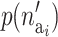 is the prior for the number of active components in the proposed risk set (i) and
is the prior for the number of active components in the proposed risk set (i) and 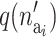 is the probability of proposing this number.
is the probability of proposing this number.
Updating missing genotypes:
The priors for the missing genotypes are calculated on the basis of the frequencies of the observed genotypes, pj, at locus j and assuming Hardy–Weinberg equilibrium
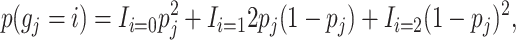 |
(A9) |
where i is the number of minor alleles and gj is the state of the genotype.
All the missing genotypes for one SNP are updated at the same time and proposed from the prior distribution so that the acceptance probability is given by (A3).
The priors for the genotypes can also be specified for each individual, which can be very efficient when the SNPs are in LD. These priors can be estimated, for example, using the posterior estimates from the fastPHASE software (Scheet and Stephens 2006).
References
- Baker, S. G., 2005. A simple loglinear model for haplotype effects in a case-control study involving two unphased genotypes. Stat. Appl. Genet. Mol. Biol. 4 14. [DOI] [PubMed] [Google Scholar]
- Bellman, R. E., 1961. Adaptive Control Processes. Princeton University Press, Princeton, NJ.
- Breiman, L., 2001. Random forest. Mach. Learn. 45 5–32. [Google Scholar]
- Breiman, L., J. H. Friedman, R. A. Olshen and C. J. Stone, 1984. Classification and Regression Trees, Ed. 1. Wadsworth, Belmont, CA.
- Brooks, S. P., and A. Gelman, 1998. General methods for monitoring convergence of iteractive simulations. J. Comp. Graph. Stat. 7 434–455. [Google Scholar]
- Cardon, L. R., and J. I. Bell, 2001. Association study designs for complex diseases. Nat. Rev. Genet. 2 91–99. [DOI] [PubMed] [Google Scholar]
- Carlborg, Ö., and C. S. Haley, 2004. Epistasis: too often neglected in complex trait studies. Nat. Rev. Genet. 5 618–625. [DOI] [PubMed] [Google Scholar]
- Carlson, C. S., M. A. Eberle, M. J. Rieder, Q. Yi, L. Kruglyak et al., 2004. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am. J. Hum. Genet. 74 106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerham, C. C., and Z. B. Zeng, 1996. Design III with marker loci. Genetics 143 1437–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell, H. J., 2002. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Hum. Mol. Genet. 11 2463–2468. [DOI] [PubMed] [Google Scholar]
- Culverhouse, R., T. Klein and W. Shannon, 2004. Detecting epistatic interactions contributing to quantitative traits. Genet. Epidemiol. 27 141–152. [DOI] [PubMed] [Google Scholar]
- Falconer, M., 1996. Introduction to Quantitative Genetics, Ed. 4. Longman, New York.
- Freund, Y., and R. E. Schapire, 1997. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 55 119–139. [Google Scholar]
- Gelman, A. F., and D. Rubin, 1992. Inference from iterative simulation using multiple sequences (with discussion). Stat. Sci. 7 457–511. [Google Scholar]
- Glazier, A. M., J. H. Nadeau and T. J. Aitman, 2002. Finding genes that underlie complex traits. Science 298 2345–2349. [DOI] [PubMed] [Google Scholar]
- Glümer, C., T. Jorgensen and K. Borch-Johnsen, 2003. Prevalences of diabetes and impaired glucose regulation in a Danish population: the Inter99 study. Diabetes Care 26 2335–2340. [DOI] [PubMed] [Google Scholar]
- Green, P. J., 1995. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika 82 711–732. [Google Scholar]
- Hirschhorn, J. N., K. Lohmueller, E. Byrne and K. Hirschhorn, 2002. A comprehensive review of genetic association studies. Genet. Med. 4 45–61. [DOI] [PubMed] [Google Scholar]
- Huang, J., A. Lin, B. Narasimhan, T. Quertermous, C. A. Hsiung et al., 2004. Tree-structured supervised learning and the genetics of hypertension. Proc. Natl. Acad. Sci. USA 101 10529–10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18 337–338. [DOI] [PubMed] [Google Scholar]
