Abstract
Bacterial genes required for proper partitioning consist of two transacting genes that encode proteins and a cis-acting gene that functions like a centromere. Plasmids actively partitioning by means of these genes migrate from midcell to the cell quarters and are tethered to these sites until the cells divide. Previously the partitioning genes were mainly found on plasmids and phages in Escherichia coli. However, progress in genome sequencing reveals that partitioning genes are ubiquitous in many bacterial plasmids and chromosomes. Each homologue of the two transacting genes belongs to a family, ParA or ParB. Moreover, phylogenic analysis of members of the ParA and ParB families indicates that each member falls into a chromosomal group or an extrachromosomal group. It is known that the parAB genes in the chromosomal group are located on relatively conserved chromosomal regions in several bacterial species. This suggests that the parAB genes were transferred from a chromosome to plasmids and phages, so the genes have diverged among bacterial species. To support this possibility, we show that the Bacillus subtilis Soj and Spo0J members of the ParAB families are responsible for the specific localization of plasmids at cell quarters in E. coli and can function as partition proteins. Host factors to tether actively partitioning plasmids at subcellular sites may be conserved in Gram-negative and Gram-positive bacteria so that phages and plasmids with the ParAB partitioning system can be stably inherited in host cells across bacterial species.
Low copy-number plasmids are stably maintained in host Escherichia coli cells because of active partitioning mechanisms; in the cases of F plasmid and P1 phage, which is a plasmid in lysogenized host cells, these are specified by the sopABC and parABS genes, respectively (1). SopA and P1/ParA are similar in amino acid sequence and function, as are SopB and P1/ParB. Both sopC and parS are cis-acting sites that function as centromere-like elements. In the case of the sopABC mechanism, the SopB protein binds to sopC. SopA interacts with the SopB-sopC complex to ensure proper segregation. Plasmids actively partitioned by the sopABC or parABS systems are localized at specific subcellular sites throughout the cell-division cycle (2–4). In newborn cells, the plasmids are localized at midcell. Replicated plasmids are then rapidly relocalized at the 1/4 and 3/4 positions of cell length. The SopAB-sopC complex probably binds to unidentified host factors that localize at the cell quarters, tethering actively partitioning plasmids at these specific subcellular sites.
Many homologues of SopAB and P1/ParAB have been identified in various plasmids and phages, and in fact several members are involved in the accurate partitioning of extrachromosomal elements (1, 5–7). We therefore call these homologues the ParA and ParB families. Recent genome searches have revealed that genes of members of both families are found in many bacterial genomes (8, 9). Homologues of the parAB genes are often located in a region near the replication origin (oriC) that is well known to conserve several gene homologues and similar gene order in Gram-positive and -negative bacteria (10, 11). Members of both families play an important role in chromosome partitioning in Caulobacter crescentus (12) and Bacillus subtilis (13). B. subtilis has the Soj and Spo0J proteins, members of the ParA and ParB families. Spo0J binds to multiple DNA sites in the origin-proximal 20% of the chromosome, which is organized into a compact chromosomal region including the replication origin (14, 15). Thus, assembled Spo0J proteins migrate with the replication origin during the cell division cycle (16–19). In addition to a role in chromosome partitioning, plasmid molecules with the Spo0J-binding site (parS) are accurately segregated into daughter cells in B. subtilis, dependent on the Soj and Spo0J proteins (14). Thus, the parS sequence functions as a cis-acting site for plasmid partitioning. On the other hand, almost all species in the proteobacterial γ subdivision, including E. coli, Shigella species, and Salmonella typhymurium, have neither parA nor parB genes on their chromosomes, at least of the genomes that have been sequenced so far. Instead, conjugative plasmids and temperate phages in these bacteria possess both parA and parB partitioning genes and a cis-acting site for stable inheritance in the host cells. In the above pathogenic bacteria, plasmids and phages are intimately related to the transfer of pathogenic and drug-resistance genes between and within species.
Here, we report a phylogenetic analysis of members of the ParA and ParB families. We also show that the ParAB partitioning mechanisms on chromosomes and plasmids may have evolved from an ancient one. To support this possibility, we also demonstrate that B. subtilis Soj and Spo0J proteins and a parS site can carry out accurate segregation of plasmids in E. coli cells. These findings indicate that host components involved in plasmid partitioning are highly conserved between Gram-negative and Gram-positive bacteria.
Materials and Methods
Bacterial Strains and Culture.
