Abstract
LoxP-Cre technology was used to remove the selenocysteine tRNA gene, trsp, in either endothelial cells or myocytes of skeletal and heart muscle to elucidate the role of selenoproteins in cardiovascular disease. Loss of selenoprotein expression in endothelial cells was embryonic lethal. A 14.5-day-old embryo had numerous abnormalities including necrosis of the central nervous system, subcutaneous hemorrhage and erythrocyte immaturity. Loss of selenoprotein expression in myocytes manifested no apparent phenotype until about day 12 after birth. Affected mice had decreased mobility and an increased respiratory rate, which proceeded rapidly to death. Pathological analysis revealed that mice lacking trsp had moderate to severe myocarditis with inflammation extending into the mediastinitis. Thus, ablation of selenoprotein expression demonstrated an essential role of selenoproteins in endothelial cell development and in proper cardiac muscle function. The data suggest a direct connection between the loss of selenoprotein expression in these cell types and cardiovascular disease.
Keywords: Cardiovascular disease, Conditional knockout, Endothelial cells, Heart muscle, Selenocysteine, Selenocysteine tRNA, Selenoproteins
1. Introduction
There are 24 known selenoproteins in rodents and 25 in humans [1]. Some, but not all of these selenoproteins, have an essential role in mammalian development. The targeted removal of different selenoprotein genes in mice have shown that glutathione peroxidase-4 (GPx4) [2], selenoprotein P [3,4], thyroid hormone deiodinase 2 [5–8], cytosolic thioredoxin reductase [9] and mitochondrial thioredoxin reductase [10] appear to have essential roles, since their knockout is lethal or results in abnormal phenotypic changes. On the other hand, glutathione peroxidase-1 (GPx1) [11] and glutathione peroxidase-2 [12] appear to have non-essential roles, since their knockout results in little or no phenotypic change. Those selenoproteins that are non-essential to the life of the animal may provide protection from environmental stress, since such animals do not cope as well as their wild type counterparts to certain traumas [13,14]. Interestingly, many selenoproteins that apparently have a role in stress-related phenomena appear to be dependent on the presence of a 2′-O-methyl group on the ribosyl moiety at position 34 (designated Um34) in selenocysteine (Sec) tRNA[Ser]Sec [15]. That is, rescue of the selenoprotein population in a standard knockout of the Sec tRNA[Ser]Sec gene (trsp) with a mutant transgene whose product cannot synthesize Um34 results in the animal’s failure to express several selenoproteins including GPx1, glutathione peroxidase-3, SelR, SelT and SelW. The rescued animals appear to be phenotypically normal with the exception of reduced fertility in males and a reduced litter size in females.
A unique feature of selenoproteins is their absolute dependence on the presence of Sec tRNA[Ser]Sec for their synthesis. Removal of trsp from the mouse genome is embryonic lethal [16,17] resulting in death within the first few days post-coitum (dpc) [16]. These data further demonstrate that selenoproteins have a major role in mammalian development. However, the mechanism of how and when selenoprotein expression is required in the development of various tissues and organs is largely unknown. Generation of a conditional knockout of trsp using loxP-Cre technology [17] has provided an important tool for carrying out a more detailed analysis of the role of selenoproteins in specific tissue and organ development. For example, targeted removal of trsp from mammary epithelium has shown that selenoprotein expression is altered when Cre is under the control of the mouse mammary tumor virus long terminal repeat promoter, but that there were no observed phenotypic changes [17]. Targeted removal of trsp from adult liver, however, demonstrated an essential requirement for selenoprotein expression in liver function following complete removal of the synthesis of this selenium-containing protein class [18]. These studies prompted us to examine the role of selenoproteins in the development of other tissue types by perturbing their expression using loxP-Cre technology to selectively remove trsp and to provide models for examining the role of selenoproteins in disease prevention.
In the present study, we have examined the targeted removal of floxed trsp with Cre recombinase in two different cell types. These studies were designed to elucidate the role of selenoproteins in cardiovascular disease since both cell types are directly related to heart development and function. In one cell type, Cre recombinase is under the control of the Tie2 promoter that is highly specific to endothelial cells [19,20], and in the other, the recombinase is under the control of the muscle creatine kinase (MCK) promoter that is highly specific to skeletal and cardiac muscle [21]. Removal of trsp in endothelial cells resulted in embryonic death, and the affected fetuses had multiple abnormalities compared to their wild type siblings at 14.5 dpc. Removal of trsp in skeletal and heart muscle resulted in abrupt death at about 12 days after birth and the abnormality appeared to be a result of the loss of selenoprotein expression in cardiac muscle. The results of these observations are described herein.
