Abstract
The tomato Leu-rich repeat receptor kinase BRASSINOSTEROID INSENSITIVE1 (BRI1) has been implicated in both peptide (systemin) and steroid (brassinosteroid [BR]) hormone perception. In an attempt to dissect these signaling pathways, we show that transgenic expression of BRI1 can restore the dwarf phenotype of the tomato curl3 (cu3) mutation. Confirmation that BRI1 is involved in BR signaling is highlighted by the lack of BR binding to microsomal fractions made from cu3 mutants and the restoration of BR responsiveness following transformation with BRI1. In addition, wound and systemin responses in the cu3 mutants are functional, as assayed by proteinase inhibitor gene induction and rapid alkalinization of culture medium. However, we observed BRI1-dependent root elongation in response to systemin in Solanum pimpinellifolium. In addition, ethylene perception is required for normal systemin responses in roots. These data taken together suggest that cu3 is not defective in systemin-induced wound signaling and that systemin perception can occur via a non-BRI1 mechanism.
INTRODUCTION
Brassinosteroids (BRs) are plant steroid hormones required for normal growth and development. Mutants in BR biosynthesis or signaling have a characteristic dramatic dwarf phenotype, indicating the importance of these hormones in plant growth and development (for reviews on BRs and BR signaling, see Bishop, 2003; Vert et al., 2005; Szekeres and Bishop, 2006). The major BR receptor in Arabidopsis thaliana is encoded by the BRASSINOSTEROID INSENSITIVE1 (BRI1) gene, which when mutated, results in a dwarf plant, similar in stature to BR biosynthesis mutants (Clouse et al., 1996). BRI1 encodes a receptor Ser/Thr kinase with a predicted extracellular domain of ∼24 Leu-rich repeats (LRRs) that is interrupted by an island domain between repeat 20 and 21 (Li and Chory, 1997; Vert et al., 2005). This is followed by a transmembrane domain and a C-terminal kinase (Li and Chory, 1997; Vert et al., 2005). BRI1 binds BRs via a novel steroid binding domain of ∼100 amino acids that encompasses LRR21 and the island domain (Kinoshita et al., 2005; Vert et al., 2005). BRI1 orthologs are known in several plant species. In tomato (Solanum lycopersicum), the curl3 (cu3) mutant has been characterized as a BR-insensitive dwarf (Koka et al., 2000), and a candidate gene approach showed that this mutant was defective in a homolog of BRI1 (Montoya et al., 2002).
Tomato BRI1 also has been purified as a systemin binding protein (Scheer and Ryan, 2002). Systemin is an 18–amino acid protein that is proteolytically cleaved from a 200–amino acid precursor (Pearce et al., 1991; McGurl et al., 1992). This peptide is able to induce the production of protease inhibitors (PINs), and plants that have reduced systemin levels are more susceptible to insect herbivory (Orozco-Cárdenas et al., 1993). Interestingly, systemin appears to be a Solanaceae-specific protein and has only been identified in certain Solanaceae tribes (e.g., solanum and capsicum) (Constabel et al., 1998). The putative receptor for systemin was isolated biochemically using a radiolabeled photoaffinity systemin that bound to a 160-kD protein named systemin receptor 160 (SR160). Cloning of SR160 revealed that it was a BRI1 homolog (Scheer and Ryan, 2002). To confirm that Sl BRI1 (SR160) was the systemin receptor, cu3 mutants were assayed for their response to systemin. Although cu3 mutants are predicted to have a null allele (nonsense mutation in LRR24; Montoya et al., 2002), they exhibit only a 50% reduction in their response to systemin via measurement of PIN gene induction (Scheer et al., 2003). Tobacco (Nicotiana tabacum), another solanaceous species, lacks a closely related systemin sequence, and transformation of SR160 into tobacco produced the predicted systemin-specific binding and alkalinization response (Scheer et al., 2003). Taken together, these data suggested that Sl BRI1 (SR160) was a dual ligand receptor for systemin and brassinolide (BL).
Here, we provide further proof that Sl BRI1 is a functional BR receptor. In addition, we demonstrate that both cu3 and the BR synthesis mutant extreme dwarf (dx) are capable of producing systemin and wound responses. Intriguingly, we found that Solanum pimpinellifolium roots elongate in response to systemin, in a BRI1-dependent fashion. However, systemin-induced root elongation was not observed in closely related species. In these species, root length was reduced, which in S. lycopersicum is both jasmonate and BR independent but is ethylene dependent. These results indicate that Sl BRI1 is not essential for systemin-induced wound response, although in certain species, it may promote root growth.
RESULTS
Genetic Complementation of the cu3 Phenotype
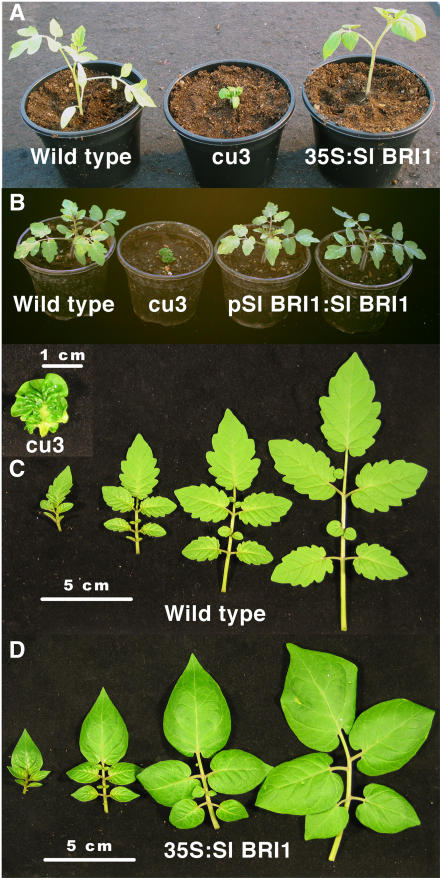
Previous analysis of the cu3 dwarf mutant demonstrated that it is insensitive to BL and that Spi BRI1 (S. pimpinellifolium BRI1) is mutated (Montoya et al., 2002). To confirm that Spi BRI1 is responsible for the cu3 phenotype, complementation was performed by introduction of Sl BRI1 (L. esculentum BRI1) under the control of the cauliflower mosaic virus (CaMV) 35S promoter (35S:Sl BRI1). Introduction of this T-DNA construct into the cu3 background generated two lines that recovered the dwarf phenotype (Figure 1). The 35S:Sl BRI1 plants exhibited additional phenotypes, including delayed germination by 1 to 2 d, compared with the wild type or cu3. Internodal distance and hypocotyl length were greater than wild-type plants, and lateral branching (side shoot length) was also reduced (see Supplemental Table 1 online). Although Arabidopsis plants overexpressing At BRI1 have elongated petioles, this was not observed in 35S:Sl BRI1 plants. The leaves were more ovoid, with reduced serration, than those of the wild type. Leaves also frequently developed necrotic patches, reminiscent of a hypersensitive response. However, no pathogen could be associated with this phenotype, and wild-type plants did not exhibit a similar phenotype, even following inoculation with 35S:Sl BRI1 leaf extracts. Complementation of cu3 with Sl BRI1 under the control of its native promoter restored plants to the wild type and did not exhibit the additional phenotypes observed with 35S:Sl BRI1 (Figure 1).
