Abstract
The photosynthetic CO2-fixing enzyme, Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase), is responsible for most of the world's biomass, but is a slow non-specific catalyst. We seek to identify and overcome the chemical and biological constraints that limit the evolutionary potential of Rubisco in Nature. Recently, the horizontal transfer of Calvin cycle genes (rbcL, rbcS and prkA) from cyanobacteria (Synechococcus PCC6301) to γ-proteobacteria (Escherichia coli) was emulated in the laboratory. Three unique Rubisco variants containing single (M259T) and double (M259T/A8S, M259T/F342S) amino acid substitutions in the L (large) subunit were identified after three rounds of random mutagenesis and selection in E. coli. Here we show that the M259T mutation did not increase steady-state levels of rbcL mRNA or L protein. It instead improved the yield of properly folded L subunit in E. coli 4–9-fold by decreasing its natural propensity to misfold in vivo and/or by enhancing its interaction with the GroES–GroEL chaperonins. The addition of osmolites to the growth media enhanced productive folding of the M259T L subunit relative to the wild-type L subunit, while overexpression of the trigger factor and DnaK/DnaJ/GrpE chaperones impeded Rubisco assembly. The evolved enzymes showed improvement in their kinetic properties with the M259T variant showing a 12% increase in carboxylation turnover rate (kccat), a 15% improvement in its KM for CO2 and no change in its KM for ribulose-1,5-bisphosphate or its CO2/O2 selectivity. The results of the present study show that the directed evolution of the Synechococcus Rubisco in E. coli can elicit improvements in folding and catalytic efficiency.
Keywords: CO2 fixation; directed evolution; metabolic engineering; protein engineering; ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco)
Abbreviations: 2-carboxyarabinitol-P2, 2′-carboxyarabinitol-1,5-bisphosphate; carboxypentitol-P2, isomeric mixture of carboxyarabinitol-P2 and 2′-carboxyribitol-1,5-bisphosphate; fructose-P2, fructose-1,6-bisphosphate; IPTG, isopropyl β-D-thiogalactoside; KJE, DnaK/DnaJ/GrpE; L, large; LB-kan, Luria–Bertani medium containing 50 μg/ml kanamycin; prkA, phosphoribulokinase; ribulose-P2, D-ribulose-1,5-bisphosphate; Rubisco, ribulose-P2 carboxylase/oxygenase; TF, 50 kDa trigger factor chaperone
INTRODUCTION
Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase) catalyses the nucleophilic carboxylation of ribulose-P2 (D-ribulose-1,5-bisphosphate). This conversion of inorganic CO2 into carbohydrate is frequently the rate-limiting step in the synthesis of most of the world's biomass [1]. In spite of its biological importance, Rubisco is an inefficient catalyst, particularly at limiting CO2 concentrations. Its turnover rate is less than one-thousandth of many other plant enzymes [2]. Moreover, the efficiency of Rubisco-catalysed carbon assimilation is further compromised by the fixation of O2, which competes with CO2 for addition to ribulose-P2. The 2-phosphoglycolate product of the oxygenation reaction is recycled by photorespiration, an energy consuming process that releases one-quarter of the incorporated carbon [3]. It is not clear why such an important enzyme is so slow and non-specific. Our goals are to understand the physical, chemical and biological constraints to the adaptive evolution of Rubisco, and to find ways to overcome them.
Natural selection generally favours enzyme variants that evince speed (high turnover, kcat), high substrate affinity (low Michaelis constant, KM) and high substrate specificity (kcat/KM primary substrate divided by kcat/KM secondary substrate). Extant Rubisco homologues from different photosynthetic organisms show significant variation in all three parameters (see [4] for a summary). Their KM in reactions with CO2 as the substrate (Kc) varies from 3–340 μM and the corresponding carboxylation turn-over rates (kccat) vary between 1 and 13 s−1. The ability of Rubiscos to discriminate CO2 from O2, defined as its specificity factor (Sc/o), varies between 10 and 240. Sc/o is determined by dividing a Rubisco's carboxylation efficiency (kccat/Kc) by its oxygenation efficiency (kocat/Ko), where kocat is the O2 saturated oxygenation rate and Ko is the KM for O2 [5]. This natural variability in Rubisco kinetics has encouraged attempts to engineer kinetically improved versions of the enzyme [1,6,7].
Phylogenetic analyses can highlight patterns that reflect horizontal gene transfer or gene duplication and selective loss [8,9]. For example, phylogenetic analyses of Rubisco L (large) subunit genes (rbcL) suggest that this gene was horizontally transferred at least four times during the evolution of proteobacteria, cyanobacteria and plastids [10]. Although natural mechanisms that effect the transfer of DNA between genomes have been identified [11], the mechanisms of gene adaptation upon transfer to new cellular micro-environments remain poorly understood. Gene sequence comparisons can sometimes identify instances of molecular adaptation, although most beneficial mutations are obscured by the accumulation of functionally neutral mutations [12]. The ‘resurrection’ and in vitro characterization of ancestral proteins can offer unambiguous evidence for molecular adaptation, but only if the investigator can correctly predict the biochemical tests to conduct [13]. Directed evolution experiments can emulate hypothetical evolutionary events, thereby ‘resurrecting’ ancestral proteins and the conditions in which they were selected.
In the present study, we investigated the horizontal transfer and subsequent adaptation of cyanobacteria (Synechococcus PCC-6301) Rubisco to a new cellular environment by imitating the process in the laboratory. Although the hexadecameric Synechococcus Rubisco structurally resembles Rubisco in all higher plants [comprising eight L and eight S (small) subunits; L8S8] it is unique in its ability to be functionally expressed in E. coli [14,15]. By simultaneously transferring the Synechococcus prkA (phosphoribulokinase), rbcL and rbcS (coding the Rubisco small subunit) genes into wild-type E. coli strain K-12 we have developed a high throughput genetic selection system to study the adaptive mechanism(s) that improves the fitness of Rubisco in this non-natural host. The selection pressure in the system is the alleviation of ribulose-P2 toxicity (produced by phosphorylation of ribulose-5-phosphate by PRK) via its oxygenation or carboxylation by Rubisco [16]. Three unique Rubisco variants (M259T, M259T/A8S and M259T/F342S; Table 1) emerged after three rounds of random mutagenesis and genetic selection, all sharing the M259T mutation. Preliminary biochemical studies showed that E. coli cells expressing the evolved Rubiscos exhibited 2–5-fold more carboxylase activity than those expressing the wild-type enzyme. Purified C-terminal-His6-tagged versions of these mutant proteins showed similar increases in specific activity [16]. Here we extend these initial observations to examine how the mutations in the horizontally transferred Synechococcus PCC6301 rbcL increased the fitness of the host E. coli cell during selection.
Table 1. Features of the Synechococcus PCC6301 Rubisco genes and proteins used in the present study.