2. Materials and methods
2.1. Mice
Floxed trsp (designated trspfl) mice in strain C57BL6 have been described [17]. Heterozygous Tie2Cre mice in strain C57BL6 wherein the Cre recombinase is driven by the endothelial-specific promoter/enhancer metallothionein-1 poly A signal sequence [20] were obtained from Frank Gonzalez at NCI/NIH with permission from M. Yanagisawa. MCKCre mice in strain FVB wherein the Cre recombinase is driven by the muscle specific promoter/enhancer of the mck gene [21] were from Taconic, USA with permission from C.R. Kahn.
2.2. Matings, embryos and offspring
Male mice that were heterozygous for either Cre gene (designated Tie2Cre+/−or MCKCre+/−) were bred to females that were homozygous for floxed trsp (designated trspfl/fl). Male offspring that were trspfl/+-Tie2Cre+/−or trspfl/+-MCKCre+/−were bred to females that were trspfl/fl. Carrying out breedings in this manner assured us of not having homozygous Cre offspring as mice encoding both copies of either Tie2Cre [20] or MCKCre [21] produce a characteristic phenotype. Since offspring with genotype trspfl/fl-Tie2Cre+/−were suspected of being embryonic lethal, females were monitored for the presence of a vaginal plug (VP) in order to determine the first day of gestation. Those females manifesting a VP on the first morning after mating were designated as dpc 1. Embryos at dpc 10.5, 12.5, 14.5, 16.5, 18.5 wereexamined by euthanizing dams with CO2 (dams carrying embryos greater than 14 days of age were anesthetized with Avertin IP prior to euthanasia), and the ventral abdomen was then opened to retrieve the uterine horn with implanted embryos. Embryos were placed in sterile 1×PBS and washed in the same solution to remove cellular debris. Prior to fixation, the limb of the fetus was removed for genotyping. Fetuses were fixed in Bouin’s solution and then transferred to 70% ethanol.
Pups that died suddenly on or about day 12 from matings between male trspfl/+-MCKCre+/−and female trspfl/fl mice were fixed in 10% neutral-buffered formalin within 5 min following death after opening the abdomen and thoracic and cranial cavities. Comparableaged siblings were euthanized and fixed in formalin following the same procedures and used as controls. Tails of 6–8 days old mice were clipped for isolating genomic DNA to determine genotypes. Necropsy and histopathologic evaluation were performed by a veterinary pathologist (GFM), board certified by the American College of Veterinary Pathologists. Tissues were routinely processed and embedded in paraffin. Five-micron-thick sections were prepared and stained with hematoxylin and eosin (H&E). Photomicrographs were taken using an Axiophot (Zeiss) microscope, and a Coolsnap HQ (Photometrics) digital camera.
Animal care was in accordance with the National Institutes of Health guidelines.
2.3. Genotyping by PCR
Genotypes of embryos from matings between trspfl/+-Tie2Cre+/−males and trspfl/fl females and of 6–8 days offspring from matings between trspfl/+-MCKCre+/−males and trspfl/fl females were analyzed by PCR using genomic DNA isolated from limbs and tails, respectively. Tie2Cre [20] and MCKCre transgenes [21] were detected by PCR as described. Genotyping of the trsp region was preformed by PCR as previously described [17].
2.4. Embryo processing/sectioning
Fixed 14.5 gestation day old embryos were processed for routine light microscopic evaluation (Histoserv, Inc., Germantown, MD) in paraffin block for sagittal sections. Five-micron-thick step sections, taken at 50 μm intervals, were cut on a microtome, and stained with H&E. Sections were examined using an Olympus BX41 microscope. Photomicrographs were taken as described above.
2.5. Neonatal processing/sectioning
Six day old, 10 day old and about 12 day old freshly deceased mutant, neonatal mice and the corresponding age-matched controls that were from matings between male trspfl/+-MCKCre+/−and female trspfl/fl mice were dissected by a veterinary pathologist (GFM), and organs were embedded in paraffin, sectioned 5- μm-thick, stained with H&E, coverslipped, and examined using an Olympus BX41 microscope. In addition, to detect occurrence of muscle fibrosis and deposition of amyloid in skeletal muscle tissue, Massons’s Trichrome and Congo red staining were carried out, respectively. The following organs were examined: heart, brain, spinal cord, liver, spleen, kidney, lung, thymus and skeletal muscle.