Figure 1.
Sl BRI1 Complements the the cu3 Phenotype.
cu3 plants were transformed with Sl BRI1 under the control of the CaMV 35S promoter or the Sl BRI1 promoter.
(A) Three-week-old seedlings of the wild type, cu3, and 35S:Sl BRI1 in the cu3 background.
(B) Three-week-old seedlings of the wild type, cu3, and pSl BRI1:Sl BRI1 (native promoter) in the cu3 background.
(C) and (D) Leaves were photographed from 6-week-old plants of cu3 ([C]; inset, wild type) and 35S:Sl BRI1 (D).
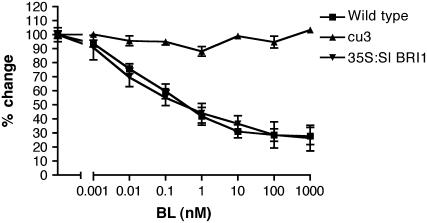
Morphological analyses showed that the introduction of 35S:Sl BRI1 restored the BL responses abolished by the cu3 mutation. To confirm this, seedlings were treated with BL and root length was recorded (Figure 2). As had been previously demonstrated, cu3 roots were insensitive to BL treatment (Koka et al., 2000; Montoya et al., 2002). The introduction of 35S:Sl BRI1 restored a BL-responsive reduction in root elongation, with the sensitivity to BL being similar to wild-type plants.
Figure 2.
Sl BRI1 Restores BL Signaling.
The root length of wild-type, cu3, and 35S:Sl BRI1 seedlings was measured in response to BL at 7 d after germination. Error bars indicate se of the mean (n = 10).
Sl BRI1 Does Not Fully Complement Arabidopsis bri1-5
The results shown here and previously strongly indicated that Sl BRI1 is the functional ortholog of At BRI1. However, when Sl BRI1 was transformed into the At BRI1 mutant bri1-5, it failed to fully complement this mutation when placed under the control of either the Arabidopsis BRI1, S. lycopersicum BRI1, or CaMV 35S promoter (Table 1; see Supplemental Figure 1 online). Partially complementing lines had longer hypocotyls, increased rosette area, and longer inflorescence stems compared with bri1-5, although they were smaller than wild-type plants. To determine if the failure of Sl BRI1 to complement the Arabidopsis mutant was specific to Sl BRI1 or common to other solanaceous species, the tobacco and potato homologs Nb BRI1 (Nicotiana benthamiana BRI1) and St BRI1 (Solanum tuberosum BRI1), respectively, were transformed into bri1-5. Both of these genes failed to fully complement the bri1-5 mutant phenotype, although both morphologically recovered the cu3 phenotype. Interestingly, At BRI1 morphologically restores the cu3 phenotype to almost the wild type, indicating that a more stringent sequence requirement in Arabidopsis is required for genetic complementation.
Table 1.
Complementation of bri1-5 with BRI1
| At BRI1 Promotera | Wild Typeb | Dwarfb |
|---|---|---|
| At BRI1 | 11 | 23 |
| Sl BRI1 | 0 | 118 |
| St BRI1 | 0 | 41 |
| Nb BRI1 | 0 | 37 |
| At LRR-LeKD | 6 | 11 |
| Sl LRR-AtKD | 0 | 17 |
| Sl BRI1 promoter | ||
| At BRI1 | 54 | 72 |
| Sl BRI1 | 0 | 312 |
| 35S promoter | ||
| Sl BRI1 | 0 | 8 |
| Empty vector | 0 | 48 |
BRI1 genes were under the control of Sl BRI1 or At BRI1 promoters.
Number of bri1-5 transformants with either wild-type or dwarf (bri1-5) morphology. Only plants that were fully complemented were scored as wild type.
To further investigate why Sl BRI1 failed to rescue bri1-5, chimeric constructs between At BRI1 and Sl BRI1 were produced. Complementation was achieved when the kinase domain of Sl BRI1 (amino acids 749 to 1207) was fused to the extracellular LRR region of At BRI1 (amino acids 1 to 739). However, the reciprocal construct failed to restore the bri1-5 mutant phenotype (Table 1).
BL Binding Is Not Detectable in cu3 Microsomes
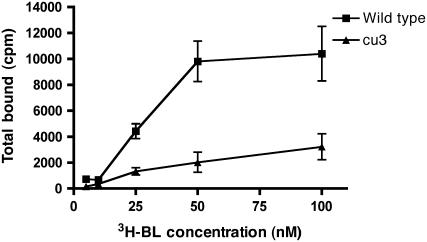
Having established that expression of Sl BRI1 restores stature and BL responsiveness to cu3, we asked whether cu3 membrane fractions contained BL binding activity (Figure 3). In [3H]-BL binding assays using total microsomal membranes, a greatly reduced BL binding activity was observed in cu3 mutants. The dissociation constant (Kd) observed in wild-type microsomal membranes was ∼28 nM, which is similar to values obtained in Arabidopsis (Wang et al., 2001). In agreement with the genetic data, these results demonstrated in vivo that wild-type tomato Sl BRI1 receptor protein is necessary for BL binding.
