Abstract
Organisms rely on a variety of regulatory architectures to control gene transcription. Whereas the functional characteristics of particular architectures are well understood, the properties of newly discovered regulatory designs cannot be easily predicted. One emerging design depends on small proteins that connect two-component regulatory systems, which constitute the dominant form of bacterial signal transduction. These connectors enable one system to respond to the signal perceived by a different system. To understand the functional properties of such connector-mediated architectures, we investigated the pathway controlled by the PhoP-dependent connector protein PmrD of Salmonella enterica and contrasted it to the circuit in which genes are regulated directly by the transcription factor PhoP. The PmrD-mediated pathway displayed both signal amplification and persistence of expression when compared with the direct pathway. Mathematical modeling of the two pathways allowed us to identify critical factors responsible for signal amplification.
Keywords: mathematical modeling, Salmonella, signal transduction, transcriptional regulatory circuit, two-component system
The levels at which a gene is transcribed depend not only on the promoter sequences that are recognized by RNA polymerase and/or DNA-binding activators and repressors, but also on the regulatory architecture that determines the amount and activity of these proteins. Despite recent progress in the description and characterization of network motifs (1), there is still limited knowledge about the quantitative behavior of different regulatory designs even for well studied microorganisms. Here, we explore a regulatory design that relies on a class of bacterial proteins, termed connectors, that have the unique property of connecting two-component systems (2–5), which constitute the most prevalent form of bacterial signal transduction (6, 7).
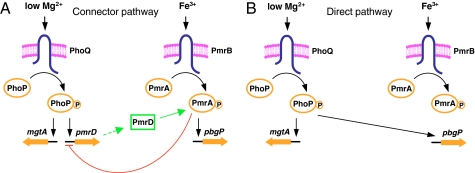
The best-characterized connector protein, designated PmrD, links the signal activating the PhoP/PhoQ two-component system with expression of the genes that are directly controlled by the PmrA/PmrB two-component system in the bacterium Salmonella enterica serovar Typhimurium (8) (Fig. 1A). The PmrD protein accomplishes this task by binding to the phosphorylated form of the DNA-binding protein PmrA, protecting it from dephosphorylation by the sensor protein PmrB (2). This activity enables binding of the PmrA protein to its regulated promoters and transcription of PmrA-regulated genes. The PmrD protein is produced when the sensor protein PhoQ responds to low extracytoplasmic Mg2+ by promoting phosphorylation of the DNA-binding protein PhoP. The phosphorylated PhoP protein then binds to a large number of target promoters, including that of the pmrD gene, and activates or represses gene transcription (9, 10). Three of the PmrA-activated loci mediate resistance to the antibiotic polymyxin B. Therefore, the PmrD-mediated connection enables S. enterica to be resistant to polymyxin B not only in response to the Fe3+ signal that is sensed by the PmrB protein (11) but also in low-Mg2+ environments, which activate the PhoP/PhoQ system (12).
Fig. 1.
Schematics for the circuit designs. (A) Model illustrating the connector-mediated regulatory design present in wild-type S. enterica in which low Mg2+ detected by the PhoQ protein activates the PhoP protein, which promotes expression of PmrD, a small posttranslational activator that binds to the phosphorylated PmrA protein (green line), resulting in transcription of the pbgP gene. The phosphorylated PmrA protein represses transcription of the pmrD gene (red line) (33). Fe3+ activates the PmrA/PmrB system directly. The PhoP protein can also activate transcription of genes, such as mgtA, directly. (B) Model illustrating a direct regulatory design where the PhoP/PhoQ system promotes transcription of the pbgP gene directly in response to low Mg2+.
In this work, we explore the quantitative differences that exist between the direct transcriptional activation carried out by the PhoP protein and the connector-mediated regulatory design that depends not only on PhoP but also on the PmrD and PmrA proteins. Using a combination of molecular genetics and modeling approaches, we are able to establish that the connector-mediated pathway promotes higher output levels than the direct pathway, and that the expression of target genes remains active for longer periods of time after an organism stops experiencing inducing conditions. A comparison with other network architectures suggests that these properties can be expected of multistage regulatory circuits.