3. Results
3.1. Targeted removal of trsp in endothelial cells
3.1.1. Phenotypes and genotypes
Examination of the offspring from matings between trspfl/+-Tie2Cre+/−male and trspfl/fl female parents revealed that none of the pups were trspfl/fl-Tie2Cre+/−. We therefore turned our attention to the embryonic stages. Embryos were retrieved at different periods of gestation. A total of 10 Tie2Cre knockout embryos and an equal number of littermate controls were analyzed. Two mice of E 10.5, three of E 12.5, three of E 14.5 and one each of E 16.5 and E 18.5 were examined. At day 10.5, the embryos were small and no phenotypic or pathological differences between wild type and knockout siblings were apparent. By day 12.5, some embryos appeared to be developing more slowly, but no significant differences were found between these and wild type embryos by pathological analysis (see below). However, by day 14.5 dpc, striking differences were found among embryos (see 14.5 dpc siblings in Fig. 1A). Affected E 14.5 embryos were smaller in size, more fragile, had a poorly developed vascular system, underdeveloped limbs and tails and smaller heads than their normal siblings. The skull of affected embryos was malformed as can be visualized in the figure. By day 16.5, affected embryos began undergoing major disintegration, and by day 18.5, they had been virtually reabsorbed.
Fig. 1.

Phenotypes and genotypes of embryos with and without trsp in endothelial cells. In (A), 14.5 day old embryos lacking trsp (left panel) or encoding trsp (right panel) were photographed (10×magnification). In (B), DNA was extracted from the limbs of embryos, digested and analyzed with either the probe for trsp (upper panel) or Tie2Cre (lower panel) as given in Section 2. The 1.1 bp fragment contains trspfl/fl while the 900 bp fragment encodes trsp. The 100 bp fragment indicates the presence of Cre.
The genotypes of affected embryos and their siblings were determined (Fig. 1B). All E 14.5 embryos homozygous for trspfl/fl and heterozygous for Tie2Cre+/−exhibited the same pathological phenotype. As Tie2Cre was not expressed in the limb tissue (which is composed of less than 2% endothelial cells) and only trsp was targeted for removal in endothelial cells, the genotype shown in the figure contained intact trspfl/fl. Normal (control) embryos that were used for pathological analysis were heterozygous for floxed trsp and Tie2Cre (Fig. 1B) or were heterozygous only for floxed trsp (data not shown). No phenotypic differences were observed among control embryos even though their genotypes were different. Pathological analysis was then carried out on affected (trspfl/fl-Tie2Cre+/−) and unaffected embryos (either trspfl/+-Tie2Cre+/−or trspfl/+ [see below]).
3.2. Pathological analysis
Three each of 12.5 and 14.5-gestation-day-old embryos, trspfl/fl-Tie2Cre+/−, trspfl/+ and trspfl/+-Tie2Cre+/−, were examined microscopically. Overall, in the case of E 12.5 embryos there were no significant differences found in the various tissues and organs examined, including brain (Fig. 2A), spinal cord (Fig. 2B), subcutaneous region (Fig. 2C), endothelial cells lining the aorta (Fig. 2D) and erythrocytes, even though this cell type was found to be nucleated in both wild type and knockout embryos at this developmental stage (Fig. 2E). Abnormal changes were found only in the E 14.5 embryos carrying trspfl/fl-Tie2Cre+/−. The parenchyma of the brain and spinal cord was largely necrotic (compare knockout and control embryo brain and spinal cord sections in Fig. 2F and G, respectively). Derivative of the central nervous system (dorsal root ganglia, eyes, optic nerves), however, were largely unaffected (not shown). There was extensive multifocal subcutaneous hemorrhage (Fig. 2H) and endothelial cells lining the aorta were hypertrophied (Fig. 2I) in affected embryos compared to control embryos (trspfl/+ and trspfl/+-Tie2Cre+/−) as shown by the arrows in the respective panels. Erythrocytes were less mature than those in control embryos (Fig. 2J). As expected, only few erythrocytes in the control embryos retained nuclei, which were small and hyperchromatic. In contrast, most erythrocytes in affected embryos had large nuclei as shown by the arrow in the knockout panel in the figure. The thymus was either not present or was disorganized in affected embryos, preventing its recognition. There was abundant cellular debris within the tongue, consistent with cell lysis (not shown). Since endothelial cells line the blood vessels, gene inactivation within this cell line will most certainly not be restricted to any specific tissue and thus will lead to widespread effects. No differences were observed between trspfl/+ and trspfl/+ -Tie2Cre+/− control embryos.