Figure 3.
BL Binding Is Diminished in cu3 Microsomal Fractions.
Mature tomato wild-type and cu3 leaves of mature plants were used to assess [3H]-BL binding in vivo. Microsomal extracts from fresh tissues were incubated in 2, 10, 25, 50, 75, or 100 nM [3H]-BL. Specific [3H]-BL binding is reduced in the cu3 microsomal extracts compared with the wild type. Data represent the average measurements of three independent experiments. Error bars indicate se of the mean.
Systemin-Induced PINII Expression Does Not Require BRI1 or BR
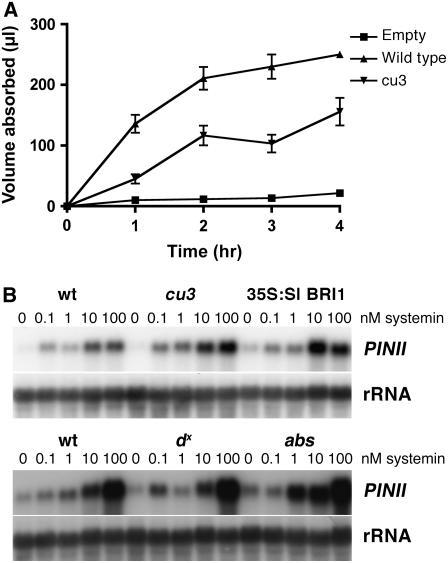
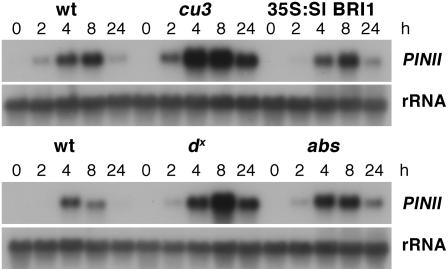
PIN gene induction and rapid alkalization of cell suspension culture medium are key assays in determining the bioactivity of systemin (Meindl et al., 1998). Scheer et al. (2003) showed that cu3 had a reduced ability to induce the accumulation of PIN protein following treatment with either methyl jasmonate or systemin. Systemin treatments of 35S:Sl BRI1, cu3, and wild-type seedlings were performed to confirm the BRI1 dependence of systemin-induced PINII expression. During these studies, we noted that the rate of systemin solution uptake in cu3 was significantly lower than the wild type (Figure 4A). This was presumably due to a lower transpiration rate in cu3 mutants because of the reduced leaf area. Systemin treatments were performed so that all seedlings were treated with the same amount of systemin, and no difference in the level of PINII transcripts or the sensitivity to systemin was observed between the wild type and mutants (Figure 4B). To confirm this result, systemin treatments were performed with another tomato BRI1 mutant, abs, and systemin-induced PINII transcript accumulation was also found to be similar to wild-type plants (Figure 4B). To test whether BRs are required for PINII transcript accumulation, the BR synthesis mutant dx, which lacks bioactive BRs in vegetative tissue (Bishop et al., 1999), was examined for its response to systemin. The dx mutants had wild-type levels of PINII expression, indicating that bioactive BRs are not required for systemin-mediated PIN gene induction.
Figure 4.
BR Mutants Can Induce PINII in Response to Systemin.
(A) Reduced transpiration rate in cu3. Seedlings were cut through the stem and placed in 250 μL of 10 nM systemin and allowed to take up the solution. Each hour seedlings were removed and the volume of solution recorded. Six seedlings were used for each time point. Evaporation of solution was also recorded (empty). Error bars indicate se of the mean.
(B) RNA gel blot analysis of PINII expression in response to systemin applied at 0, 0.1, 1, 10, and 100 nM. Leaf samples were taken 8 h after systemin treatment. Membranes were probed with PINII (top) and reprobed with an 18S rRNA probe (bottom) as a loading control. The top PINII blot was exposed for 4 h and the bottom blot for 16 h. S. pimpinellifolium (wt, top panel), cu3, 35S:Sl BRI1, S. lycopersicum (wt, bottom panel), dx, and abs.
cu3 Is Not Compromised in Wound-Induced PINII Expression
Systemin induction is associated with mechanical wounding, and it may be expected that mutants in the systemin receptor would be defective in their wound response. cu3 mutants were mechanically wounded, and this treatment induced systemic PINII expression (Figure 5). In addition, systemic accumulation of PINII transcripts was found in the Sl BRI1 mutant abs and in the BR synthesis mutant dx. PINII transcripts were detectable between 2 and 4 h after wounding, and no significant difference was observed in the timing of the induction or the duration of response in any of the mutants. The level of PINII accumulation in all mutants was greater than the wild type, which may be a consequence of leaf morphology rather than alteration in wound responsiveness.
Figure 5.
Wound-Induced PINII Expression Is Not Compromised in BR Mutants.
Wounding was performed on S. pimpinellifolium, cu3, 35S:Sl BRI1, S. lycopersicum, dx, and abs plants (see Methods). RNA was extracted from systemic unwounded leaves at 0, 2, 4, 8, and 24 h after wounding. Membranes were probed with PINII (top) and reprobed with an 18S rRNA probe (bottom) as a loading control.