CSH50 [ara, Δ(lac-pro), strA, thi] (20) cells harboring miniF plasmids were grown in LB with 20 μg/ml ampicillin at 37°C, and cells in mid-log phase were transferred to nonselective LB without ampicillin at zero time. After 6 and 12 h, an aliquot of cells was spread on LB plates. To determine the percentage of cells containing plasmids that have an ampicillin-resistant gene (bla), 300 colonies that had grown on the plate were streaked on LB plates containing 20 μg/ml ampicillin. Generation time (doubling time) was determined by turbidity of cultures.
Plasmid Construction.
The plasmid pUC25E contains a B. subtilis chromosomal DNA segment including the soj and spo0J-parS genes. After digestion of pUC25E by EcoRI, the digested fragment including the soj and spo0J-parS genes was inserted into the miniF plasmid pXX705 (2) at EcoRI sites, resulting in pXX762. A Spo0J-binding sequence, parS (TGTTTCACGTGAAACA) (14), was synthesized as the parS DNA fragment (49 bp) with BamHl sites at both ends. The parS DNA fragment was inserted into pXX762 and pXX705 at BamHI sites, resulting in pXX765 and pXX764, respectively. Plasmid pXX762 was digested with EcoRI to delete the DNA fragment including soj and spo0J-parS. Digested DNA fragments were circularized by self ligation, resulting in pXX766.
Homology Search and Phylogenetic Tree.
blast searches with the default parameters were performed by using amino acid databases including Protein Identification Resource, SWISS-PROT, Protein Data Bank, and DAD at the DNA Data Bank of Japan (DDBJ; http://crick.genes.nig.ac.jp/homology/blast-e.shtml). Preliminary sequence data were obtained from the Institute for Genomic Research web site at http://www.tigr.org. Alignments of the ParAB members were analyzed by a clustalw program at DDBJ (http://crick.genes.nig.ac.jp/homology/clustalw-e.shtml). On the basis of parameters obtained by using the clustalw method (21), phylogenetic trees were drawn by using treeview software (http://taxonomy.zoology.gla.ac.uk/rod/rod.html). According to the annotation (source organism) in the description of registered amino acid sequences, we sorted members of the ParAB families into chromosome-derived (genes located on chromosomes) and plasmid-derived (on plasmid and phages) groups. A chromosome is defined by a replicon that has housekeeping genes (22, 23).
Copy Number and Multimer Resolution.
Total DNA was prepared from exponentially growing cells containing plasmids. DNA (250 μg) was digested with BamHI and EcoRI, separated on a 0.8% agarose gel, and transferred to a positively charged nylon membrane. Hybridization was carried out by using both digoxigenin-labeled plasmid (pXX705) and oriC DNA-specific probes simultaneously. The oriC probe DNA segment was amplified from E. coli chromosomal DNA by PCR (24). The hybridized probes were immunodetected and visualized with a chemiluminescence substrate CSPD (Roche Diagnostics). The intensity of detected bands was determined by lumino-image analyzer LAS-1000 (Fuji).
Plasmid DNA was extracted from exponentially growing cells by using a Qiagen (Chatsworth, CA) plasmid purification kit. Plasmid DNA was separated on a 0.6% agarose gel and detected bands by using the chemiluminescence reaction described above after Southern hybridization.
Fluorescence in Situ Hybridization.
E. coli cells harboring pXX762 or pXX764 were grown in M9-minimal medium with proline (50 μg/ml), glucose (0.5%), and thiamine (2 μg/ml). To fix cells, an equal volume of fixation solution [methanol/acetic acid (3:1)] was added directly to a bacterial culture growing exponentially in M9 medium at 37°C. The fixed cells were collected by centrifugation and suspended in 1/10 volume of fixation solution. To detect plasmid DNA in fixed cells by in situ hybridization, we used the miniF plasmid DNA of pXX704 according to the procedure described previously (2, 24). All images were recorded with a cooled charge-coupled device camera, 05810–01 (Hamamatsu Photonics, Hamamatsu City, Japan), by using a phase contrast and fluorescence microscope (Nikon).
Results
Phylogenetic Trees of the ParAB Families.