Fig. 2.
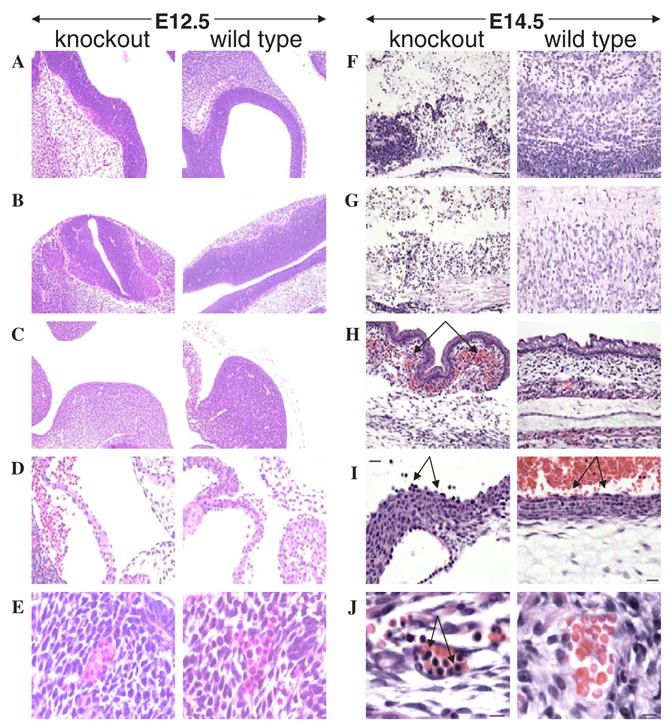
Pathological analysis of embryos with and without trsp in endothelial cells. Left panels show tissues of affected embryos (designated knockout) and right panels the corresponding tissues of unaffected embryos (designated wild type). In E 12.5 embryos, tissues and organs analyzed and the fold magnifications of each were: (A) brain (20×); (B) spinal cord (20×); (C) subcutaneous region (20×); (D) endothelial cells lining the aorta (40×); and (E), RBC’s (100×). In E 14.5 embryos, tissues and organs analyzed and the fold magnifications of each were: (F) brain (20×); (G) spinal cord (20×); (H) subcutaneous region (20×); (I) endothelial cells lining the aorta (40×); and (J) RBC’s (100×). Panel descriptions are given in the text. Staining was carried out as given in Section 2.
3.3. Targeted removal of trsp in skeletal and heart muscle
3.3.1. Phenotypes and genotypes
The offspring from matings between trspfl/+-MCK-Cre+/−male and trspfl/fl female parents did not manifest any apparent differences phenotypically (i.e., no external morphological differences) until about day 12, when some of the neonatal mice died suddenly. The ill mice exhibited labored breathing and reduced mobility (weakness) 4–5 h before death.
Genotyping 6–8 days old offspring from trspfl/+ -MCKCre+/−and trspfl/fl matings revealed that the litters contained each of the expected genotypes in targeted tissues and in control organs and tissues (Fig. 3). trspfl/fl was partially removed in skeletal and heart muscle DNA, while it was unaffected in tail, kidney, liver, spleen, brain and lung. Only partial loss of trsp was expected in targeted tissues since the MCKCre is specific to myocytes and both of these tissues are made up of less than 50% myocytes. Each tissue contained the MCKCre transgene (Fig. 3). The only neonatal mice to become ill were those containing the trspfl/fl-MCKCre+/−genotype, and we therefore took 10 day old mice of all genotypes for pathological analysis. Neonates with genotype trspfl/fl or genotype trspfl/+-MCKCre+/−showed no differences in the tissues and organs examined.
Fig. 3.
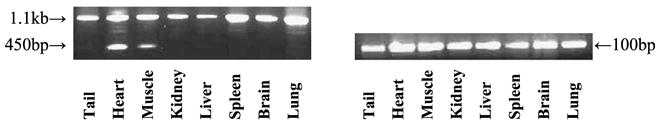
Genotyping of neonatal mice. The 1.1 kb fragment from the various tissues or appendages shown encodes trspfl/fl, while the 450 bp fragment in heart and muscle indicates the loss of trsp. The 100 bp fragment indicates the presence of Cre.