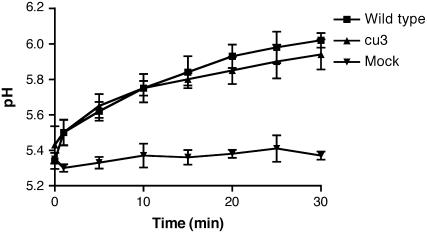
Medium Alkalinization in Response to Systemin Is Not Altered in cu3
The alkalinization of cell suspension medium has been used to show the activity of systemin (Meindl et al., 1998). This response is not affected by BL, although overexpression of tomato BRI1 in tobacco suspension enables systemin-responsive alkalinization (Scheer et al., 2003). Suspension cultures of the wild type and cu3 were generated, and the pH of the medium was recorded after the addition of systemin (Figure 6). Both mutant and wild-type cultures responded at similar times and with similar amplitudes (P > 0.1).
Figure 6.
Medium Alkalinization Induced by Systemin.
Suspension cells of the wild type and cu3 were treated with 100 pM systemin and the pH of the suspension recorded. Mock treatment of the wild type had water added instead of systemin. Data represent the average measurements of three samples from one representative experiment. Error bars indicate se of the mean.
Tomato Roots Respond to Systemin
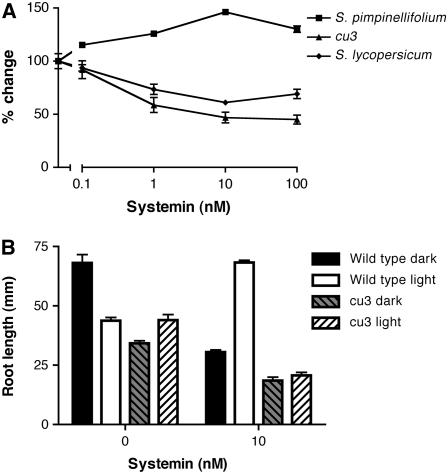
To discern the possible involvement of systemin in the BR response, we used root elongation to monitor the bioactivity of various systemin and hormone treatments. Root elongation was used because it is known to be a sensitive method to measure BR bioactivity in tomato (Roddick, 1994) and because cu3 mutants are known to be defective in the inhibition of root elongation in response to BRs (Koka et al., 2000; Montoya et al., 2002). Wild-type and cu3 seedlings were grown on medium containing systemin, and root length was recorded (Figure 7). When wild-type S. pimpinellifolium seedlings were grown on systemin, roots were longer than those of untreated seedlings (P < 0.001 for all systemin concentrations). In addition, lateral branching and root hair growth were inhibited. Interestingly, cu3 seedlings exhibited a dose-dependent inhibition of root elongation (P < 0.001 for systemin concentrations >0.1 nM). Root hair growth in these mutants was not suppressed by the addition of systemin. In addition, an anthocyanin-like coloration was observed in cu3 roots at systemin concentrations >10 nM. The involvement of BRI1 in this response was confirmed by systemin treatment of cu3 plants transformed with Sl BRI1 (Figure 8). These plants showed a restoration of wild-type systemin response. As a control for these experiments, the biologically inactive Ala-17 systemin was used and did not exhibit any significant bioactivity in roots (P > 0.05; see Supplemental Figure 2 online).
Figure 7.
Root Elongation in Response to Systemin.
(A) Seedlings were grown on medium containing 0, 0.1, 1, 10, or 100 nM systemin for 7 d (S. pimpinellifolium and cu3) or 5 d (S. lycopersicum cv Moneymaker). Root length was then measured. Data are expressed as percentage change of untreated seedlings.
(B) S. pimpinellifolium and cu3 seedlings were light or dark grown on medium with or without 10 nM systemin for 7 d. Error bars indicate se of the mean (n = 10).
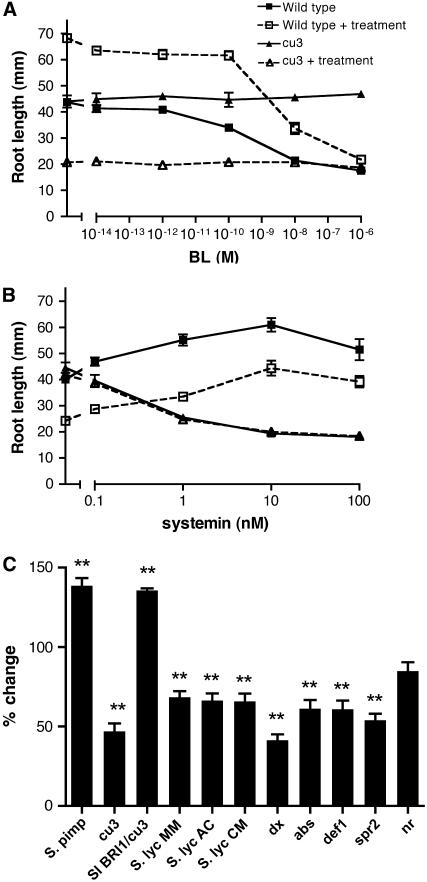
Figure 8.
Root Elongation in Response to Systemin and BL.
Seedlings were grown with or without treatment and root length was recorded after 7 d.
(A) Seedlings grown with varying concentrations of BL with or without 10 nM systemin treatment.
(B) Seedlings were grown on medium containing different levels of systemin with or without 1 nM BL.
(C) Response of mutant and Sl BRI1 transformed plants to systemin. Seedlings were grown on medium with or without the addition of 10 nM systemin and root length recorded. Plants analyzed are S. pimpinellifolium, cu3, pSl BRI1:Sl BRI1 transformed into cu3, S. lycopersicum cv Moneymaker (MM), S. lycopersicum cv Ailsa Craig (AC), S. lycopersicum cv Castlemart (CM), dx, abs, def1, spr2, and Nr. Asterisks indicate P < 0.001. Error bars indicate se from the mean (n = 10).
The inhibition of root growth in cu3 indicates that BRI1 is required for a wild-type response to systemin in this assay. To further investigate the role of BR in this systemin response, treatments with systemin in the presence of BL were performed (Figure 8). Not surprisingly, the response to systemin in the BR-insensitive mutant cu3 was not altered by the addition of BL. In wild-type seedlings, systemin partially suppressed the BL-induced reduction in root length. This response was dose dependent and required >1 nM systemin to elongate roots in the presence of 1 nM BL. This indicated that BRI1 plays a role in root elongation and that its function impacts both the systemin and BL signaling pathways.