Results
PmrA-Activated Genes Are Induced to Higher Levels than Those Regulated Directly by the PhoP Protein.
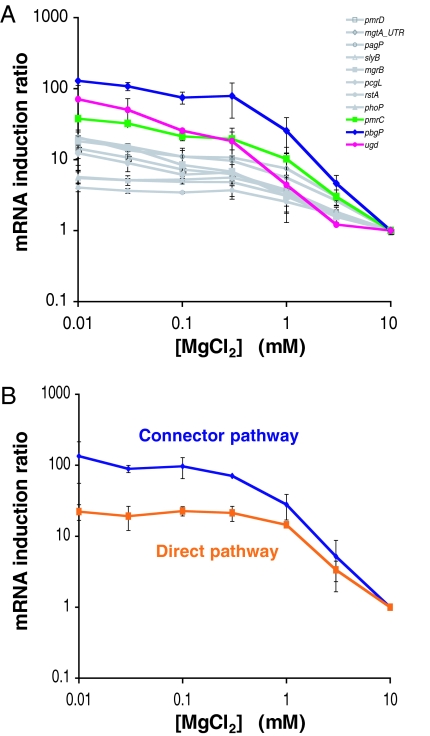
We examined the expression levels of PhoP-activated genes in bacteria that were grown for 4 h in 10 mM Mg2+, which are repressing conditions for the PhoP/PhoQ system, and then shifted to media with different Mg2+ concentrations and incubated for 2 h. We determined the induction ratio (i.e., the mRNA levels for organisms grown in an inducing Mg2+ concentration divided by the mRNA levels corresponding to organisms grown in 10 mM Mg2+) for three genes that are regulated via the PhoP–PmrD–PmrA pathway and for eight genes that are directly controlled by the PhoP protein. [The latter genes harbor prototypical PhoP-dependent promoters (13–16) such as that corresponding to the mgtA gene, which does not require proteins other than PhoP and RNA polymerase for in vitro transcription (ref. 17; J. C. Perez and E.A.G., unpublished data). The induction ratio ranged from 38 to 128 for the genes regulated via the connector-mediated pathway, which is much higher than the 4 to 20 induction ratio for the genes regulated via the direct pathway (Fig. 2A).
Fig. 2.
The connector-mediated pathway exhibits higher output levels than the direct pathway. (A) Induction ratio of mRNA levels of genes that are regulated by the PhoP protein directly (i.e., pagP, pmrD, mgtA5′UTR, slyB, mgrB, pcgL, phoP, and rstA) or via the PhoP–PmrD–PmrA pathway (i.e., pbgP, ugd, and pmrC) in organisms grown for 2 h in N-minimal medium with the indicated concentrations of MgCl2. The ratio shown in the y axis corresponds to the mRNA levels after growth at the indicated MgCl2 concentrations to the mRNA level after growth in 10 mM MgCl2. (B) Induction ratio of mRNA levels of pbgP gene in wild-type S. enterica and in the engineered strain with the direct pathway (EG17344) grown for 2 h in N-minimal medium with the indicated concentration of MgCl2. The ratio shown in the y axis corresponds to the mRNA levels after growth at the indicated MgCl2 concentration to the mRNA level after growth in 10 mM MgCl2.
To avoid the potentially confounding issue of comparing the induction ratio for different genes, we investigated the induction ratio of the same gene in isogenic strains that differed only in whether the gene was activated directly by the PhoP protein or via the connector-mediated pathway. We created an S. enterica strain that was deleted for the pmrD gene and harboring a four-nucleotide substitution in the pbgP promoter that replaced the original PmrA-binding site with a PhoP-binding site (Fig. 1B). The induction ratio for the pbgP gene was only 22 for the latter strain, which is one-sixth the induction ratio of 134 exhibited by the strain with the connector-mediated pathway (Fig. 2B). This difference is due to both higher levels of pbgP expression in the strain with the connector-mediated pathway and leakiness of expression in the strain with the direct activation pathway [supporting information (SI) Fig. 6]. We designate signal amplification the property of the connector pathway to promote higher induction ratios than the direct pathway.