- Jiang, Y. D., C. J. Yen, W. L. Chou, S. S. Kuo, K. C. Lee et al., 2005. Interaction of the G182C polymorphism in the APOA5 gene and fasting plasma glucose on plasma triglycerides in type 2 diabetic subjects. Diabet. Med. 22 1690–1695. [DOI] [PubMed] [Google Scholar]
- Kao, J.-T., H.-C. Wen, K.-L. Chien, H.-C. Hsu and S.-W. Lin, 2003. A novel genetic variant in the apolipoprotein A5 gene is associated with hypertriglyceridemia. Hum. Mol. Genet. 12 2533–2539. [DOI] [PubMed] [Google Scholar]
- Kilpikari, R., and M. J. Sillanpaa, 2003. Bayesian analysis of multilocus association in quantitative and qualitative traits. Genet. Epidemiol. 25 122–135. [DOI] [PubMed] [Google Scholar]
- Klos, K. L. E., S. Hamon, A. G. Clark, E. Boerwinkle, K. Liu et al., 2005. APOA5 polymorphisms influence plasma triglycerides in young, healthy African Americans and whites of the CARDIA study. J. Lipid Res. 46 564–571. [DOI] [PubMed] [Google Scholar]
- Kooperberg, C., and I. Ruczinski, 2005. Identifying interacting SNPs using Monte Carlo logic regression. Genet. Epidemiol. 28 157–170. [DOI] [PubMed] [Google Scholar]
- Lai, C.-Q., E.-S. Tai, C. E. Tan, J. Cutter, S. K. Chew et al., 2003. The APOA5 locus is a strong determinant of plasma triglyceride concentrations across ethnic groups in Singapore. J. Lipid Res. 44 2365–2373. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., and N. J. Schork, 1994. Genetic dissection of complex traits. Science 265 2037–2048. [DOI] [PubMed] [Google Scholar]
- Merkel, M., B. Loeffler, M. Kluger, N. Fabig, G. Geppert et al., 2005. Apolipoprotein AV accelerates plasma hydrolysis of triglyceride-rich lipoproteins by interaction with proteoglycan-bound lipoprotein lipase. J. Biol. Chem. 280 21553–21560. [DOI] [PubMed] [Google Scholar]
- Nelson, M. R., S. L. Kardia, R. E. Ferrell and C. F. Sing, 2001. A combinatorial partitioning method to identify multilocus genotypic partitions that predict quantitative trait variation. Genome Res. 11 458–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva, C. P., L. Pisciotta, G. Li Volti, M. P. Sambataro, A. Cantafora et al., 2005. Inherited apolipoprotein A-V deficiency in severe hypertriglyceridemia. Arterioscler. Thromb. Vasc. Biol. 25 411–417. [DOI] [PubMed] [Google Scholar]
- Pennacchio, L. A., M. Olivier, J. A. Hubacek, J. C. Cohen, D. R. Cox et al., 2001. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science 294 169–173. [DOI] [PubMed] [Google Scholar]
- Richardson, S., and P. J. Green, 1997. On Bayesian analysis of mixtures with an unknown number of components (with discussion). J. R. Stat. Soc. Ser. B 59 731–792. [Google Scholar]
- Risch, N., and K. Merikangas, 1996. The future of genetic studies of complex human diseases. Science 273 1516–1517. [DOI] [PubMed] [Google Scholar]
- Ritchie, M. D., L. W. Hahn, N. Roodi, L. R. Bailey, W. D. Dupont et al., 2001. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am. J. Hum. Genet. 69 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid, D. J., C. M. Rowland, D. E. Tines, R. M. Jacobson and G. A. Poland, 2002. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 70 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet, P., and M. Stephens, 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78 629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuren, T., 2000. Hepatic lipase and HDL metabolism. Curr. Opin. Lipidol. 11 277–283. [DOI] [PubMed] [Google Scholar]
- Weiss, K. M., 1994. Genetic Variation and Human Disease. Cambridge University Press, Cambridge/London/New York.
- World Health Organisation, 1999. Definition, diagnosis and classification of diabetes mellitus and its complications: report of a WHO consultation. Part 1: diagnosis and classification of diabetes mellitus. Technical Report. World Health Organisation, Geneva.