The interaction between systemin and BL responses was examined further by investigating the effect of BL on the root growth of seedlings expressing reduced levels of prosystemin. Roots of antisense prosystemin plants responded in a similar way as the wild type (see Supplemental Figure 3 online). This was not unexpected since systemin is not expressed in roots (McGurl et al., 1992; N. Holton, unpublished data).
The antisense line used was in an S. lycopersicum cultivar, and this species exhibited reduced root length in response to systemin (P < 0.001 for systemin concentrations >0.1 nM) (Figure 7). This result was opposite to that observed in S. pimpinellifolium roots. However, similar to S. pimpinellifolium roots, those of S. lycopersicum had decreased root hair growth and lateral branching in the presence of systemin.
Since BR mutants exhibit defective etiolation, we analyzed the root response to systemin in both light- and dark-grown seedlings. When systemin treatments of S. pimpinellifolium and cu3 were performed on etiolated seedlings, root length was decreased in the presence of systemin (Figure 7B). We also noticed that the root length of untreated etiolated S. pimpinellifolium seedlings was significantly longer than light-grown seedlings; however, this was not the case for cu3 (P < 0.001 and P > 0.1, respectively). These data indicated that the response to systemin was different between S. lycopersicum and S. pimpinellifolium species (i.e., S. lycopersicum seedlings lacked root elongation). However, when grown in the dark, S. pimpinellifolium seedlings responded in a similar manner to light-grown S. lycopersicum seedlings, in that both sets of seedlings have reduced root length in the presence of systemin.
Further investigation of the role of BRI1 in the response to systemin was performed using the weak BRI1 mutant abs (S. lycopersicum). In this mutant, systemin induced a response similar to wild-type roots. In addition, the BL synthesis mutant dx produced a response equivalent to the wild type. These data suggested that the systemin-induced root inhibition of S. lycopersicum seedlings is not influenced by BR signaling, but it is in S. pimpinellifolium seedlings.
Overexpression of prosystemin results in jasmonate-dependent increased hypocotyl elongation (Howe and Ryan, 1999). To determine if the systemin-induced changes in root length are also jasmonate dependent, the responses of jasmonate synthesis mutants def1 and spr2 (Li et al., 2003) (S. lycopersicum mutants) were investigated. Both mutants had a wild-type reduction in root length when grown on systemin (Figure 8), which indicated that the reduced root growth was independent of jasmonate signaling. Ethylene is also known to be involved in systemin responses. Root lengths in the ethylene-insensitive mutant Never ripe (Nr) were not significantly different in response to systemin (P > 0.05). This indicates that in S. lycopersicum, the reduction in root length is dependent on ethylene signaling.
DISCUSSION
The role of BRI1 in BR perception in Arabidopsis is well established. As such, the purification of tomato BRI1 by binding to systemin has been an enigma (Yin et al., 2002; Bishop, 2003; Szekeres, 2003; Boller, 2005). It was therefore important to show that Sl BRI1 was a functional BR receptor using both genetic and biochemical approaches. Genetic restoration of the cu3 mutant phenotype using the CaMV 35S promoter to express the tomato BRI1 generated nondwarf plants that had elongated stems, indicating a role for tomato BRI1 in growth. The rescued lines also exhibited less serration of the leaf margins, indicating a role for BRs in leaf margin development. This is not surprising since Arabidopsis mutants in BR biosynthesis and signaling have been identified in screens for leaf phenotypes (Kim et al., 1998; Pérez-Pérez et al., 2002). Transformation experiments using the endogenous Sl BRI1 promoter to express the Sl BRI1 gene resulted in plants that have phenotypes very similar to those of the wild type, providing further proof that the cu3 mutant is defective in the LRR receptor kinase Sl BRI1.
Sl BRI1, Nb BRI1, and St BRI1 are able to phenotypically rescue the cu3 phenotype to the wild type. However, these genes fail to fully complement the Arabidopsis BRI1 mutant bri1-5 when expressed under the control of the Arabidopsis BRI1 promoter. When the extracellular domain of the Arabidopsis gene is fused to the tomato kinase domain, complementation is achieved; this suggests that the extracellular LRR region from Arabidopsis is required for full function in Arabidopsis.
The extracellular region of BRI1 is poorly conserved between species when compared with the kinase domain. However, several amino acids are conserved in this region in all known BRI1 homologs. Of particular interest is the NGSM motif located in the region that has recently been shown to bind BRs (Kinoshita et al., 2005; see Supplemental Figure 4 online). This motif is found in all known BR binding proteins, BRI1 and related BRL1 and BRL3 (Caño-Delgado et al., 2004), but not in the closely related At BRL2 and Os BRL2. At BRL2 and Os BRL2 do not bind BR or restore BR responsiveness (Nakamura et al., 2006). This motif is found in Sl BRI1; therefore, it is not surprising that the cu3 mutants defective in Sl BRI1 had reduced BL binding.
To study the putative interaction of BR and systemin signaling, we sought confirmation that cu3 mutants were defective in systemin signaling. In previous studies, the cu3 mutant was shown to have only ∼50% of wild-type PIN gene induction (Scheer et al., 2003), even though this is a null mutant. To help clarify, we assayed PINII mRNA accumulation using a range of systemin concentrations and altering the length of time for systemin uptake in the mutant lines to allow equal systemin loading. Following such systemin treatments, bri1 mutants showed PINII induction equivalent to the wild type, indicating that the previously reported reduced level of PIN accumulation may have been the consequence of the absolute amount of systemin that the mutant was exposed to. It could be argued that due to the longer exposure of the mutants to systemin that the relative concentration in the cu3 mutants may be higher than that in wild-type plants. Without measuring systemin levels in the plants this will be difficult to resolve. However, this does not detract from the fact that the tomato bri1 mutants are able to respond to systemin and that this suggests the presence of an additional mechanism for systemin perception.