3.4. Pathological analysis of a3ected and una3ected neonatal offspring
To investigate the cause of death, affected neonatal mice were stored in formalin shortly after death and pathological analysis performed. Unaffected siblings of the same age were sacrificed and also examined pathologically as controls. No gross changes were found in the fixed carcasses. Samples of heart, lung, liver, spleen, thymus, pancreas, kidneys, skeletal muscle and bone were collected and examined microscopically. In affected mice, the myocardium was multifocally infiltrated by mixed inflammatory cells (∼90% neutrophils, ∼5% lymphocytes and ∼5% macrophages), with degeneration and loss of myofibers as compared to controls (see Fig. 4A). In several mice, the inflammation extended into the mediastinum. Cause of death was apparently acute myocardial failure. No degenerative or inflammatory changes were found in other examined tissues or in the control mice. To examine the pathogenesis of these inflammatory and degenerative changes, a total of three muscle-specific trsp knockout pups, one 6-day and two 10-day old with an equal number of control pups were euthanized and a full necropsy performed. Gross changes were not found in the following tissues examined microscopically: thymus, kidney, adrenal, spleen, liver, stomach, intestine, pancreas, skeletal muscle, bone, lung, trachea, esophagus, tongue, brain and skull. The mediastinum of the 6-day-old muscle specific knockout mouse was infiltrated, however, by small numbers of inflammatory cells, primarily neutrophils (see arrows in left panel, Fig. 4B), while no changes were found in the age-matched control (right panel, Fig. 4B). Myocarditis, myocardiofiber degeneration, pulmonary arteritis/ periarteritis, and mediastinitis were found in the 10-day-old muscle-specific trsp knockout mouse (see arrows in left panel, Fig. 4C), but no changes were found in the age-matched control (right panel, Fig. 4C). No significant differences were found in the skeletal muscle tissue of wild type and knockout animals on H&E staining (Fig. 4D). In addition, Masson’s Trichrome staining for increased muscle fibrosis and Congo red for deposition of amyloid in skeletal muscle tissues did not show any significant difference between wild type and knockout animals (data not shown).
Fig. 4.
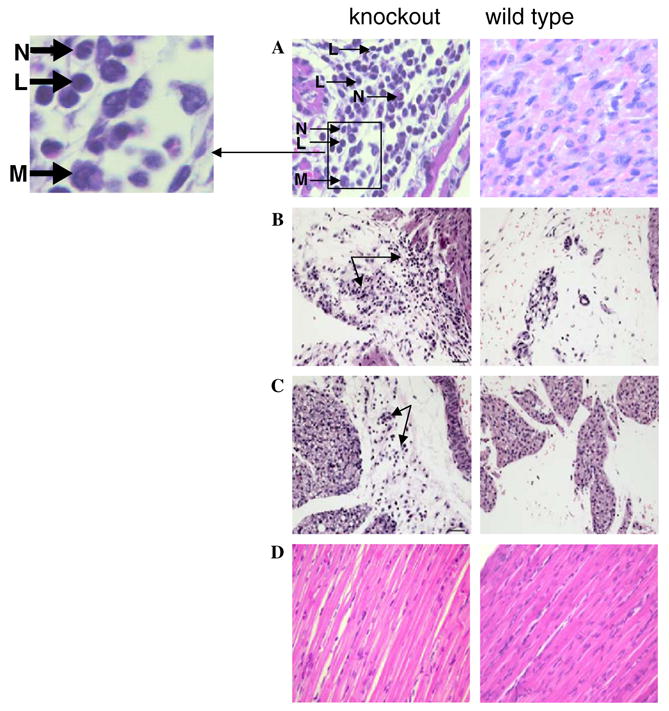
Pathological analysis of neonatal mice with and without trsp in muscle. Left panels show tissues and organ from affected pups (designated knockout) and right panels, the corresponding tissues and organ from unaffected pups (designated wild type). Tissue analyzed, ages of neonatal mice, and the fold magnifications were: (A) heart, 10 days (40×); and letters with arrows designate: L, lymphocyte; N, neutrophil; M, macrophage; (B) mediastinum, 6 days (20×); (C) mediastinum, 10 days (20×) and (D) skeletal muscle, 10 days (40×). Panel descriptions are given in the text. Staining was carried out as given in Section 2.