The Connector-Mediated Pathway Promotes Transcription with Slower Kinetics than the Direct Pathway.
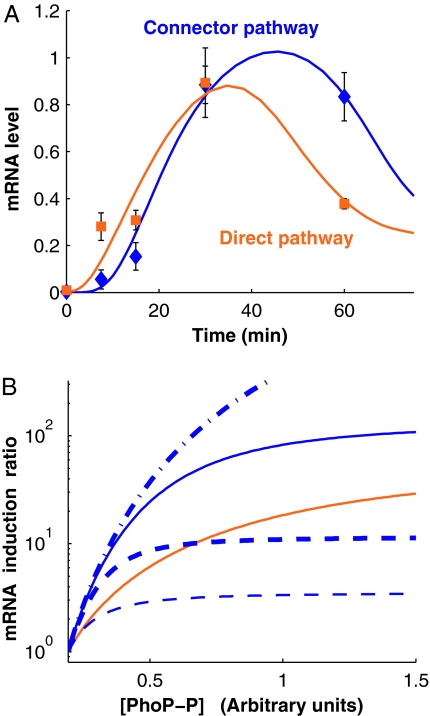
We examined the transcription kinetics of the pbgP gene in the two isogenic strains described above that were grown under repressing conditions (i.e., 10 mM Mg2+) for 4 h and then incubated under inducing conditions (i.e., 20 μM Mg2+) for different extents of time before harvesting the bacteria. The pbgP mRNA was detected immediately after the shift to inducing conditions in the strain in which PhoP regulates pbgP transcription directly, which is in contrast to the delay in the appearance of the pbgP mRNA exhibited by the strain with the connector-mediated pathway (Fig. 3A). The pbgP transcript reached maximum levels by ≈30 min in the two strains. The lag exhibited by the strain with the connector-mediated pathway is likely due to the need to transcribe the pmrD gene and translate the corresponding mRNA so the resulting PmrD protein can protect phospho-PmrA (PmrA-P) from PmrB-promoted dephosphorylation, thereby allowing PmrA-P to bind to the pbgP promoter and activate gene transcription.
Fig. 3.
Modeling of the connector-mediated and direct pathways. (A) The connector-mediated pathway of wild-type S. enterica exhibits an initial delay for activation compared with the direct pathway. mRNA levels of pbgP gene in wild-type S. enterica and in the engineered strain with the direct pathway (EG17344) after bacteria were grown in noninducing (i.e., 10 mM MgCl2) conditions for 4 h and then grown for the indicated times after they were switched to inducing conditions (i.e., 20 μM MgCl2). The value at time 0 corresponds to the time the organisms were switched to inducing conditions. The experimental data corresponding to the direct and connector-mediated pathways are presented as orange squares and blue diamonds, respectively, whereas solid lines indicate trajectories for models fitted as described in Materials and Methods. (B) Expected steady-state induction ratios for the pbgP mRNA as a function of phospho-PhoP concentration ([PhoP-P]) for the two pathways computed by using the fitted models (solid lines; color coding as in A). The key roles played by the rates of PmrD production and PmrA-P–PmrD complex formation is demonstrated by their distinct behavior upon modification of the values for these parameters. The curves corresponding to scenarios with a 10-fold increase in the rates of PmrD production and PmrA-P–PmrD complex formation are shown by thin and thick dashed lines, respectively. The curves corresponding to scenarios with a 10-fold decrease in the rates of PmrD production and PmrA-P/PmrD complex formation are shown by thin and thick dash-dot lines, respectively; in the figure, the latter two lines coincide.
Mathematical Modeling of the Connector-Mediated and Direct-Activation Pathways.
We modeled the two regulatory pathways discussed above as described in Materials and Methods. We focused on the transcriptional regulatory modules (as opposed to the complete signal transduction cascade) because the architectural differences in the modules appear to be directly responsible for the differences in dynamic behavior of the two pathways. The models were fitted to the activation kinetics data (Fig. 3A) assuming that the initial concentrations of the chemical components involved in the pathways were equal to their steady-state concentrations under repressing conditions. The models allowed us to obtain estimates for the rate/equilibrium constants and the initial concentrations of the chemical components.