These results are consistent with the observation that dx, cu3, and abs mutants exhibit a wound response. This response is greater than in wild-type plants, and this is most likely the consequence of altered plant morphology. However, the possibility exists that BRI1 and BRs are involved in the regulation of wound-induced PINII induction. Taken together, these data suggested that a functional BRI1 receptor is not essential for the systemin-induced wound response. Furthermore, systemin treatments of suspension cultures from cu3 mutants and the wild type confirmed that medium alkalinization in response to systemin was normal. Thus, Sl BRI1 is not essential for the systemin-induced alkalinization response. However, Scheer et al. (2003) demonstrated that the introduction of Spv BRI1 into tobacco suspension cultures resulted in alkalinization in response to systemin. This response does not occur in untransformed cultures and therefore indicates that in tobacco, Spv BRI1 facilitates alkalinization in response to systemin. In S. pimpinellifolium, however, an additional systemin receptor is present that enables the alkalinization response in cu3 mutants.
BRs cause cell elongation, and although the overexpression of prosystemin results in plants with jasmonate-dependent elongated hypocotyls (Howe and Ryan, 1999), further clarification of systemin-induced elongation was sought. Systemin was found to increase S. pimpinellifolium root length, but in cu3 mutants, it inhibited root elongation, indicating the presence of both BRI1-dependent and -independent systemin signaling. In addition, treatment of wild-type roots with both systemin and BL suggests some antagonism.
When etiolated S. pimpinellifolium or light-grown S. lycopersicum roots were investigated, root length was reduced in response to systemin, suggesting that both environmental and species-specific factors influence the way in which roots respond to systemin. The reduction in root length observed in S. lycopersicum was not altered in jasmonate synthesis mutants, unlike the response observed in hypocotyls of plants overexpressing prosystemin (Howe and Ryan, 1999). However, Nr root length was not significantly altered by the addition of systemin, suggesting that ethylene perception is required for the reduction in root length observed. Taken together, these results imply that an interaction between ethylene signaling and signaling via BRI1 plays a key role in the systemin-induced root growth observed. Environmental (e.g., light) and species-specific factors influence the interaction between the two pathways determining whether there is inhibition or stimulation of root elongation.
The interaction between ethylene and BR signaling is not well understood; however, an interaction has been observed. BRs can stimulate ethylene biosynthesis, and ethylene induces expression of BR synthesis genes (Schlagnhaufer and Arteca, 1985; De Grauwe et al., 2005). BR synthesis mutants fail to develop the characteristic apical hook in response to the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (De Grauwe et al., 2005); however, BRI1 and BR synthesis mutants are not altered in ethylene sensitivity (Clouse et al., 1996; Mussig et al., 2003). Given the complexity of BR/ethylene interactions, further investigation of the influence of systemin in these pathways is required.
Systemin is a member of the growing number of peptides that have been shown to have biological activity in plants that include tobacco systemins, CLAVATA3, S-locus protein 11, phytosulfokine, and RALFs (Matsubayashi and Sakagami, 2006). One newly identified Arabidopsis peptide, At Pep1 (Huffaker et al., 2006), has been shown to elicit plant defense responses. Interestingly, At PEP1 binds an LRR kinase, PEPR1. While systemin does not compete with binding of At PEP1 to PEPR1 (Yamaguchi et al., 2006), the fact that both systemin and At PEP1 bind to an LRR Se/Thr kinase suggests that their mode of action is similar. These findings indicate that further investigation into systemin perception is required to clarify whether conserved mechanisms of peptide hormone signaling exist in plants and may also define systemin's role in plant development.
METHODS
T-DNA Construction and Generation of Complemented Lines
Sl BRI1 was PCR amplified from Solanum lycopersicum cv Moneymaker using primers 5′-CATCAAGAGCTCAAGCTATAGATTCAAG-3′ to introduce an SstI site in the 5′ untranslated leader of Sl BRI1 and 5′-TGGATGGGAACTAGTGGTACATAC-3′ that introduced an SpeI site in the 3′ untranslated tail. The PCR product was cloned and sequenced. The resulting plasmid was digested with SstI and SpeI, and Sl BRI1 was cloned into T-DNA vector GB1421 (Bishop et al., 1999) cut with SstI and XbaI to generate 35S:Sl BRI1.
To produce BRI1 constructs under the control of the S. lycopersicum promoter, a 2-kb promoter region was amplified from the tomato BAC LeHBa28J23 (Clemson University Genomics Institute) using the primers 5′-GAATTCTAGAGAGGGAAACATCGT-3′ and 5′-GGTACCGAAACTTTATAGCTTAAATGGTG-3′. The promoter region was then cloned into the EcoRI/KpnI sites of pCA2302 (pCAMBIA2300 containing the EcoRI/BstUI green fluorescent protein fragment from pCAMBIA1302 cloned into the EcoRI/PmeI sites). A 1.8-kb Arabidopsis thaliana promoter was amplified using PCR primers (5′-CAATTGGAGCGCGTGTAGACCACG-3′ and 5′-GGTACCTTGTGAGAGAGAAAAGTGTG-3′) and cloned as an MfeI/KpnI fragment into EcoRI/KpnI of pCA2302. BRI1 gene regions were then cloned into either vector as KpnI/SpeI fragments following amplification with the following PCR primers: Sl BRI1, 5′-GGTACCTTTGAAGATGAAAGC-3′ and 5′-ACTAGTACCTCCAAGGTGTTTGCTCAG-3′; At BRI1, 5′-GGTACCTTGAGAAATGAAGAC-3′ and 5′-ACTAGTTCCACCTAATTTTCCTTCAGG-3′. The Sl BRI1/At BRI1 fusion constructs were produced by amplifying both the LRR and kinase regions and introducing an internal Kpn2I restriction site. This was performed using terminal primers described above and the following internal primers: Sl BRI1, 5′-TCATTCCGGAGAGATTATTGTTTGACAGGTCAATC-3′ and 5′-AATCTCTCCGGAATGATTCCTGAATCTGCACC-3′; At BRI1, 5′-TTGTCCGGACCGATTCCTGAGATGGGTC-3′ and 5′-TCGGTCCGGACAAATTATTATTCG-3′. These fragments were then sequentially ligated into the binary vectors containing either promoter. The Nicotiana benthamiana and Solanum tuberosum cv Maris Peer BRI1 genes were amplified by PCR (5′-TCGGTACCTTTGAAGATGAAACCTCACAAGAG-3′ and 5′-TCACTAGTACCTCCTAGGTGTTTGCTCAGCTCATTA-3′; 5′-GGTACCTTTGAAGATGAAAGC-3′ and 5′-ACTAGTACCTCCAAGGTGTTTGCTTAGCTCATTGCC-3′, respectively) and cloned as KpnI/SpeI fragments under the control of the Arabidopsis promoter. All constructs were sequenced to ensure no mutations were introduced during PCR.