4. Discussion
The standard knockout of trsp in mammals is embryonic lethal demonstrating that selenoprotein expression is essential in mammalian development [16,17]. Removal of trsp, therefore, does not permit us to assess the manner in which selenoprotein expression is essential. By producing the conditional knockout of trsp [17] and then targeting specific tissues and organs, we can begin to decipher how these tissues and organs are dependent on selenoprotein expression in development and proper function and the possible role of selenoproteins in disease prevention. In the present study, we find that endothelial cells, which play a major role in the development of the vascular system, are dependent on selenoprotein expression for development and proper function. The Cre recombinase that is responsible for targeting the removal of trspfl/fl is under the control of the Tie2 promoter. The Tie2 promoter normally governs the expression of the tie gene that encodes the receptor tyrosine kinase, which is specific to endothelial cells and is first detected at day 8 of embryogenesis ([19] and references therein). Embryos from matings between male and female mice with genotypes trspfl/+-Tie2Cre+/−and trspfl/fl, respectively, showed no differences in phenotype at day 10. However, by 12 dpc, some of the embryos were slower developing, and by day 14.5, these embryos were much smaller with numerous phenotypic abnormalities. Pathological analysis showed several severely affected organs and tissues. For example, necrosis of the parenchyma of the brain and spinal cord was evident and endothelial cells lining the aorta were hypertrophic. Erythrocytes in affected embryos were less mature and had large nuclei. There was extensive multifocal subcutaneous hemorrhage in the affected embryos.
Studies involving mammalian endothelial cells in culture have provided insights into which selenoproteins are expressed in this cell type that include GPx1, GPx4, thioredoxin reductase-1 and SelP [22,23]. These studies were prompted by the fact that selenium appears to protect endothelial cells from oxidative damage which has been implicated in cardiovascular diseases, including atheroma ([22,23] and references therein). In the present study, our data provide strong evidence that selenoproteins play a major role in the development and proper function of endothelial cells. It seems likely, therefore, that the beneficial effect of selenium in protecting endothelial cells from oxidative damage is through the action of selenoproteins.
We also examined the role of selenoproteins in skeletal and heart muscle development and function. The Cre recombinase was under the control of the MCK promoter that normally encodes the mck gene which is highly specific to skeletal and cardiac muscle [21]. Interestingly, neonatal mice that carried a knockout of trsp manifested no phenotypic differences compared to their siblings carrying wild type trsp until about day 12 after birth. Those neonates with the trsp knockout became severely ill manifesting rapid breathing 4–5 h before dying. Pathological analysis suggested that the mice died from acute myocardial failure and that they manifested moderate to severe lesions in heart, but not in the skeletal muscle. In mice, MCK expression is complete by day eight after birth in heart muscle, whereas its expression at maximal levels in skeletal muscle is not as clearly defined (C.R. Kahn, personal communication). Thus, it seems that in heart the loss of selenoprotein expression presumably by day eight after birth requires additional time to manifest itself as the lack of selenoproteins in heart muscle is severe leading to sudden cardiac arrest at day 12. Possible explanations for skeletal muscle being normal is that the animals do not live long enough to exhibit gross abnormalities in skeletal muscle tissue or that MCKCre is not fully expressed as early in skeletal as in heart muscle. The effect of the lack of selenoproteins in heart muscle is severe leading to sudden cardiac arrest and the animal dies before effects can be manifested in skeletal muscle. As noted in Section 3, the heart lesions consisted predominantly of neutrophils, macrophages and lymphocytes as an inflammatory infiltrate. The most likely cause of inflammation in the heart muscle of the trsp knockout mice is oxidative stress due to the depletion of selenium-containing antioxidant enzymes in the myocardium [24] as removal of trsp will deplete the entire class of selenoproteins in myocytes. Several selenoproteins are known to be important antioxidants and redox regulators in mammals protecting cells from endogenous and exogenous reactive oxygen species. Selenium at low doses has been reported to provide significant protection of the human coronary artery endothelium against damage by oxidative stress [24]. Various chemotactic factors released by such damaged cells could lead to widespread inflammation. It is common during acute inflammation that the major invading cell type is the neutrophil followed by monocytes, both of which respond to chemotactic signals such as the release of cytokines, e.g., interleukin-1. Thus we presume that necrosis of the myocytes occurs first and further leads to inflammation.
Characterization of the Sec tRNA[Ser]Sec population in endothelial cells and myocytes was not possible in the present study due to the low abundance of the targeted trsp knockout cell type. Furthermore, there are technical limitations for demonstrating reduced selenoprotein expression as antibodies against many selenoproteins are not available and many of those antibodies that are available cross-react with other proteins yielding substantial backgrounds rendering them unusable for immuno-staining. It should also be noted that only small amounts of the total cell population in embryos consist of endothelial cells and that less than 50% of the cell population in cardiac muscle consists of myocytes. Thus, acquiring a pure population of these cell types is a limiting factor. Genotyping affected embryos demonstrated that they contained floxed trsp and the appropriate Cre, while genotyping cardiac muscle in affected animals demonstrated that less than 50% of floxed trsp was removed by the encoded Cre. In both cases, the observed level of trsp removal was expected from the known amounts of the targeted cell type.