The models reproduce the main features of the activation behavior exhibited by the strains with the connector-mediated and the direct-regulation pathways (Fig. 3A). As previously demonstrated (10), low Mg2+ increases the level of the phosphorylated PhoP protein (i.e., PhoP-P), which is the key regulator in both pathways. The parameter sets obtained via fitting were used to generate steady-state curves for the induction ratios (Fig. 3B). We chose an interval of PhoP-P concentrations ([PhoP-P]) so that the computed mRNA levels (SI Fig. 6) and induction ratios at the ends of the interval would be close to the experimental measurements. Consistent with the experimental data (Fig. 2B), our model demonstrated signal amplification (Fig. 3B).
Analytical investigation of the models' steady-state equations showed that if the affinity of PmrD for PmrA-P is high and the production rate of PmrA-P is much lower than its depletion rate (which is the case for the fitted model of the connector-mediated pathway), then signal amplification should take place at sufficiently high PhoP-P concentration (SI Text, Cascade-like Properties of the Connector-Mediated Pathway). We could establish that this property results from the difference in the architectures of the two pathways. The connector-mediated pathway can be viewed as a two-stage regulatory cascade with each stage having a sigmoidal signal–response curve (with Hill coefficient 2), which is in contrast to the direct pathway, where there is a single stage (also with sigmoidal response curve and Hill coefficient 2). Therefore, the signal–response curve for the connector-mediated pathway has a Hill coefficient 4, and the steady-state concentration of pbgP mRNA for the cascade approaches 0, [PhoP-P] → 0, much faster than that for the direct pathway model (SI Text Eqs. 15 and 16), which leads to signal amplification at high PhoP-P concentrations (SI Text Eq. 17).
The models' steady-state equations (SI Text Eqs. 7 and 8) showed that the induction ratio curves are insensitive to changes in pbgP promoter activity. On the other hand, our computations demonstrated that the curves are quite sensitive to changes in other parameters, such as the rates at which the PmrD protein is produced and the PmrA-P/PmrD complex is formed (Fig. 3B).
The Levels of the Connector Protein, PmrD, Are Critical for the Output of the Connector-Mediated Pathway.
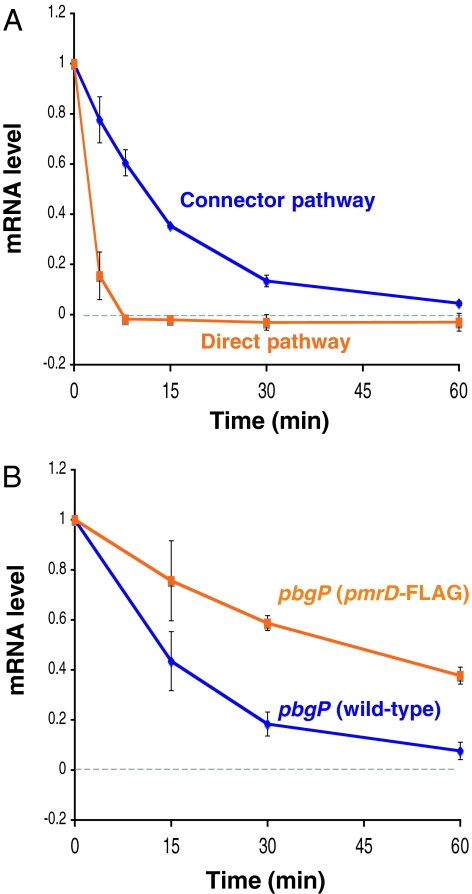
Our model (SI Text Eqs. 9–12) predicts that an increase in the amount of PmrD protein (such as one resulting from a longer half-life) leads to heightened levels of pbgP mRNA. To test this prediction, we created a strain that expressed a PmrD variant (termed PmrD-FLAG) with eight additional amino acids at the C terminus from the normal pmrD promoter and chromosomal location. Because many proteases recognize amino acids in the C terminus of a protein, we anticipated that the PmrD-FLAG protein might have a different half-life than the wild-type protein, thus impacting the pbgP mRNA levels. Indeed, the half-life of the wild-type PmrD protein was ≈11 min, whereas that of the PmrD-FLAG protein was ≈37 min (SI Fig. 7). The levels of the pbgP mRNA were approximately twofold higher in the strain expressing PmrD-FLAG than in the one with the wild-type PmrD protein; this was true at both inducing and repressing Mg2+ concentrations (SI Table 1).