WT, wild-type rbcL-rbcS operon.
| Rubisco name | rbcLS mutant allele number* | Silent rbcL mutations |
|---|---|---|
| (−)WT | − | nil |
| Revertant WT | − | c489t, c783t, t799c |
| (−)M259T | − | nil |
| M259T | 2.29 | c489t, c783t, t799c |
| A8S/M259T | 2.24 | c783t, t799c |
| M259T/F342S | 3.54 | t702c, c783t, t799c |
* Clone number described in [16].
EXPERIMENTAL PROCEDURES
Materials
The construction of the rbcLS-pET30a+ expression vectors containing the wild-type Synechococcus PCC6301 Rubisco rbcL–rbcS operon and those encoding the single (M259T) and double (M259T/A8S, M259T/F342S) amino acid mutations (with and without C-terminal His6 tags fused to the large subunit; Table 1), and most of the materials used for the present study have been described previously [16]. The Sephadex® G-50 Fine and the radiolabelled NaH14CO3 were from Pharmacia (GE Healthcare). The 0.2 micron nylon filters were from Alltech. All other chemicals were from Sigma Chemicals.
Site-directed mutagenesis
PCR-mediated site-directed mutagenesis was applied to change Thr259 of Rubisco 2.29 back to a methionine residue (to create an otherwise wild-type rbcLS with only silent mutations), and to introduce the M259T (t776c) mutation into the wild-type rbcLS gene [to create the (−)M259T Rubisco without silent mutations].
Expression of wild-type and artificially evolved Rubiscos
E. coli BL21(DE3) cells were transformed with the rbcLS-pET30a+ plasmids encoding the wild-type and mutant Rubisco variants. The transformed cells were grown separately at 23 °C to mid-logarithmic phase in LB-kan (Luria–Bertani medium containing 50 μg/ml kanamycin) and Rubisco expression was induced for 16 h with 0.5 mM IPTG (isopropyl β-D-thiogalactoside). The cells were harvested by centrifugation (5 min at 5000 g at 4 °C), resuspended in extraction buffer (100 mM Hepps/NaOH, pH 8.0, 1 mM EDTA, 2 mM dithiothreitol and 0.05% E. coli protease inhibitor cocktail) and lysed by passage through a French pressure cell (140 MPa). A 50 μl aliquot of lysate (total cellular protein) was removed for SDS/PAGE analysis and the remainder centrifuged at 38000 g for 10 min at 4 °C to remove the insoluble material. Aliquots of the supernatant were either assayed for protein content (1 μl), Rubisco content (100 μl; see below) or processed for SDS/PAGE (12% gels) and non-denaturing PAGE (4–12% acrylamide gradient) analysis as described previously [17].
Measuring Rubisco content by [14C]2-carboxyarabinitol-P2 (2′-carboxyarabinitol-1,5-bisphosphate) binding
The concentration of Rubisco active sites was determined by the stoichiometric binding of the inhibitor [14C]2-carboxyarabinitol-P2. Ribulose-P2 was synthesized and purified as described previously [18] and used to make 14C-labelled carboxypentitol-P2 (97000 cpm/nmol) according to the method used by Pierce et al. [19]. Carboxypentitol-P2 is an isometric mixture of carboxyribitol-P2 and carboxyarabinitol-P2. The latter is a transition-state analogue that binds preferentially to activated Rubisco in an almost irreversible manner [20]. Samples containing Rubisco were activated with 25 mM NaHCO3 and 20 mM MgCl2 at 25 °C for 30 min before incubating with 27 μM [14C]2-carboxypentitol-P2 for 15–45 min. The Rubisco–[14C]2-carboxyarabinitol-P2 complex was separated from unbound [14C]2-carboxypentitol-P2 by size-exclusion chromatography and Rubisco content calculated as described previously [21].
Purification of native (untagged) Rubisco variants
The Rubisco variants were expressed in 1 litre cultures of E. coli rbcLS-pET30/BL21(DE3) cells as described above. After 16 h of induction with 0.5 mM IPTG, the cells were harvested by centrifugation (5 min at 6000 g at 4 °C), resuspended in 45 ml ice-cold extraction buffer, and lysed with a French pressure cell. The cellular debris was removed by centrifugation at 35000 g for 15 min at 4 °C. The Rubiscos were purified by heating the extracts at 50 °C, followed by precipitation with ammonium sulfate, and then ion-exchange chromatography using a Mono-Q (10/10) column (GE Healthcare) as described previously [22]. Collected fractions (5 ml) were assayed for substrate-saturated ribulose-P2 carboxylase activity using NaH14CO3 (2 Ci/mol; GE Healthcare) [23]. Appropriate fractions were pooled and a saturated ammonium sulfate solution (pH 7) was slowly added to 60% (w/v) final concentration. The Rubisco precipitate was collected by centrifugation at 22000 g for 15 min at 4 °C, dissolved in 3 ml storage buffer (15 mM Hepps/NaOH, pH 8.0, 1 mM EDTA and 50 mM NaCl) and then dialysed using a Slide-a-lyzer cassette (Pierce Chemical Co.) against 1 litre of storage buffer overnight at 4 °C. The dialysis was repeated against fresh storage buffer for 2–4 h and then against storage buffer containing 20% (v/v) glycerol for 4 h. The purified Rubisco was frozen in liquid nitrogen and stored at −70 °C. SDS/PAGE and non-denaturing PAGE analysis showed that these preparations were >95% pure.
Kinetic assays
The purified Rubisco preparations were used to measure the CO2/O2 specificity (Sc/o) at pH 8.3 as described previously [24]. Sc/o was measured in reactions equilibrated with an atmosphere of O2 accurately mixed with 0.1% (v/v) CO2 using Wostoff pumps. The Michaelis constants for substrates CO2 (Kc) and ribulose-P2 (KRuBP) were measured in 14CO2-fixation assays at 25 °C and pH 8, according to the method described previously [23]. The purified enzyme was pre-incubated at 25 °C for 30 min in buffer containing 20 mM MgCl2 and 25 mM NaHCO3, and Kc measurements were performed in nitrogen-sparged septum-capped scintillation vials. The reactions were initiated by the addition of 10 μl of purified enzyme to 1 ml of N2-equilibrated assay buffer (100 mM Hepps/NaOH, pH 8, 20 mM MgCl2, 0.8 mM ribulose-P2 and 0.1 mg/ml carbonic anhydrase) containing various concentrations of NaH14CO3. KRuBP measurements were performed in air-equilibrated assay buffer containing different concentrations of ribulose-P2 (0–2 mM). The 14CO2-assays were stopped after 1.5 or 2 min with 0.5 vol. of 25% (v/v) formic acid and dried at 80 °C. The residue was dissolved in 0.5 ml of water before the addition of 1 ml of scintillant (UltimaGold XR; Perkin Elmer Life Sciences) for scintillation counting. The Michaelis constants were determined by fitting the data to the Michaelis–Menten equation. From the Kc measurements, the catalytic turnover rate (kccat) was calculated by dividing the extrapolated maximal carboxylase activity by the concentration of Rubisco active sites in the assay (measured by [14C]2-carboxyarabinitol-P2 binding, as described above).