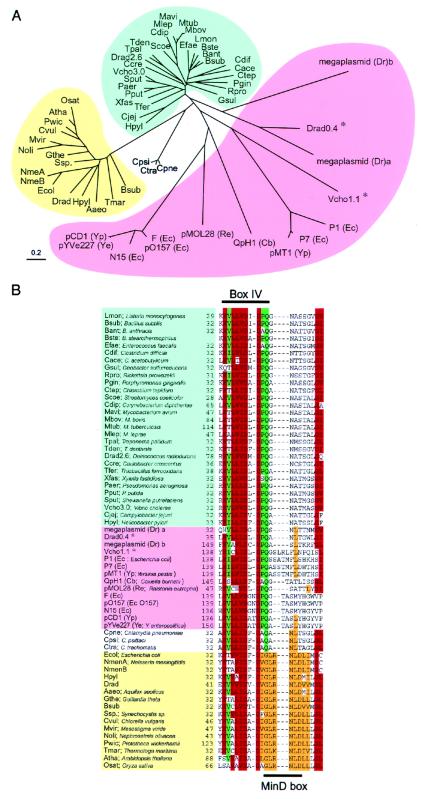
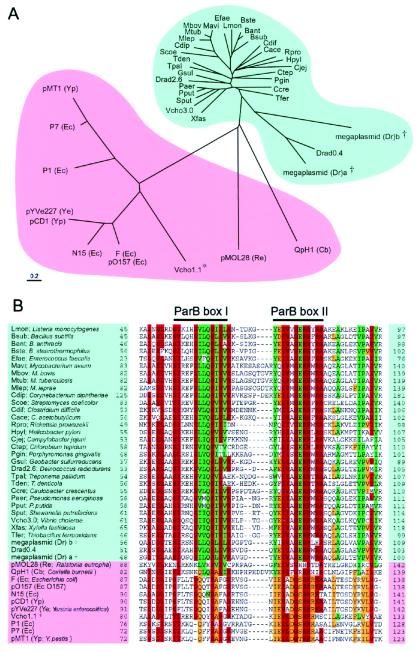
On the basis of amino acid sequences, homologues of SopA, SopB, Soj, and Spo0J proteins were sought by blast search. More than 50 homologues of SopA and Soj or SopB and Spo0J have been detected so far (Table 2, which is published as supplemental data on the PNAS web site, www.pnas.org). For a given pair of partition genes, sopA-sopB, parA-parB, and soj-spo0J consist of an operon in that order. For present purposes, we therefore selected members of ParAB families from the blast results that show this gene organization. Amino acid sequence comparison of members of the ParA and ParB families appears to fall into two groups (Figs. 1A and 2A and supplemental Figs. 7 and 8, which are published as supplemental data on the PNAS web site). Members in one group are mainly derived from chromosomes, which we call the chromosomal group. Members in the other are mainly derived from plasmids, which we call the extrachromosomal group. Some amino acid residues were mainly conserved in the chromosomal group, whereas others were mainly conserved in the extrachromosomal group (Figs. 2B, 7, and 8). Phylogenetic trees of ParAB members revealed that the amino acid sequences of ParA and ParB proteins are well conserved when encoded by the chromosomes (Figs. 1A and 2A). In contrast, those of ParAB on plasmids or phages were diverse, having only limited regions of similarity. However, several conserved motifs were found in all members of each family (Figs. 7 and 8). In Fig. 1B, one of the conserved motifs in the ParB family is indicated as the ParB box.
Figure 1.
Phylogenetic tree (A) and alignment of partial amino acid sequences (B) of ParA and MinD homologues in various bacteria. Forty-three ParA and sixteen MinD homologues were found in databases by using the blast algorithm. Homologues of the E. coli MinD protein are shown when the blast score was over 65. The ParA homologues that belong to the chromosomal group are indicated in blue and those to the extrachromosomal group in pink. The MinD members are indicated in yellow. In the aligned sequences, conserved residues and conservative substitutions (V/I/L, T/S, D/E, N/Q, Y/F, R/K) are shaded in red if present in 48 or more sequences of 60 ParA/MinD homologues, in green if present in 34 or more sequences of 43 ParA homologues, and in orange if present in 13 or more sequences of 16 MinD homologues. Amino acid residues are indicated with the one-letter code. *, parA genes in the extrachromosomal group were located on chromosomes.
Figure 2.