All constructs were transformed into cu3 as described previously (Bishop et al., 1999; Montoya et al., 2005). Constructs were transformed into Arabidopsis bri1-5 by floral dipping as described by Clough and Bent (1998).
3H-BL Binding to Microsomal Fractions
The microsomal fractions for 3H-BL experiments were extracted from fresh leaves of wild-type and cu3 mature plants as described by Kinoshita et al. (2005). Protein concentration was determined (Protein Assay; Bio-Rad) and samples resuspended to 1 mg/mL in MES binding buffer (10 mM Mes-KOH, pH 5.7, 5 mM MgCl2, 0.1 mM CaCl2, and 0.25 M mannitol). Each BL binding assay contained 200 μL membrane suspensions in 2, 10, 25, 50, 75, or 100 nM [3H]-BL, without or with 100-fold excess unlabeled BL or indicated amount of unlabeled steroids, 1 mg mL−1 BSA, and BL binding buffer in 1000 μL total volume. The binding reactions were incubated for 30 min at 25°C. The bound and free [3H]-BL were separated by filtering the mixture through a glass-fiber filter (Whatman; GF/F) and washing three times with 50 mL ice-cold BL binding buffer. Bound BL was quantified by scintillation counting as described by Wang et al. (2001).
Systemin Application and Wounding
Systemin treatments were performed by cutting the stem of 3- to 4-week-old greenhouse-grown seedlings with a single-edge razor blade and then placing them in a systemin solution (peptides synthesized by Afiniti Research Products) or water for 1 h for the wild type (Solanum pimpinellifolium or S. lycopersicum), abs, and dx or 3 h for cu3. After systemin treatment, seedlings were transferred to water and samples taken 8 h later. At least six seedlings were sampled for each treatment, and only the two uppermost leaves were sampled. Treatments were performed in continuous light.
Mechanical wounding was performed on 4- to 5-week-old seedlings by crushing terminal leaflets with forceps three times. Samples were taken from the leaf immediately above that which had been wounded. At least six seedlings were used for each sample. RNA was extracted from systemin- and wound-treated samples, and RNA gel blotting was performed using either PINII or tomato 18S rRNA derived probes. Wounding and systemin treatments were performed at least three times with similar results.
Alkalinization Experiments
To analyze the alkalinization response of suspension cultures, S. pimpinellifolium (wild type) and cu3 suspension cultures were produced from leaf-derived callus. Callus was maintained on Murashige and Skoog medium (Duchefa) containing 3% (w/v) sucrose, 3 mg/L 1-naphthalene acetic acid, and 1.5 mg/L 6-benzylaminopurine. Callus was transferred to liquid culture, in the same medium used for callus maintenance, and subcultured every 7 d. Experiments were performed using 5 mL of culture 5 to 7 d after subculture. The pH of the culture medium was recorded prior to and following the addition of systemin or water, in a volume of <10 μL. Experiments were repeated a total of three times. Two independent culture lines of the wild type and cu3 were used.
Root Elongation Assay
Seeds were surface sterilized with 10% commercial bleach, rinsed in water, and plated on one half-strength Murashige and Skoog medium containing 1% (w/v) sucrose. At radical emergence, seeds were transferred to 120-mm square Petri dishes containing the same medium as above with the addition of systemin or hormones. The plates were then placed vertically and seedlings grown for 5 d for S. lycopersicum lines or 7 d for S. pimpinellifolium lines. Seedlings were grown in a controlled environment at 24°C with a 16-h daylength. Root length was then recorded for at least 10 seedlings per treatment. Experiments were repeated at least twice, with similar results.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EF471736 to EF471738.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotypes of bri1-5 Transgenic Plants.
Supplemental Figure 2. Ala-17–Substituted Systemin Is Biologically Inactive.
Supplemental Figure 3. BL Response of Antisense Prosystemin Roots.
Supplemental Figure 4. Sequence Alignment of the BL Binding Region of BRI1 and Related Proteins.
Supplemental Table 1. Characterization of cu3 Transformants.
Supplementary Material
Acknowledgments
We thank the glasshouse staff at both the Institute of Biological Sciences, Aberystwyth, and the Division of Biology, Wye, for plant care. This work has been generously supported by the Biotechnology and Biosciences Research Council (Grant P19961 to G.J.B.), by the Human Frontier Research Program (G.J.B. and J.C.), and by the USDA (J.C.). A.C.-D. was a long-term fellow of the Human Frontier Research Program. We also thank C. Ryan and G. Howe for seeds of the tomato antisense systemin line and jasmonate mutants, respectively, and J. Giovannoni for the tomato BAC clone.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Gerard J. Bishop (g.bishop@imperial.ac.uk).
Online version contains Web-only data.