PAGE and immunoblot analyses of protein expression in wild-type and artificially evolved Rubiscos
SDS/PAGE, non-denaturing PAGE and immunoblot analyses were performed according to methods described previously [17]. Immunoblot analyses using polyclonal antisera raised in rabbits against purified Synechococcus PCC6301 Rubisco or tobacco Rubisco were performed.
Reverse transcriptase–real time PCR
E. coli strain DH5Δlac(DE3) [25] was separately transformed with the rbcLS-pET30a+ plasmids coding for wild-type and M259T Rubisco. The transformed cells were propagated to mid-logarithmic stage in LB-kan. Rubisco expression was induced with 0.5 mM IPTG overnight at 23 °C. Total RNA was extracted using the RNeasy kit (Qiagen) and its concentration determined by absorbance measurements at 260 nm using a Shimadzu UV-1601 spectrophotometer. The RNA (30 or 0.3 ng) and various amounts of the rbcLS-pET30a+ plasmids were used in conjuction with the iScript one-step real time PCR kit (Bio-Rad) using the rbcL-specific primer 5′-TGGTGACCGACCCTTCTTC-3′ (reverse) to synthesize cDNA. Real time PCR reactions were performed in duplicate using rbcL-specific reverse and forward (5′-GCTGACCGACATGGATCGGTACAA-3′) primers according to manufacturer's instructions. Melt curve analysis confirmed the stability, accuracy and specificity of the primer pair. The PCR amplification reactions were monitored with an iCycler real time PCR machine (Bio-Rad) and the amount of rbcL mRNA in each sample quantified using the accompanying software.
Analysis of wild-type and M259T Rubisco expression with the addition of osmolites and chaperones
E. coli BL21(DE3) cells were transformed with pET30a+ (negative control) or the rbcLS-pET30a+ plasmids (encoding either the wild-type or M259T Rubisco). The cells were grown in LB-kan in the presence or absence of 0.3 M NaCl and 20 mM proline at 37 °C to a D600 of 0.8 before the induction of Rubisco expression with 0.5 mM IPTG at 23 °C for 5 h. Each cell type was separately co-transformed with plasmids pGro7 (encoding GroEL/ES), pKJE7 (encoding DnaJ/DnaK/GrpE), or pTF16 (encoding the trigger factor) (a set of chaperone expression vectors have been described previously [26,27]) and similarly grown in LB-kan containing chloramphenicol (30 μg/ml). Protein expression was similarly induced at 23 °C for 5 h with 0.5 mM IPTG (Rubisco), 0.05% (w/v) L-arabinose (pGro7, pKJE7, pTF16 and pG-KJE8) or/and 5 μg/ml tetracycline (pG-KJE8 and pG-Tf2). Measurement of Rubisco content by SDS/PAGE, non-denaturing PAGE and [14C]carboxyaranitol-P2 binding was standardized relative to cell density by measurement of the D600 immediately prior to harvesting 15 ml of the cells by centrifugation (5 min at 4000 g). For BL21(DE3) cells transformed with pET30 or rbcLS-pET30 a D600 of 1.0 was equivalent to 2.7×108 cells/ml.
CD spectra analyses
Far UV spectra and thermal denaturation were measured using a Jasco-810 spectropolarimeter. Far UV spectra were measured at 20 °C between 185 and 260 nm in 0.5 nm increments using a 0.01-cm path-length cell, a 2 nm bandwidth, a 4 s response time and a scanning speed of 20 nm/min. Thermal denaturation (unfolding) curves were measured at 2 °C increments at a rate of 1 °C/min between 20 and 80 °C in a 0.1-cm path-length cell at 222 nm.
RESULTS
Our objective was to elucidate the biochemical mechanism(s) by which the Synechococcus PCC6301 Rubisco adapted to our E. coli-based selection system (Table 1). Previous analyses indicated that compared with wild-type controls the artificially evolved Rubisco variants were expressed 2–5-fold more efficiently in E. coli and the specific carboxylase activities of purified His6-tagged enzymes were approx. 5-fold greater [16]. In the present study, we have carefully characterized the steady-state kinetics of the native (not tagged) versions of the wild-type and evolved Rubisco variants, and examine the mechanisms that account for the expression differences.
Catalytic properties
The untagged wild-type Synechococcus PCC6301 Rubisco and the M259T, A8S/M259T and M259T/F342S variant enzymes were purified from recombinant E. coli by ammonium sulfate fractionation and ion exchange chromatography. Affinity tag-fusions at either the N- or C-terminus of the Rubisco L subunit were avoided since modifications to these regions can influence the enzymes kinetics [28,29]. The CO2/O2 specificity (Sc/o), the Michaelis constants of the Rubisco/substrate interactions (Kc, KM for CO2; KRuBP, KM for ribulose-P2), and the substrate saturated carboxylation rate (kccat) of the wild-type PCC6301 Rubisco were similar to values reported previously (Table 2). The replicate Sc/o measurements were highly reproducible and revealed no change for the M259T enzyme and modest decreases (<15%) for the A8S/M259T and M259T/F342S Rubisco variants.
Table 2. Kinetic properties of purified wild-type and mutant Synechococcus PCC6301 Rubiscos identified by genetic selection.
WT, wild-type.
| Rubisco type | Sc/o, CO2/O2 specificity* | kccat carboxylation turnover rate (s−1) | Kc Michaelis constant for CO2 (μM) | kccat/Kc Carboxylation efficiency (M−1·s−1) | KRuBP Michaelis constant for pure ribulose-P2 (μM) |
|---|---|---|---|---|---|
| WT rbcL–rbcS operon | 43.9±1.8 | 11.4±0.1* | 273±10* | 4.2×104 | 63.9±4.5* |
| 43.0±1.0† | 11.6† | 284† | 4.1×104† | 54±3‡ | |
| M259T | 42.8±3.9 | 12.8±0.2* | 237±10* | 5.4×104 | 67.0±3.2* |
| A8S/M259T | 40.5±2.6 | 11.4±0.1§ | 216±9§ | 5.3×104 | 47.8±4.6§ |
| M259T/F342S | 38.4±0.7 | 10.0±0.1§ | 207±8§ | 4.8×104 | 33.7±5.9§ |
The M259T variant evinced a 12% increase in kccat, while the A8S/M259T and M259T/F342S double mutants exhibited slightly diminished kccat values (wild-type-like and 12% less than wild-type respectively). The Kc for the M259T, A8S/M259T and M259T/F342S Rubisco reactions were 13%, 21% and 24% lower than the comparable wild-type value respectively. Likewise, KRuBP values of the A8S/M259T and M259T/F342S enzyme-catalysed reactions were reduced 25% and 47% respectively, while the KRuBP for M259T remained comparatively unchanged. The three evolved Rubiscos exhibited 15–28% improvements in carboxylation efficiency (kccat/Kc) relative to the wild-type enzyme (Table 2).