Phylogenetic tree (A) and alignment of partial amino acid sequences (B) of ParB in various bacteria. Forty-three ParB members were found in databases by using the blast algorithm. The ParB homologues that belong to the chromosomal group are indicated in blue and those from the extrachromosomal group in pink. In the aligned sequences, conserved residues and conservative substitutions (V/I/L, T/S, D/E, N/Q, Y/F, R/K) are shaded in red if present in 34 or more sequences of 43 ParB homologues, in green if present in 26 or more sequences of 31 ParB homologues on the chromosomes, and in orange if present in 8 or more sequences of 12 ParB homologues on extrachromosomal elements. * parB genes in the extrachromosomal group were located on chromosomes; †parB genes in the chromosomal group were located on plasmids.
Interestingly, the genomes of Vibrio species including V. cholerae consist of two circular chromosomes (25, 26). Two sets of parAB genes are located on the V. cholerae chromosomes. The ParAB proteins specified by the smaller chromosome (Vcho 1.1) showed more similarity to ParAB members of the extrachromosomal group according to the alignment of amino acid sequences, whereas the ParAB proteins specified by the larger chromosome (Vcho 3.0) were closer to the chromosomal group (Figs. 1A and 2A). The genome of Deinococcus radiodurans R1 consists of two chromosomes, a megaplasmid, and a small plasmid (27, 28). This organism has four different ParAB pairs: two are located on the two chromosomes (Drad2.6 and Drad0.4), and the others are located on the megaplasmid. The ParAB pairs on the 2.6 Mb chromosome belonged to the chromosomal group. On the other hand, ParA homologues on the 0.4 Mb chromosome, and the megaplasmid belonged to the chromosomal group according to conserved amino acids residues, whereas ParB homologues belonged to the extrachromosomal group (Figs. 1B, 2B, 7, and 8).
The ParA/MinD Superfamily Is an ATPase Family.
MinD proteins, which regulate nucleation sites for the cell division protein FtsZ, show sequence homology to members of the ParA family (29). A phylogenetic tree shows that MinD proteins belong to a subgroup of the ParA/MinD superfamily (Fig. 1A). Both the MinD and ParA members share the Walker type ATPase domains (30). ATPase activity is essential for biological function in MinD (29), SopA (31), and P1/ParA (32, 33). Sequences in box IV of the ATPase motifs, which include the catalytic carboxylate for ATP hydrolysis, are relatively diverse in the various subgroups of the Walker type ATPases (34). However, the sequences in box IV are highly conserved in ParA and MinD proteins, suggesting that the two proteins have similar functions involving ATP hydrolysis (Fig. 1B). A conserved motif unique to MinD proteins was located adjacent to box IV and is indicated as the MinD box (six amino acids: GLRNLD).
B. subtilis ParAB Members Can Function Biologically in E. coli Cells.
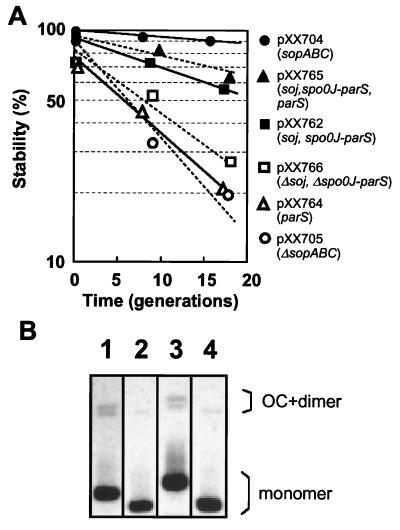
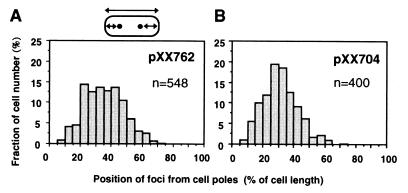
We examined whether the Soj and Spo0J partitioning proteins can function in E. coli cells. Eight parS sites are located on the B. subtilis chromosome, and one of them is located within the coding region of spo0J gene (14). We refer to the spo0J gene including parS as spo0J-parS. If Soj and Spo0J partition proteins and a parS site can function normally in E. coli, then a plasmid with the soj and spo0J-parS genes would be actively partitioned into daughter cells and stably maintained. The DNA segment including soj and spo0J-parS was inserted into an unstable miniF plasmid that had the sopABC segment deleted. In E. coli, this miniF plasmid (pXX762) was stably maintained under nonselective growth conditions (Fig. 3A). However, the stability of the miniF plasmid with soj and spo0J-parS was less than that of a miniF plasmid with sopABC (pXX704). A miniF plasmid with only parS (pXX764) was as unstable as the miniF plasmid without sopABC (pXX705). As several parS sites are located on the B. subtilis chromosome (14), the stability of a plasmid with multiple parS sites was tested. However, the presence of an extra parS site (pXX762) was not significantly effective in improving the stability of the miniF plasmid. When the DNA segment including soj-spo0J was deleted from the pXX762 plasmid, the deleted plasmid (pXX766) again became unstable in cells. These results suggest that the soj and spo0J-parS genes are able to ensure stable maintenance of the miniF plasmids in E. coli. We showed that the soj and spo0J-parS genes do not significantly affect copy number or multimer resolution of the plasmid, as is also true for the sopABC genes (Fig. 3B and Table 1).