Selenium has been known for many years to have a role in heart disease prevention [25] and it seems from the present study that selenoproteins are the effective agents. It should also be noted that a link between selenium deficiency, Coxsackie virus infection, myocarditis and the development of dilatative myocardopathy (Keshan disease) has been observed in children residing in selenium deficient regions in China [25]. Selenium supplementation in the diet of individuals residing in areas of China deficient in this trace element have virtually eradicated Keshan’s disease. Interestingly, Beck and collaborators [26] have shown that mice fed a selenium-deficient diet developed myocarditis when inoculated with a strain of Coxsackie virus that was non-pathogenic in mice fed a diet with adequate selenium providing an excellent model for studying the molecular biology of the role of selenoproteins in viral expression and heart disease. Interestingly, another selenoprotein, selenoprotein N, has been found to be associated with three congenital myopathies including desmin-related myopathy with Mallory body-like inclusions, rigid spine muscular dystrophy and multiminicore disease ([27] and references therein) providing additional links between selenium and muscle disorders.
Since the targeted removal of selenoproteins in endothelial cells is embryonic lethal and their removal in myocytes results in premature death due to cardiac failure in newborn mice, it is unlikely that these approaches will provide a model for specifically identifying those selenoproteins that play a major role in cardiovascular disease prevention. However, expression of the selenoprotein population can be perturbed using specific Sec tRNA[Ser]Sec mutant isoforms [28] in such a manner that only a portion of the protein population can be rescued [15] or partially replaced in tissues in which the selenoprotein population has been targeted for removal (B.A. Carlson, M.E. Moustafa, V.N. Gladyshev, D.L. Hatfield, unpublished). By preserving expression of selenoproteins involved in housekeeping and other essential functions, and disrupting gene expression of selenoproteins involved in stress-related phenomena, we can begin assessing the roles of the latter selenoproteins in cardiovascular disease.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health, NCI, Center for Cancer Research and by Grants GM065204 and CA080946 (to V.N.G.).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.nmd.2006.10.006.
References
- 1.Kryukov GV, Castellano S, Novoselov SV, et al. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–43. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 2.Yant LJ, Ran Q, Rao L, et al. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic Biol Med. 2003;34:496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- 3.Hill KE, Zhou J, McMahan WJ, et al. Deletion of selenoprotein P alters distribution of selenium in the mouse. J Biol Chem. 2003;278:13640–6. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- 4.Schomburg L, Schweizer U, Holtmann B, Flohe L, Sendtner M, Kohrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christoffolete MA, Linardi CCG, De Jesus L, et al. Mice with targeted disruption of the Dio2 gene have cold-induced overexpression of the uncoupling protein 1 gene but fail to increase brown adipose tissue lipogenesis and adaptive thermogenesis. Diabetes. 2004;53:577–84. doi: 10.2337/diabetes.53.3.577. [DOI] [PubMed] [Google Scholar]
- 6.De Jesus LA, Carvalho SD, Ribeiro MO, et al. The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue. J Clin Invest. 2001;108:1379–85. doi: 10.1172/JCI13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng L, Goodyear RJ, Woods CA, et al. Hearing loss and retarded cochlear development in mice lacking type 2 iodothyronine deiodinase. Proc Natl Acad Sci USA. 2004;101:3474–9. doi: 10.1073/pnas.0307402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider MJ, Fiering SN, Pallud SE, Parlow AF, Germain DL, Galton VA. Targeted disruption of the type 2 selenodeiodinase gene (DIO2) results in a phenotype of pituitary resistance to T4. Mol Endocrinol. 2001;15:2137–48. doi: 10.1210/mend.15.12.0740. [DOI] [PubMed] [Google Scholar]