Rubisco synthesis and assembly in E. coli
To examine whether the mutations in the evolved Rubisco variants influenced their production in E. coli, the total and soluble cellular proteins were analysed by SDS/PAGE. For the six Rubisco variants examined, the L band was the predominant protein in the total cellular protein samples (Figure 1A). To more accurately assess the relative amounts of the L subunit expressed, each sample was diluted 100-fold and re-analysed by SDS/PAGE (Figure 1B). We observed little difference in the amount of L subunit synthesized, apart from a modest (<2-fold) increase in the amount of total L in the E. coli expressing the A8S/M259T Rubisco. Likewise, the nucleotide substitutions did not influence the steady state rbcL mRNA levels. Reverse transcriptase–real time PCR measurements revealed similar amounts of rbcL message in cells expressing the wild-type or M259T enzyme (Table 1); both alleles showed an approx. 10-fold increase in the amount of rbcL mRNA upon induction with IPTG (results not shown). These results showed that the nucleotide substitutions in rbcL (non-silent mutations, and the c783t and t799c silent mutations) had little, if any, influence on the transcriptional or translational processing of the gene variants.
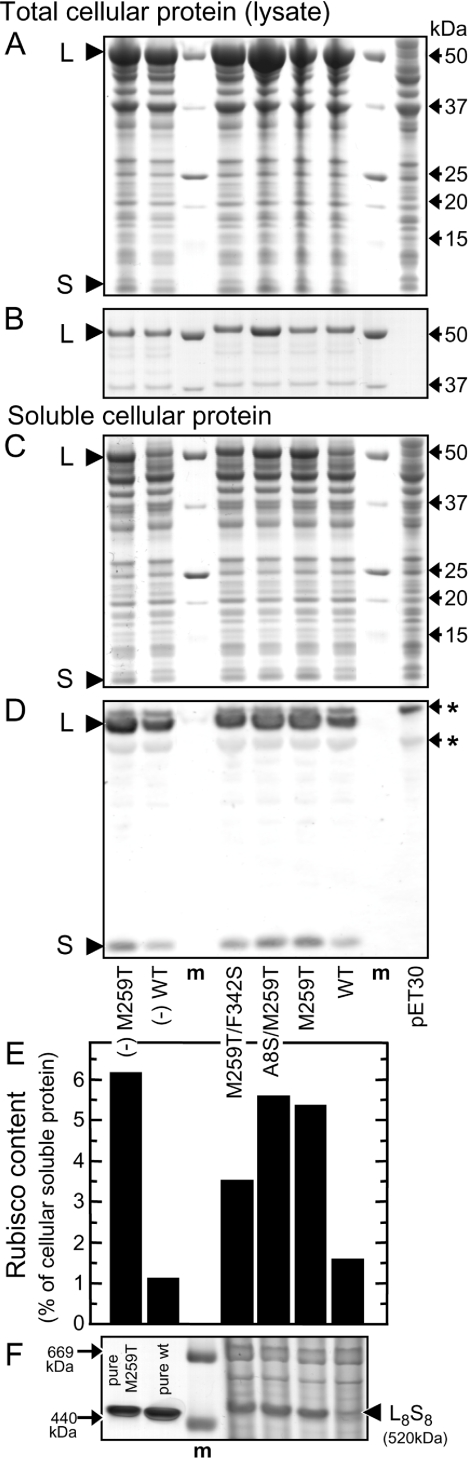
Figure 1. Evolved Rubisco variants fold and assemble more efficiently in E. coli.
Rubisco expression was induced with IPTG in BL21(DE3) E. coli cells transformed with rbcLS-pET30a+ plasmids coding for wild-type (WT) and the evolved Synechococcus PCC6301 Rubisco variants (see Table 1). To compare the amount and partitioning of Rubisco L (52 kDa) and S (13 kDa) subunits in the soluble and insoluble protein cellular fractions, comparable samples of total cellular protein (lysate) containing 10 μg (A) and 0.1 μg (B) or only the soluble cellular protein fraction (10 μg) (C) were separated by SDS/PAGE and visualized with Coomassie Blue staining. (D) Detection of the L and S subunit expression levels in the soluble cellular protein by immunoblotting using antibodies against Synechococcus sp PCC7942 Rubisco. (E) The amount of functional Rubisco measured by [14C]carboxyarabinitol-P2 binding, expressed as a percentage of the soluble cellular protein assuming a molecular mass of 65 kDa for each L subunit active site (assuming one S subunit for each L subunit that binds the [14C]carboxyarabinitol-P2). (F) Coomassie Blue staining of soluble cellular protein (30 μg) and purified L8S8 Rubisco separated by non-denaturing PAGE. m, molecular mass marker (sizes shown: 669 kDa, thyroglobulin and 440 kDa, ferritin); *, non-Rubisco E. coli proteins recognized by the antibody.
SDS/PAGE analysis of comparable soluble cellular protein samples showed that >90% of the L subunits were insoluble (Figure 1C). Immunodetection indicated that the relative levels of soluble L and S subunits were higher in cell extracts containing evolved Rubisco variants than in control extracts containing the wild-type protein (Figure 1D). These 2–4-fold differences in the amount of soluble L correlated with the different concentrations of functional L8S8 Rubisco measured by carboxyarabinitol-binding (Figure 1E) and non-denaturing PAGE (Figure 1F). They are consistent with previous findings that most, if not all, of the soluble L subunit detected in E coli extracts assembled into functional L8S8 enzyme [17,30]. In contrast, the production of the S subunit in E. coli does not limit assembly of PCC6301 Rubisco, since as much as 50% of the soluble S subunit is not assembled in L8S8 complexes, but can readily do so when supplied with L8 cores [31]. Taken together, these data suggested the M259T mutation present in the L subunit of all three artificially evolved Rubisco variants improves the ability of the L subunit to fold and/or assemble correctly with the S subunit into the functional enzyme within E. coli. Notably the additional F342S mutation compromised the yield of functional Rubisco produced by approx. 2-fold, consistent with previous results [16].
E. coli chaperone-assisted folding
We examined whether the improved folding and assembly of the M259T L in E. coli resulted from an increased compatibility between the mutated L subunits and several key E. coli chaperone complexes. Indeed, 10-fold improvements in the amount of cyanobacterial Rubisco folded correctly and assembled in E. coli can be obtained by overexpressing the GroES/GroEL chaperonins [14,32]. The 70 kDa chaperone DnaK that works in concert with its co-chaperones DnaJ (a chaperone activating protein) and GrpE (a nucleotide exchange factor) is also requisite for productive folding of bacterial Rubiscos in E. coli [15]. The 50-kDa TF (trigger factor) chaperone similarly co-translationally binds nascent peptide chains to prevent their misfolding and aggregation [27,33]. E. coli BL21(DE3) cells expressing the wild-type and M259T Rubiscos were therefore co-transformed with plasmids directing the expression of GroES–GroEL, KJE (DnaK/DnaJ/GrpE) or TF. PAGE and carboxyarabinitol-binding analyses were performed on the total and soluble cellular E. coli proteins to confirm chaperone expression and measure the differences in Rubisco content and functional assembly (Figure 2). A comparison of the relative banding intensities in the total and soluble protein samples analysed by SDS/PAGE (Figures 2A and 2B respectively) showed the chaperones were highly expressed and soluble, while the majority of the abundant L subunit produced was prone to aggregation and was predominantly insoluble. Only the GroEL and its co-chaperonin GroES stimulated assembly of the M259T and wild-type PCC6301 Rubisco in E. coli.