Figure 3.
Stability and multimer resolution of various miniF plasmids. (A) Cells harbored miniF plasmid pXX704 (sopABC; ●), pXX765 (soj, spo0J-parS, parS; ▴), pXX762 (soj, spo0J-parS; ■), pXX766 (Δsoj, Δspo0J-parS; □), pXX764 (parS; Δ), and a miniF plasmid pXX705 (ΔsopABC; ○). (B) Purified plasmid DNA (100 ng) was separated on a 0.6% agarose gel and detected with a chemiluminescence reaction after Southern hybridization. OC, open circular. Lanes: 1, pXX704 (sopABC); 2, pXX705 (ΔsopABC); 3, pXX762 (soj, spo0J-parS); 4, pXX764 (parS).
Table 1.
Copy number comparison of plasmid with and without the ParABC partitioning system
| Plasmid | Fraction of cells with plasmid | Plasmid/oriC in total population of cells | Plasmid/oriC per* plasmid-containing cell |
|---|---|---|---|
| pXX704 (sopABC) | 0.89 | 0.47 | 0.52 |
| pXX705 (ΔsopABC) | 0.29 | 0.16 | 0.56 |
| pXX762 (soj, spo0J-parS) | 0.78 | 0.38 | 0.49 |
| pXX764 (parS) | 0.50 | 0.27 | 0.55 |
The plasmid/oriC ratio was divided by the fraction of cells containing a plasmid.
Plasmids with B. subtilis ParAB Members Can Localize at the Cell Quarters in E. coli.
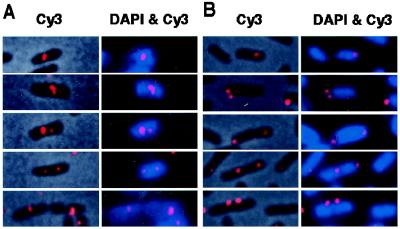
We next analyzed the subcellular localization of the actively partitioning miniF plasmid with soj and spo0J-parS by using fluorescence in situ hybridization and pXX704 DNA as probe. One or two fluorescence foci of pXX762 with soj and spo0J-parS and pXX764 with a parS site were detected in cells under the growth conditions used (Fig. 4). However, there was a remarkable difference in the subcellular localization between the two plasmids. The foci of pXX762 (soj, spo0J-parS) were located primarily in the area occupied by host chromosomal DNA, whereas the foci of pXX764 (parS) tended to be located at or near cell poles. In addition, the foci of pXX764 (parS) were often located at the interstice between two nucleoids in long cells.
Figure 4.
Localization of fluorescent foci of the miniF plasmids, pXX762 (A) and pXX764 (B). In A and B, overlaid phase contrast and fluorescence in situ hybridization (FISH) (Cy3) images of the same field are shown (Left), and overlaid Cy3 (FISH) and 4′,6-diamidino-2-phenylindole (DNA staining) fluorescence images of the same field are shown (Right). Bar = 2 μm.