References
- Bishop, G.J. (2003). Brassinosteroid mutants of crops. J. Plant Growth Regul. 22 325–335. [DOI] [PubMed] [Google Scholar]
- Bishop, G.J., Nomura, T., Yokota, T., Harrison, K., Noguchi, T., Fujioka, S., Takatsuto, S., Jones, J.D.G., and Kamiya, Y. (1999). The tomato DWARF enzyme catalyses C-6 oxidation in brassinosteroid biosynthesis. Proc. Natl. Acad. Sci. USA 96 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, T. (2005). Peptide signalling in plant development and self/non-self perception. Curr. Opin. Cell Biol. 17 116–122. [DOI] [PubMed] [Google Scholar]
- Caño-Delgado, A., Yin, Y.H., Yu, C., Vafeados, D., Mora-Garcia, S., Cheng, J.C., Nam, K.H., Li, J.M., and Chory, J. (2004). BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131 5341–5351. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Clouse, S.D., Langford, M., and McMorris, T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constabel, C.P., Yip, L., and Ryan, C.A. (1998). Prosystemin from potato, black nightshade, and bell pepper: Primary structure and biological activity of predicted systemin polypeptides. Plant Mol. Biol. 36 55–62. [DOI] [PubMed] [Google Scholar]
- De Grauwe, L., Vandenbussche, F., Tietz, O., Palme, K., and Van Der Straeten, D. (2005). Auxin, ethylene and brassinosteroids: Tripartite control of growth in the Arabidopsis hypocotyl. Plant Cell Physiol. 46 827–836. [DOI] [PubMed] [Google Scholar]
- Howe, G.A., and Ryan, C.A. (1999). Suppressors of systemin signaling identify genes in the tomato wound response pathway. Genetics 153 1411–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker, A., Pearce, G., and Ryan, C.A. (2006). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 103 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, G.-T., Tsukaya, H., and Uchimiya, H. (1998). The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 12 2381–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, T., Caño-Delgado, A.I., Seto, H., Hiranuma, S., Fujioka, S., Yoshida, S., and Chory, J. (2005). Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433 167–171. [DOI] [PubMed] [Google Scholar]
- Koka, C.V., Cerny, R.E., Gardner, R.G., Noguchi, T., Fujioka, S., Takatsuto, S., Yoshida, S., and Clouse, S.D. (2000). A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol. 122 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., Liu, G., Xu, C., Lee, G.I., Bauer, P., Ling, H.Q., Ganal, M.W., and Howe, G.A. (2003). The tomato suppressor of prosystemin-mediated responses2 gene encodes a fatty acid desaturase required for the biosynthesis of jasmonic acid and the production of a systemic wound signal for defense gene expression. Plant Cell 15 1646–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.M., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90 929–938. [DOI] [PubMed] [Google Scholar]
- Matsubayashi, Y., and Sakagami, Y. (2006). Peptide hormones in plants. Annu. Rev. Plant Biol. 57 649–674. [DOI] [PubMed] [Google Scholar]
- McGurl, B., Pearce, G., Orozco-Cárdenas, M., and Ryan, C.A. (1992). Structure, expression, and antisense inhibition of the systemin precursor gene. Science 255 1570–1573. [DOI] [PubMed] [Google Scholar]
- Meindl, T., Boller, T., and Felix, G. (1998). The plant wound hormone systemin binds with the N-terminal part to its receptor but needs the C-terminal part to activate it. Plant Cell 10 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya, T., Nomura, T., Farrar, K., Kaneta, T., Yokota, T., and Bishop, G.J. (2002). Cloning the tomato curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/SR160 in plant steroid hormone and peptide hormone signaling. Plant Cell 14 3163–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya, T., Nomura, T., Yokota, T., Farrar, K., Harrison, K., Jones, J.G.D., Kaneta, T., Kamiya, Y., Szekeres, M., and Bishop, G.J. (2005). Patterns of Dwarf expression and brassinosteroid accumulation in tomato reveal the importance of brassinosteroid synthesis during fruit development. Plant J. 42 262–269. [DOI] [PubMed] [Google Scholar]
- Mussig, C., Shin, G.H., and Altmann, T. (2003). Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 133 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, A., et al. (2006). The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol. 140 580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Cárdenas, M., Mcgurl, B., and Ryan, C.A. (1993). Expression of an antisense prosystemin gene in tomato plants reduces resistance toward Manduca sexta larvae. Proc. Natl. Acad. Sci. USA 90 8273–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce, G., Strydom, D., Johnson, S., and Ryan, C.A. (1991). A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253 895–898. [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez, J.M., Ponce, M.R., and Micol, J.L. (2002). The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev. Biol. 242 161–173. [DOI] [PubMed] [Google Scholar]
- Roddick, J.G. (1994). Comparative root-growth inhibitory activity of 4 brassinosteroids. Phytochemistry 37 1277–1281. [Google Scholar]
- Scheer, J.M., Pearce, G., and Ryan, C.A. (2003). Generation of systemin signaling in tobacco by transformation with the tomato systemin receptor kinase gene. Proc. Natl. Acad. Sci. USA 100 10114–10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer, J.M., and Ryan, C.A. (2002). The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc. Natl. Acad. Sci. USA 99 9585–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlagnhaufer, C.D., and Arteca, R.N. (1985). Brassinosteroid-induced epinasty in tomato plants. Plant Physiol. 78 300–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres, M. (2003). Brassinosteroid and systemin: Two hormones perceived by the same receptor. Trends Plant Sci. 8 102–104. [DOI] [PubMed] [Google Scholar]
- Szekeres, M., and Bishop, G.J. (2006). Integration of brassinosteroid biosynthesis and signaling. Ann. Plant Rev. 24 67–92. [Google Scholar]
- Vert, G., Nemhauser, J.L., Geldner, N., Hong, F.X., and Chory, J. (2005). Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21 177–201. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., Seto, H., Fujioka, S., Yoshida, S., and Chory, J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410 380–383. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, Y., Pearce, G., and Ryan, C.A. (2006). The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl. Acad. Sci. USA 103 10104–10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y.H., Wu, D.Y., and Chory, J. (2002). Plant receptor kinases: Systemin receptor identified. Proc. Natl. Acad. Sci. USA 99 9090–9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.