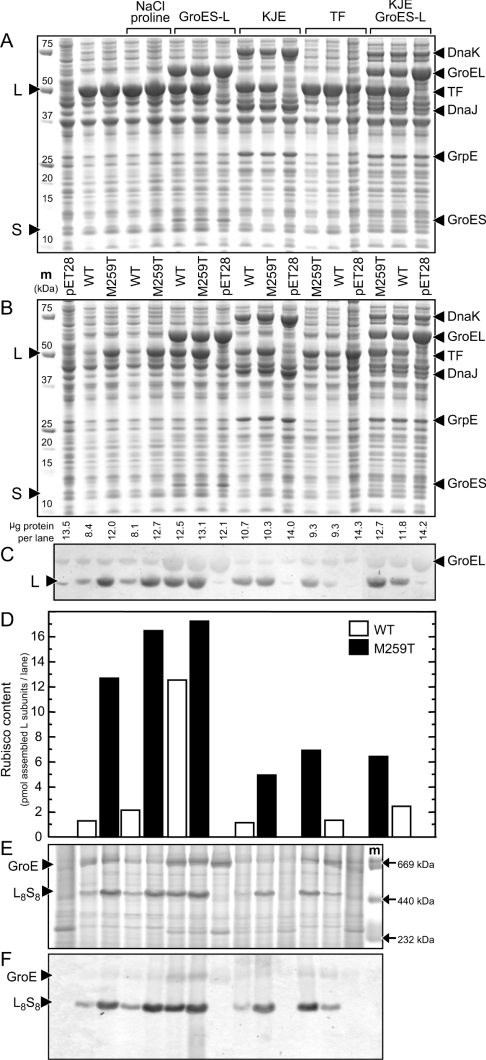
Figure 2. Influence of osmolites and E. coli chaperones on the expression and assembly of wild-type (WT) and the M259T Synechococcus PCC6301 Rubisco variants in BL21(DE3) E. coli.
Total (A) and soluble (B) cellular protein from 1.5×106 cells separated by SDS/PAGE and visualized by Coomassie Blue staining. The relative amount of soluble protein loaded per lane is shown. (C) Detection of the L subunit in the soluble cellular protein by immunoblotting using an antibody against tobacco Rubisco that also recognizes the GroEL chaperonin and an unknown 50 kDa E. coli protein. (D) The amount of L subunit assembled into functional Rubisco measured by [14C]carboxyarabinitol-P2 binding. Soluble protein [twice that loaded in (B)] separated by non-denaturing PAGE, and the position of correctly assembled L8S8 Rubisco and the GroES–GroEL chaperonin complex (GroE) identified by Coomassie Blue staining (E) and immunodetection (F) with the tobacco Rubisco antibody. m, molecular mass markers (sizes shown). GroES–L, GroES (10.4 kDa) and GroEL (57.3 kDa) chaperonins; KJE, DnaK (70 kDa)-DnaJ (41 kDa)-GrpE (22 kDa) chaperones; TF, trigger factor (50 kDa).
Differences in the amount of soluble L subunit produced in the E.coli BL21(DE3) cells after 5 h of induction with IPTG at 23 °C (compared with 16 h in Figure 1) were confirmed by immunoblot analysis (Figure 2C) and correlated with the amount of L8S8 enzyme measured by [14C]-carboxyarabinitol-P2 binding (Figure 2D), and non-denaturing PAGE (Figures 2E and 2F). Without additional GroES–GroEL the amount of functional M259T Rubisco produced was approx. 9-fold that of the wild-type. Overexpression of GroES–GroEL enhanced the yield of functional wild-type Rubisco by approx. 9-fold, whereas the excess chaperonins evinced only a 30% improvement in the amount of assembled M259T enzyme. In contrast, overexpression of the KJE and TF molecular chaperones reduced the amount of soluble wild-type and M259T L subunits produced (and accordingly the amount of L8S8 enzyme) by 20% and 40–60% respectively, indicating that incompatibilities between these chaperones and the L subunits hampered their productive folding (Figures 2C–2F). The overexpression of GroES–GroEL and KJE effected modest improvements of 30% and 70% in the amount of functional M259T and wild-type Rubisco produced respectively (Figure 2); no stimulation in functional Rubisco production by GroES–GroEL was evident in cells producing excess TF (results not shown). This suggests that under elevated levels of KJE and TF, assembly of L8S8 enzyme is limited primarily by the co-translational processing of both the wild-type and mutated Synechococcus L subunits.
Osmolite-assisted folding
Addition of the osmolite, proline, to the growth medium suppressed self-aggregation of the L subunits within E. coli. High concentrations of salt cause E. coli cells to concentrate proline in vivo and decrease the propensity of some proteins to aggregate [34]. In cells grown in LB supplemented with 20 mM proline and 300 mM NaCl, the solubility of both aggregation-prone L subunits for wild-type and M259T Rubiscos were both enhanced by 35% (Figure 2). The amount of additional soluble M259T L subunit exceeded the amount of extra wild-type soluble L subunit by a factor of approx. 5, indicating that aggregation of the mutant L subunit is more readily suppressed by proline.
In vitro thermostability and solubility
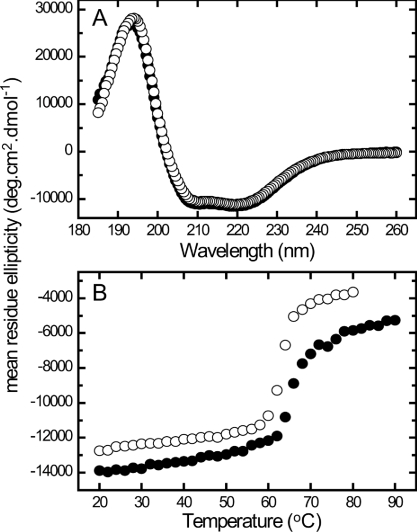
The structural integrity of the wild-type and M259T Rubiscos were compared by CD spectroscopy. Both enzymes showed matching CD spectra, indicating that their secondary structures were identical (Figure 3A). Thermally induced unfolding of the enzymes was also monitored by CD at 222 nm (Figure 3B). At this wavelength the melting temperature (TM) for M259T Rubisco (TM=63.5 °C) was slightly lower than the wild-type enzyme (TM=65 °C), suggesting that the M259T mutation may slightly compromise the structural integrity of the L8S8 complex. Using the same enzyme preparations, no intrinsic improvement in the solubility of the M259T Rubisco relative to wild-type was evident by ammonium sulfate precipitation analyses (results not shown).