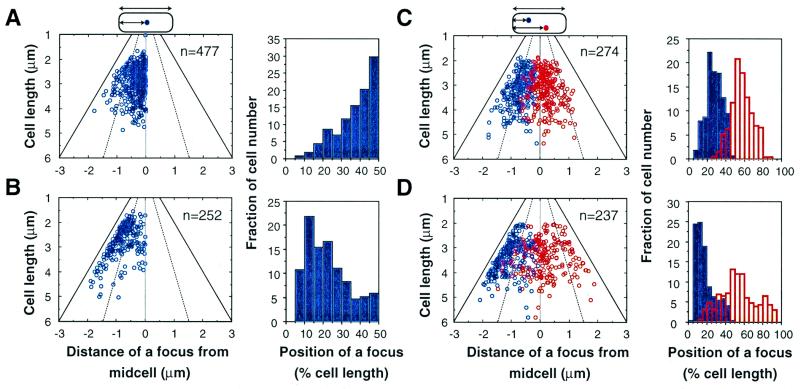
Statistical analysis of cells containing pXX762 (soj, spo0J-parS) showed that fluorescent foci in cells with a single focus were mainly localized at midcell (50.1% of the foci were localized at 40–50% of cell length; Fig. 5A). In cells with two foci, the two were clearly separated (Fig. 5C). The focus nearest to a cell pole (blue) was mainly located at or near the 25% position of cell length, and the other focus (red) was localized within the central half of the cell, most often at midcell. As shown in Fig. 6A, the foci of pXX762 (soj, spo0J-parS) were broadly distributed at 25–50% of cell length when distance was measured from each cell pole to the nearest focus in cells with two foci. These results suggest that the foci tend to be localized at the cell quarters. In contrast, cells with pXX764 (parS) had a single fluorescent focus located mainly near a cell pole (48% of the foci were localized at 0–20% of cell length; Fig. 5B). In cells with two foci of pXX764 (parS), one focus was localized at or near a cell pole, and the other was localized either near the same pole, near the opposite pole, or at midcell (Fig. 5D). It is known that miniF plasmids without an active partitioning system are often located at midcell, at the interstice between two nucleoids in long dividing cells (2). Thus, the localization patterns of a plasmid actively partitioning via soj and spo0J-parS were remarkably similar to those of plasmids actively partitioning by parABS of P1 phage and sopABC of F plasmid (2–4). Although the foci of pXX762 (soj, spo0J-parS) were broadly distributed around the 25% position of cell length, the foci of the miniF plasmid with sopABC (pXX704) were more precisely localized at the 25% position of cell length (Fig. 6). As shown in Fig. 3, pXX762 (soj, spo0J-parS) was less stable than pXX704 (sopABC).
Figure 5.
Analysis of subcellular localization of the miniF plasmid with soj-spo0J-parS. Scatter diagrams and histograms of localization of fluorescent foci in cells with a single focus of pXX762 (A) or pXX764 (B). The distance of each focus from midcell was measured from the nearest cell pole to the center of the focus. Scatter diagrams and histograms of the localization of fluorescent foci in cells with two foci carrying pXX762 (C) or pXX764 (D). The focus closest to a cell pole is shown in blue. The distance of the other focus from the same pole was measured (red). The broken lines indicate the 1/4 and 3/4 positions of cell length, and the solid lines indicate the position of a cell pole. The thin lines indicate midcell.
Figure 6.
Histogram showing the localization of fluorescent foci in cells with miniF plasmid pXX762 (soj-spo0J-parS; A) or pXX704 (sopABC; B). In cells with two foci, the distance of one focus from the nearest cell pole was measured, then that of the other focus from the other pole.
Discussion
The B. subtilis soj, spo0J-parS partitioning mechanism functions in E. coil cells, despite the fact that the two species are well separated from each other in prokaryote phylogeny, diverging at least 1,200 million years ago (35). It is plausible then that the basic mechanism used to determine subcellular positions (the cell quarters) for anchoring the ParAB partition complex is conserved and common among bacteria. Perhaps the mechanism is related to the cell division site that leads to the assembly of FtsZ protein, because the next cell division site is midcell, and 1/4 and 3/4 sites of the cell length are future cell division sites. In addition to conservation in almost all eubacteria, FtsZ homologues are widely distributed in eubacteria and plant species. It seems likely that a fundamental mechanism of determining midcell for cell division and partitioning appeared in an ancient unicellular organism.
The plasmids with soj and spo0J-parS tend to be localized at midcell in E. coli. However, the replicated plasmids were not symmetrically distributed in growing cells. The plasmids were broadly distributed around quarter positions of cell length. Presumably migration from midcell toward the quarters is inefficient, and a significant fraction of migrating plasmids do not reach the proper positions. Alternatively, active tethering of plasmids to cell quarters, which may require binding of the ParB-plasmid DNA to specific subcellular components that are localized in the cell membrane at the cell quarters, may be inefficient. Although such components may be encoded in the chromosomes of both E. coil and B. subtilis, the amino acid sequences may not be optimally similar. This is likely because the ParB proteins are a diverse family. The lower stability of pXX762 (soj, spo0J-parS) probably reflects inaccurate positioning of foci at the 1/4 and 3/4 positions of the cells.