- 9.Jakupoglu C, Przemeck GK, Schneider M, et al. Cytoplasmic thioredoxin reductase is essential for embryogenesis but dispensable for cardiac development. Mol Cell Biol. 2005;25:1980–8. doi: 10.1128/MCB.25.5.1980-1988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conrad M, Jakupoglu C, Moreno SG, et al. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol Cell Biol. 2004;24:9414–23. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng WH, Ho YS, Ross DA, Valentine BA, Combs GF, Lei XG. Cellular glutathione peroxidase knockout mice express normal levels of selenium-dependent plasma and phospholipid hydroperoxide glutathione peroxidases in various tissues. J Nutr. 1997;127:1445–50. doi: 10.1093/jn/127.8.1445. [DOI] [PubMed] [Google Scholar]
- 12.Esworthy RS, Aranda R, Martin MG, Doroshow JH, Binder SW, Chu FF. Mice with combined disruption of Gpx1 and Gpx2 genes have colitis. Am J Physiol Gastrointest Liver Physiol. 2001;281:G848–55. doi: 10.1152/ajpgi.2001.281.3.G848. [DOI] [PubMed] [Google Scholar]
- 13.Chu FF, Esworthy RS, Doroshow JH. Role of Se-dependent glutathione peroxidases in gastrointestinal inflammation and cancer. Free Radic Biol Med. 2004;36:1481–95. doi: 10.1016/j.freeradbiomed.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Lei XG. Glutathione peroxidase-1 gene knockout on body antioxidant defense in mice. Biofactors. 2001;14:93–9. doi: 10.1002/biof.5520140113. [DOI] [PubMed] [Google Scholar]
- 15.Carlson BA, Xu XM, Gladyshev VN, Hatfield DL. Selective rescue of selenoprotein expression in mice lacking a highly specialized methyl group in selenocysteine tRNA. J Biol Chem. 2005;280:5542–8. doi: 10.1074/jbc.M411725200. [DOI] [PubMed] [Google Scholar]
- 16.Bösl MR, Takaku K, Oshima M, Nishimura S, Taketo MM. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp) Proc Natl Acad Sci. 1997;94:5531–4. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumaraswamy E, Carlson BA, Morgan F, et al. Selective removal of the selenocysteine tRNA[Ser]Sec gene (Trsp) in mouse mammary epithelium. Mol Cell Biol. 2003;23:1477–88. doi: 10.1128/MCB.23.5.1477-1488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson BA, Novoselov SV, Kumaraswamy E, et al. Specific excision of the selenocysteine tRNA[Ser]Sec (Trsp) gene in mouse liver demonstrates an essential role of selenoproteins in liver function. J Biol Chem. 2004;279:8011–7. doi: 10.1074/jbc.M310470200. [DOI] [PubMed] [Google Scholar]
- 19.Gustafsson E, Brakebusch C, Hietanen K, Fassler R. Tie-1-directed expression of Cre recombinase in endothelial cells of embryoid bodies and transgenic mice. J Cell Sci. 2001;114:671–6. doi: 10.1242/jcs.114.4.671. [DOI] [PubMed] [Google Scholar]
- 20.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–42. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 21.Bruning JC, Michael MD, Winnay JN, et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol Cell. 1998;2:559–69. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 22.Hara S, Shoji Y, Sakurai A, Yuasa K, Himeno S, Imura N. Effects of selenium deficiency on expression of selenoproteins in bovine arterial endothelial cells. Biol Pharm Bull. 2001;24:754–9. doi: 10.1248/bpb.24.754. [DOI] [PubMed] [Google Scholar]
- 23.Miller S, Walker SW, Arthur JR, et al. Selenoprotein expression in endothelial cells from different human vasculature and species. Biochim Biophys Acta. 2002;1588:85–93. doi: 10.1016/s0925-4439(02)00143-6. [DOI] [PubMed] [Google Scholar]
- 24.Miller S, Walker SW, Arthur JR, et al. Selenite protects human endothelial cells from oxidative damage and induces thioredoxin reductase. Clin Sci. 2001;100:543–50. [PubMed] [Google Scholar]
- 25.Burke MP, Opeskin K. Fulminant heart failure due to selenium deficiency cardiomyopathy (Keshan disease) Med Sci Law. 2002;42:10–3. doi: 10.1177/002580240204200103. [DOI] [PubMed] [Google Scholar]
- 26.Beck MA, Handy J, Levander OA. Host nutritional status: The neglected virulence factor. Trends Microbiol. 2004;12:417–23. doi: 10.1016/j.tim.2004.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tajsharghi H, Darin N, Tulinius M, Oldfors A. Early onset myopathy with a novel mutation in the Selenoprotein N gene (SEPN1) Neuromusc Disorders. 2005;12:299–302. doi: 10.1016/j.nmd.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Moustafa ME, Carlson BA, El-Saadani MA, et al. Selective inhibition of selenocysteine tRNA maturation and selenoprotein synthesis in transgenic mice expressing isopentenyladenosine-deficient selenocysteine tRNA. Mol Cell Biol. 2001;21:3840–52. doi: 10.1128/MCB.21.11.3840-3852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