Figure 3. Unfolding of the M259T and wild-type Rubiscos.
(A) CD spectra of equimolar amounts (0.31 μM) of His6-tagged wild-type (WT) (●) and M259T (○) Rubisco are indistinguishable in the far-UV region indicating equivalent structural folding. (B) Thermally induced unfolding transitions of the Rubiscos monitored by CD at 222 nm.
DISCUSSION
The expression of the prkA gene by E. coli is detrimental to cell viability because the bacterium does not ordinarily metabolize its catalytic product, ribulose-P2 [35]. In the present study, we demonstrated that the adaptive M259T mutation overcame ribulose-P2 toxicity during selection by increasing the pool of correctly folded Rubisco molecules. To our knowledge, this is the first reported mutation that confers an improvement to productive Rubisco assembly within E. coli.
Our results suggest that the M259T mutation may improve the capacity for productive folding of the L subunits by either reducing their propensity to form kinetically trapped misfolded intermediates and/or by improving their post translational processing by GroES–GroEL chaperonins. The increased assembly of the M259T L8S8 holoenzyme (relative to the wild-type control) in cells treated with the osmolite proline (Figure 2) confirmed that the mutant L subunits were less prone to unproductive aggregation. Overexpression of both the TF and KJE chaperones clearly constrained productive assembly of both wild-type and M259T Rubiscos, and this limitation was not overcome by the overproduction of GroES–GroEL. This implies there are incompatibilities between the L subunits and the co-translational activities of the TF and the KJE chaperone machinery, resulting in an increased susceptibility of the L subunit to form misfolded intermediates. However, at normal physiological levels of TF and KJE excess levels of GroES–GroEL enhance wild-type L8S8 production 9-fold, indicating under these physiological conditions productive folding of L subunits was limited by post-translational processing by the GroES–GroEL chaperonin complex, which is consistent with previous observations [14,32]. Similar yields of the M259T Rubisco were obtained at normal physiological chaperone levels. This implies the post-translational assembly limitation seen for wild-type L subunit is not as severe for the mutated L subunit, suggesting the M259T mutation improves productive interaction between the L subunit and the GroES–GroEL chaperonin complex. Under saturating levels of GroES–GroEL, the marginal improvements in M259T Rubisco production (0.3-fold yield increase; Figure 2) suggests productive folding of the M259T L subunits become limited prior to their transfer to GroEL, possibly stemming from incompatibilities with the TF and KJE chaperone machineries.
The propensity for the Synechococcus PCC6301 L subunits to misfold and aggregate in E. coli indicates that its chaperones (particularly TF and KJE [27,33]) have not been optimized for this protein. These incompatibility problems suggest that wild-type Rubisco assembly might be improved by the transplantation of the analogous chaperone genes from Synechococcus PCC6301 into E. coli or by directed evolution of the E. coli chaperones towards optimum L8S8 assembly. A limitation to both strategies is the possibility that the folding and assembly of native E. coli proteins may be unfavourably impaired, as observed previously when increasing GFP (green fluorescent protein) expression in E. coli through directed evolution of GroES–GroEL [36].
Previous amino acid replacements in Rubiscos, introduced through site-directed or random mutagenesis, have sometimes conferred improvement in one or more catalytic properties, but always to the detriment of another [1,6,29,37]. Such catalytic trade-offs are apparently commonplace for Rubiscos in both natural and artificial selection. However, the M259T Rubisco characterized here is novel in that the improvements in kccat and KM for CO2 occurred with little, or no, detriment to its specificity (Sc/o) or its KM for ribulose-P2 (Table 2). Notably, the 12% improvement in kccat for the M259T Rubisco contrasts with the 5-fold difference in specific activity measured previously for purified His6-tagged wild-type and M259T Rubisco [16]. Previous mutagenic studies have highlighted that subtle modifications to the length and amino acid composition of the Synechococcus Rubisco L subunit C-terminus significantly perturbs the kinetics of the enzyme [17,28,29]. The extent to which the C-terminally appended GHHHHHH sequence affected the kinetics of the wild-type and M259T Rubiscos relative to each other and the native (non-tagged) enzymes was not examined.
Curiously Met259 is positioned close to the surface of the protein, within the solvent channel that traverses the hexadecamer along its fourfold axis (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/404/bj4040517add.htm). At this position Met259 does not interact closely with any active site residues or with residues from other subunits in the L8S8 complex. Other mutagenic studies on form I Rubiscos, particularly those targeting modifications to residues at the L and S subunit interface, have also shown that perturbations to residues distant from the active site have significant influences on the kinetic properties of enzyme [6,37]. The changes in enzyme kinetics are caused by subtle changes to the active site geometry, and/or changes in the conformational dynamics of the L8S8 holoenzyme (either in mobile structural elements such as loop 6 [1,4,38] or in larger scale conformational changes [39]). Curiously the same M259T mutation has been selected previously using a trans-complementation screen of an XL1-Red generated Synechococcus PCC6301 Rubisco mutant library using a Rubisco deficient strain of the bacterium Rhodobacter capsulatus [40]. Notably the expression level, and kinetics, of the selected M259T variant in R. capsulatus were not quantified and therefore it remains to be assessed whether, as in E. coli, the M259T Rubisco supported growth of the mutated bacterium due to its improved tendency toward productive assembly.
Modest changes were observed in the kinetic properties of the two other Rubisco variants selected in E. coli, both of which shared the M259T mutation. The additional mutations in the A8S/M259T and M259T/F342S enzymes resulted in further improvements in their Kc and KRuBP, with slight reductions in Sc/o and kccat (compared with the parental M259T enzyme; Table 2). Like the M259T mutation, it is not readily evident from the positioning of both these amino acids in the Synechococcus Rubisco crystal structure how they impart the changes in kinetics. Ala8 is within the highly divergent N-terminal region of the L subunit exposed to the external solvent that, in the crystal structure, resides at the interface between an L and an S subunit (see Supplementary Figure 1). Despite the high variability at the N-termini between different L subunits, this region has a significant influence on the catalytic prowess of both plant and bacterial Rubiscos (see [29] for a summary). Phe342 is more highly conserved, presumably due to its close location downstream of the highly conserved loop 6 region (Gly326–Gly334 in PCC6301, which corresponds to residues 329–337 in the spinach L subunit), which plays an integral part during catalysis [1]. When compared with other cyanobacteria L subunit sequences (see below) only the aromatic phenylalanine and tyrosine amino acids are found at the equivalent position to codon 342. Again, two other Phe324 mutants were identified in the aforementioned R.capsulatus-based selection of Synechococcus Rubisco; the kinetics of the F324V, but not the F324I, enzyme were characterized [40]. Comparable to that measured here for the M259T/F342S Rubisco, the KRuBP for the F342V enzyme was reduced by approx. 50%, the kccat was approx. 30% lower and the Sc/o marginally compromised. However, in contrast to the 24% decrease in Kc for the M259T/F342S Rubisco the Kc was approx. 50% higher for the F342V enzyme. Since the individual A8S and F342S substitutions did not emerge in the selection, we did not examine their effects upon the kinetic properties of Synechococcus PCC6301 Rubisco.