The parAB genes are conserved in chromosomal segments in the vicinity of the replication origin in various genomes, suggesting that the ParAB member may have been transferred from a chromosome to plasmids and phages. Therefore, plasmids and phages possessing a ParAB partitioning mechanism are easily established in the new host cells. It is thought that ParAB members in the extrachromosomal group are divergent because of the transfer of plasmids and phages among bacterial species.
In phylogenetic trees, a few exceptions in the grouping of ParAB proteins suggest a significant diversity of bacterial genomes. The ParAB sets in each chromosome of V. cholerae suggest that one of the two chromosomes is derived from a plasmid, i.e., a plasmid replicon had acquired essential genes for host growth so that the plasmid eventually became a heritable part of the genome. Interestingly, V. cholerae belongs to the proteobacterial γ subdivision, most of whose members do not appear to have parA or parB homologues on their chromosomes. V. cholerae may have diverged phylogenetically earlier than other species in the proteobacterial γ subdivision. Alternatively, parAB genes in the chromosomal group may have been laterally transferred into V. cholerae from other bacteria. In the case of D. radiodurans R1, it seems that parAB genes on the smaller chromosome and the megaplasmid are intermediates between the chromosomal and extrachromosomal groups, although the largest chromosome (Drad2.6) has parAB genes of the chromosomal group. These results support the hypothesis that the smaller chromosome and the megaplasmid were separately acquired from the largest chromosome (28). Alternatively, duplicated parAB genes may have been transferred into each chromosome or plasmid and then evolved independently.
The sequence of the MinD box is highly conserved in all bacterial MinD homologues examined so far. Additionally, MinD boxes were founded in eukaryotic homologues in Guillardia theta, Chlorella vulgaris, Mesostigma viride, Nephroselmis olivacea, Prototheca wickerhamii, Arabidopsis thaliana, and Oryza sativa, suggesting that these residues are very important for their function. In Chlamydia pneumoniae and Chlamydia trachomatis, members of the ParA/MinD superfamily, were described as MinD homologues in the sequence annotation. However, both members seem to belong to the ParA family because of loss of the sequence of the MinD box (Fig. 1B).
Recently, dynamic movement of the Soj protein was observed by using a fusion with green fluorescent protein in the B. subtilis cells (15, 36). In the presence of Spo0J, Soj–green fluorescent protein oscillates from one pole to the other on a time scale of minutes. Previously, more rapid oscillation of MinD was observed in E. coli (37). It is therefore expected that other members of the ParA family will show similar dynamic movement in cells. We are interested in the localization of SopA and Soj in living E. coli cells. The localization of these proteins fused with a fluorescent protein awaits further characterization.
Supplementary Material
Acknowledgments
We thank K. Kobayashi and S. Moriya for plasmid pUC25E, and Y. Akiyama, R. D'Ari, L. Harry, T. Ogura, and anonymous reviewers for critical reading and comments. This work was supported by a grant from the Japan Science and Technology Corporation and by a Grant in Aid for Scientific Research on Priority Areas (C) “Genome Biology” from the Ministry of Education, Science, Sports, and Culture of Japan. Y.Y. was supported by a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hiraga S. Annu Rev Biochem. 1992;61:283–306. doi: 10.1146/annurev.bi.61.070192.001435. [DOI] [PubMed] [Google Scholar]
- 2.Niki H, Hiraga S. Cell. 1997;90:951–957. doi: 10.1016/s0092-8674(00)80359-1. [DOI] [PubMed] [Google Scholar]
- 3.Gordon G S, Sitnikov D, Webb C D, Teleman A, Straight A, Losick R, Murray A W, Wright A. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 4.Niki H, Hiraga S. Mol Microbiol. 1999;34:498–503. doi: 10.1046/j.1365-2958.1999.01611.x. [DOI] [PubMed] [Google Scholar]
- 5.Ludtke D N, Eichorn B G, Austin S J. J Mol Biol. 1989;209:393–406. doi: 10.1016/0022-2836(89)90005-3. [DOI] [PubMed] [Google Scholar]