Alignment of other cyanobacterial L subunits shows that many natural homologues possess a threonine residue at position 259. The catalytic improvement in the Synechococcus PCC6301 Rubisco imparted by the M259T mutation raises the question: why wasn't Thr259 fixed in Nature? Comparison of 21 different cyanobacterial genomes available from the JGI (Joint Genome Institute) and NCBI (National Center for Biotechnology Information) databases found four species contained threonine, five glutamine and the remainder methionine residues at the position equivalent to codon 259 (see Supplementary Figure 2 at http://www.BiochemJ.org/bj/404/bj4040517add.htm). The kinetic improvements associated with the M259T mutation might not be sufficient to warrant selection of the M259T Rubisco in Nature. Alternatively, the mutation, although kinetically better, may be unfavourable in other ways, such as reducing the compatibility of the L subunit with cyanobacterial chaperone complexes or other interacting proteins, such as those involved in carboxysome formation. Clearly, future studies will have to evaluate the extent to which Rubiscos evolved in E. coli affect the fitness of their native hosts. The construction of a Synechococcus PCC6301 Rubisco mutant, similar to Synechocystis PCC6803 Syn6803Δrbc mutant [41], would enable such substitution studies.
Using transition-biased whole-gene mutagenesis, we have shown that the directed evolution of Synechococcus PCC6301 Rubisco in E. coli can lead to improvements in its biophysical and kinetic parameters. As indicated previously [16], selection for further kinetic improvements should be possible using Rubisco libraries constructed by more sophisticated mutagenic techniques. Modification of the selection process to favour the selection of mutants with improvements in other kinetic traits, such as its specificity for CO2 over O2 may also be possible. Importantly, the E. coli-based Rubisco selection may allow us to explore alternative sequence space not accessible to photosynthetic organisms whose survival is dictated by contingent physiological constraints.
Online data
Acknowledgments
We thank Ms Harley Jenks (Emony University) for assistance with reverse transcriptase–real time PCR, Dr Monica Gerth (Emony University) for help with CD, Dr Heather Kane (ANU Molecular Plant Physiology) for her help with the CO2/O2 specificity assays, Ms Emily Beck (ANU Molecular Plant Physiology) for access to her Rubisco alignment database and Dr Lisa Gloss (Washington State University) for helpful suggestions. This research was supported by NIH (National Institutes of Health)/NIAID (National Institute of Allergy and Infectious Diseases) (1 R21AI054602-01) and NIH/NIGMS (National Institute of General Medical Sciences) (1 R01 GM074264-01) grants awarded to D.G. and I.M.M. and by a Discovery grant (DP0450564) awarded to S.N.W. by the Australian Research Council.
References
- 1.Parry M. A., Andralojc P. J., Mitchell R. A., Madgwick P. J., Keys A. J. Manipulation of Rubisco: the amount, activity, function and regulation. J. Exp. Bot. 2003;54:1321–1333. doi: 10.1093/jxb/erg141. [DOI] [PubMed] [Google Scholar]
- 2.Morell M. K., Paul K., Kane H. J., Andrews T. J. Rubisco: maladapted or misunderstood? Aust. J. Bot. 1992;40:431–441. [Google Scholar]
- 3.Wingler A., Lea P. J., Quick W. P., Leegood R. C. Photorespiration: metabolic pathways and their role in stress protection. Phil. Trans. R. Soc. Lond. 2000;355:1517–1529. doi: 10.1098/rstb.2000.0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tcherkez G. G. B., Farquhar G. D., Andrews T. J. Despite slow catalysis and confused substrate specificity, all ribulose bisphosphate carboxylases may be nearly perfectly optimized. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7246–7251. doi: 10.1073/pnas.0600605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jordan D. B., Ogren W. L. Species variation in the specificity of ribulose bisphosphate carboxylase/oxygenase. Nature. 1981;291:513–515. [Google Scholar]
- 6.Spreitzer R. J., Salvucci M. E. Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Ann. Rev. Plant Biol. 2002;53:449–475. doi: 10.1146/annurev.arplant.53.100301.135233. [DOI] [PubMed] [Google Scholar]
- 7.Andrews T. J., Whitney S. M. Manipulating ribulose bisphosphate carboxylase/oxygenase in the chloroplasts of higher plants. Arch. Biochem. Biophys. 2003;414:159–169. doi: 10.1016/s0003-9861(03)00100-0. [DOI] [PubMed] [Google Scholar]
- 8.Koonin E. V. Horizontal gene transfer: the path to maturity. Mol. Microbiol. 2003;50:725–727. doi: 10.1046/j.1365-2958.2003.03808.x. [DOI] [PubMed] [Google Scholar]
- 9.Kurland C. G., Canback B., Berg O. G. Horizontal gene transfer: A critical view. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9658–9662. doi: 10.1073/pnas.1632870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delwiche C. F., Palmer J. D. Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol. Biol. Evol. 1996;13:873–882. doi: 10.1093/oxfordjournals.molbev.a025647. [DOI] [PubMed] [Google Scholar]
- 11.Thomas C. M., Nielsen K. M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 12.Kimura M. Cambridge: Cambridge University Press; 1983. The Neutral Theory of Molecular Evolution. [Google Scholar]
- 13.Thornton J. W. Resurrecting ancient genes: experimental analysis of extinct molecules. Nat. Rev. Genet. 2004;5:366–375. doi: 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- 14.Goloubinoff P., Gatenby A. A., Lorimer G. H. GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature. 1989;337:44–47. doi: 10.1038/337044a0. [DOI] [PubMed] [Google Scholar]
- 15.Checa S. K., Viale A. M. The 70-kDa heat-shock protein/DnaK chaperone system is required for the productive folding of ribulose-bisphosphate carboxylase subunits in Escherichia coli. Eur. J. Biochem. 1997;248:848–855. doi: 10.1111/j.1432-1033.1997.00848.x. [DOI] [PubMed] [Google Scholar]
- 16.Parikh M. R., Greene D. N., Woods K. K., Matsumura I. Directed evolution of Rubisco hypermorphs through genetic selection in engineered E. coli. Prot. Eng. Des. Sel. 2006;19:113–119. doi: 10.1093/protein/gzj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitney S. M., Sharwood R. E. Linked Rubisco subunits can assemble into functional oligomers without impeding catalytic performance. J. Biol. Chem. 2007;282:3809–3818. doi: 10.1074/jbc.M610479200. [DOI] [PubMed] [Google Scholar]
- 18.Kane H. J., Wilkin J. M., Portis A. R., Andrews T. J. Potent inhibition of ribulose-bisphosphate carboxylase by an oxidized impurity in ribulose-1,5-bisphosphate. Plant Physiol. 1998;117:1059–1069. doi: 10.1104/pp.117.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierce J., Tolbert N. E., Barker R. Interaction of ribulosebisphosphate carboxylase/oxygenase with transition-state analogues. Biochemistry. 1980;19:934–942. doi: 10.1021/bi00546a018. [DOI] [PubMed] [Google Scholar]
- 20.Schloss J. V. Comparative affinities of the epimeric reaction-intermediate analogs 2- and 4-carboxy-D-arabinitol 1,5-bisphosphate for spinach ribulose 1,5-bisphosphate carboxylase. J. Biol. Chem. 1988;263:4145–4150. [PubMed] [Google Scholar]
- 21.Ruuska S., Andrews T. J., Badger M. R., Hudson G. S., Laisk A., Price G. D., von Caemmerer S. The interplay between limiting processes in C3 photosynthesis studied by rapid-response gas exchange using transgenic tobacco impaired in photosynthesis. Aust. J. Plant Physiol. 1998;25:859–870. [Google Scholar]
- 22.Morell M. K., Paul K., O'shea N. J., Kane H. J., Andrews T. J. Mutations of an active site threonyl residue promote β elimination and other side reactions of the enediol intermediate of the ribulosebisphosphate carboxylase reaction. J. Biol. Chem. 1994;269:8091–8098. [PubMed] [Google Scholar]
- 23.Andrews T. J. Catalysis by cyanobacterial ribulose-bisphosphate carboxylase large subunits in the complete absence of small subunits. J. Biol. Chem. 1988;263:12213–12219. [PubMed] [Google Scholar]
- 24.Kane H. J., Viil J., Entsch B., Paul K., Morell M. K., Andrews T. J. An improved method for measuring the CO2/O2 specificity of ribulosebisphosphate carboxylase-oxygenase. Aust. J. Plant Physiol. 1994;21:449–461. [Google Scholar]
- 25.Matsumura I., Ellington A. D. In vitro evolution of β-glucuronidase into a β-galactosidase proceeds through non-specific intermediates. J. Mol. Biol. 2001;305:331–339. doi: 10.1006/jmbi.2000.4259. [DOI] [PubMed] [Google Scholar]
- 26.Nishihara K., Kanemori M., Kitagawa M., Yanagi H., Yura T. Chaperone coexpression plasmids: differential and synergistic roles of DnaK–DnaJ–GrpE and GroEL–GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl. Environ. Microbiol. 1998;64:1694–1699. doi: 10.1128/aem.64.5.1694-1699.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishihara K., Kanemori M., Yanagi H., Yura T. Overexpression of trigger factor prevents aggregation of recombinant proteins in Escherichia coli. Appl. Environ. Microbiol. 2000;66:884–889. doi: 10.1128/aem.66.3.884-889.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gutteridge S. Mutagenesis and expression of cloned Rubisco (ribulose-bisphosphate carboxylase) Bioch. Soc. Trans. 1986;14:28–29. doi: 10.1042/bst0140028. [DOI] [PubMed] [Google Scholar]
- 29.Kellogg E. A., Juliano N. D. The structure and function of Rubisco and their implications for systematic studies. Am. J. Bot. 1997;84:413–428. [PubMed] [Google Scholar]
- 30.Emlyn-Jones D., Woodger F. J., Price G. D., Whitney S. M. RbcX can function as a Rubisco chaperonin, but is non-essential in Synechococcus PCC7942. Plant Cell. Physiol. 2006;47:1630–1640. doi: 10.1093/pcp/pcl028. [DOI] [PubMed] [Google Scholar]
- 31.Andrews T. J., Ballment B. A rapid, sensitive method for quantitating subunits in purified ribulose bisphosphate carboxylase preparations. Plant Physiol. 1984;75:508–510. doi: 10.1104/pp.75.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larimer F. W., Soper T. S. Overproduction of Anabaena 7120 ribulose-bisphosphate carboxylase/oxygenase in Escherichia coli. Gene. 1993;126:85–92. doi: 10.1016/0378-1119(93)90593-r. [DOI] [PubMed] [Google Scholar]
- 33.Kaiser C. M., Chang H.-C., Agashe V. R., Lakshmipathy S. K., Etchells S. A., Hayer-Hartl M., Hartl F. U., Barral J. M. Real-time observation of trigger factor function on translating ribosomes. Nature. 2006;444:455–460. doi: 10.1038/nature05225. [DOI] [PubMed] [Google Scholar]
- 34.Ignatova Z., Gierasch L. M. Inhibition of protein aggregation in vitro and in vivo by a natural osmoprotectant. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13357–13361. doi: 10.1073/pnas.0603772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hudson G. S., Morell M. K., Arvidsson Y. B. C., Andrews T. J. Synthesis of spinach phosphoribulokinase and ribulose 1, 5-bisphosphate in Escherichia coli. Aust. J. Plant Physiol. 1992;19:213–221. [Google Scholar]
- 36.Wang J. D., Herman C., Tipton K. A., Gross C. A., Weissman J. S. Directed evolution of substrate-optimized GroEL/S chaperonins. 2002;111:1027–1039. doi: 10.1016/s0092-8674(02)01198-4. [DOI] [PubMed] [Google Scholar]
- 37.Spreitzer R. J., Peddi S. R., Satagopan S. Phylogenetic engineering at an interface between large and small subunits imparts land-plant kinetic properties to algal Rubisco. Proc. Natl. Acad. Sci. U.S.A. 2005;102:17225–17230. doi: 10.1073/pnas.0508042102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cleland W. W., Andrews T. J., Gutteridge S., Hartman F. C., Lorimer G. H. Mechanism of Rubisco: the carbamate as general base. Chem. Rev. 1998;98:549–562. doi: 10.1021/cr970010r. [DOI] [PubMed] [Google Scholar]
- 39.Schlitter J., Wildner G. F. The kinetics of conformation change as determinant of Rubisco's specificity. Photosynth. Res. 2000;65:7–13. doi: 10.1023/A:1006425607995. [DOI] [PubMed] [Google Scholar]
- 40.Smith S. A., Tabita F. R. Positive and negative selection of mutant forms of prokaryotic (cyanobacterial) ribulose-1,5-bisphosphate carboxylase/oxygenase. J. Mol. Biol. 2003;331:557–569. doi: 10.1016/s0022-2836(03)00786-1. [DOI] [PubMed] [Google Scholar]
- 41.Amichay D., Levitz R., Gurevitz M. Construction of a Synechocystis PCC6803 mutant suitable for the study of variant hexadecameric ribulose bisphosphate carboxylase/oxygenase enzymes. Plant Mol. Biol. 1993;23:465–476. doi: 10.1007/BF00019295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.