- 6.Ravin N, Lane D. J Bacteriol. 1999;181:6898–6906. doi: 10.1128/jb.181.22.6898-6906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Youngren B, Radnedge L, Hu P, Garcia E, Austin S. J Bacteriol. 2000;182:3924–3928. doi: 10.1128/jb.182.14.3924-3928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerdes K, Moller-Jensen J, Bugge Jensen R. Mol Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 9.Hayes F. Mol Microbiol. 2000;37:528–541. doi: 10.1046/j.1365-2958.2000.02030.x. [DOI] [PubMed] [Google Scholar]
- 10.Ogasawara N, Yoshikawa H. Mol Microbiol. 1992;6:629–634. doi: 10.1111/j.1365-2958.1992.tb01510.x. [DOI] [PubMed] [Google Scholar]
- 11.Gal-Mor O, Borovok I, Av-Gay Y, Cohen G, Aharonowitz Y. Gene. 1998;217:83–90. doi: 10.1016/s0378-1119(98)00357-6. [DOI] [PubMed] [Google Scholar]
- 12.Mohl D A, Gober J W. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 13.Ireton K, Gunther N W I, Grossman A D. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin D C, Grossman A D. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 15.Marston A L, Errington J. Mol Cell. 1999;4:673–682. doi: 10.1016/s1097-2765(00)80378-0. [DOI] [PubMed] [Google Scholar]
- 16.Lewis P J, Errington J. Mol Microbiol. 1997;25:945–954. doi: 10.1111/j.1365-2958.1997.mmi530.x. [DOI] [PubMed] [Google Scholar]
- 17.Lin D C-H, Levin P A, Grossman A D. Proc Natl Acad Sci USA. 1997;94:4721–4726. doi: 10.1073/pnas.94.9.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teleman A A, Graumann P L, Lin D C H, Grossman A D, Losick R. Curr Biol. 1998;8:1102–1109. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- 19.Webb C D, Teleman A, Gordon S, Straight A, Belmont A, Lin D C-H, Grossman A D, Wright A, Losick R. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 20.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 21.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolsto A B. Mol Microbiol. 1997;24:241–248. doi: 10.1046/j.1365-2958.1997.3501715.x. [DOI] [PubMed] [Google Scholar]
- 23.Kolsto A B. Trends Microbiol. 1999;7:223–226. doi: 10.1016/s0966-842x(99)01519-x. [DOI] [PubMed] [Google Scholar]
- 24.Niki H, Hiraga S. Genes Dev. 1998;12:1036–1045. doi: 10.1101/gad.12.7.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trucksis M, Michalski J, Deng Y K, Kaper J B. Proc Natl Acad Sci USA. 1998;95:14464–14469. doi: 10.1073/pnas.95.24.14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamaichi Y, Iida T, Park K S, Yamamoto K, Honda T. Mol Microbiol. 1999;31:1513–1521. doi: 10.1046/j.1365-2958.1999.01296.x. [DOI] [PubMed] [Google Scholar]
- 27.Lin J, Qi R, Aston C, Jing J, Anantharaman T S, Mishra B, White O, Daly M J, Minton K W, Venter J C, Schwartz D C. Science. 1999;285:1558–1562. doi: 10.1126/science.285.5433.1558. [DOI] [PubMed] [Google Scholar]
- 28.White O, Eisen J A, Heidelberg J F, Hickey E K, Peterson J D, Dodson R J, Haft D H, Gwinn M L, Nelson W C, Richardson D L, et al. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Boer P A, Crossley R E, Hand A R, Rothfield L I. EMBO J. 1991;10:4371–4480. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walker J E, Saraste M, Runswick M J, Gay N J. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe E, Wachi M, Yamasaki M, Nagai K. Mol Gen Genet. 1992;234:346–352. doi: 10.1007/BF00538693. [DOI] [PubMed] [Google Scholar]
- 32.Davis M A, Radnedge L, Martin K A, Hayes F, Youngren B, Austin S J. Mol Microbiol. 1996;21:1029–1036. doi: 10.1046/j.1365-2958.1996.721423.x. [DOI] [PubMed] [Google Scholar]
- 33.Bouet J Y, Funnell B E. EMBO J. 1999;18:1415–1424. doi: 10.1093/emboj/18.5.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muneyuki E, Noji H, Amano T, Masaike T, Yoshida M. Biochem Biophys Acta. 2000;1458:467–481. doi: 10.1016/s0005-2728(00)00095-5. [DOI] [PubMed] [Google Scholar]
- 35.Olsen G J, Woese C R, Overbeek R. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quisel J D, Lin D C, Grossman A D. Mol Cell. 1999;4:665–672. doi: 10.1016/s1097-2765(00)80377-9. [DOI] [PubMed] [Google Scholar]
- 37.Raskin D M, de Boer P A. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.