The Connector-Mediated Pathway Promotes Persistence of the pbgP mRNA Levels After a Shift from Inducing to Repressing Conditions.
We analyzed the pbgP transcript levels in organisms that had been grown under inducing conditions (i.e., 20 μM Mg2+) for 2 h and then subjected to repressing conditions (i.e., 10 mM Mg2+). There was a rapid decrease in the levels of pbgP mRNA in the strain with direct regulation, which was in contrast to the persisting higher levels of pbgP mRNA displayed by the strain with the connector-mediated pathway: the pbgP mRNA levels decreased to one-half of the original values in ≈3 and ≈11 min in the strains with the direct and connector-mediated pathways, respectively (Fig. 4A). This difference is not due to the particular response regulator protein that is directly promoting pbgP transcription (i.e., PhoP for the direct pathway and PmrA for the connector-mediated pathway), because the pbgP mRNA level decreased to one-half of its original value in only ≈1 min in the strain with the connector-mediated pathway when pbgP transcription was stimulated by Fe3+ [which activates the PmrA/PmrB system in a PmrD-independent manner (11) (Fig. 1A)], and then subjected to Fe3+ deprivation (SI Fig. 8). The prolonged persistence of the pbgP mRNA depends on the amount of the PmrD protein because the pbgP mRNA level decreased to one-half of the original value in ≈43 min in the strain expressing the more stable PmrD-FLAG protein (Fig. 4B); this is nearly four times longer than that displayed by the strain with the wild-type PmrD protein. Cumulatively, these data demonstrate that the connector-mediated pathway is characterized by increased mRNA persistence after a switch from inducing to noninducing conditions.
Fig. 4.
The connector-mediated pathway of wild-type S. enterica demonstrates increased persistence of pbgP expression after activation in low Mg2+ and then repression in high Mg2+. (A) pbgP mRNA levels in wild-type S. enterica (14028s) and the engineered strain (EG17344) with direct pathway after bacteria were grown in inducing conditions (20 μM MgCl2) for 2 h and then grown for the indicated times after the addition of 10 mM MgCl2. The value at time 0 corresponds to the time the organisms were switched to repressing (i.e., 10 mM MgCl2) conditions. (B) pbgP mRNA levels in wild-type (14028s) and pmrD-FLAG (EG13654) strains grown as described in A.
The Connector-Mediated Pathway Stimulates Increased Polymyxin B Resistance.
PmrA-regulated resistance to the antibiotic polymyxin B results from modification of the lipopolysaccharide with 4-aminoarabinose and phosphoethanolamine (18–20). The biosynthesis of 4-aminoarabinose and its incorporation into the lipopolysaccharide is mediated by proteins encoded by the PmrA-activated ugd gene and pbgP operon (21), whereas the pmrC gene product is responsible for the phosphoethanolamine incorporation into the lipopolysaccharide. To explore whether the pathway architecture affected the levels of polymyxin B resistance, we constructed two isogenic strains that were deleted for the PmrA-activated pmrC gene, which is a minor contributor to polymyxin B resistance in S. enterica (20), so as to focus exclusively on the ugd- and pbgP-mediated polymyxin B resistance. One of the strains retained the wild-type pmrD gene and promoters for the pbgP operon and ugd gene, whereas the other strain was deleted for the pmrD gene and harbored synthetic PhoP boxes at the pbgP and ugd promoters, which enabled direct PhoP control of transcription. Over a broad range of Mg2+ concentrations, the ΔpmrC S. enterica strain displayed higher levels of polymyxin B resistance than the strain with direct control of pbgP and ugd expression (Fig. 5), which is in agreement with the transcriptional data (Fig. 2B).
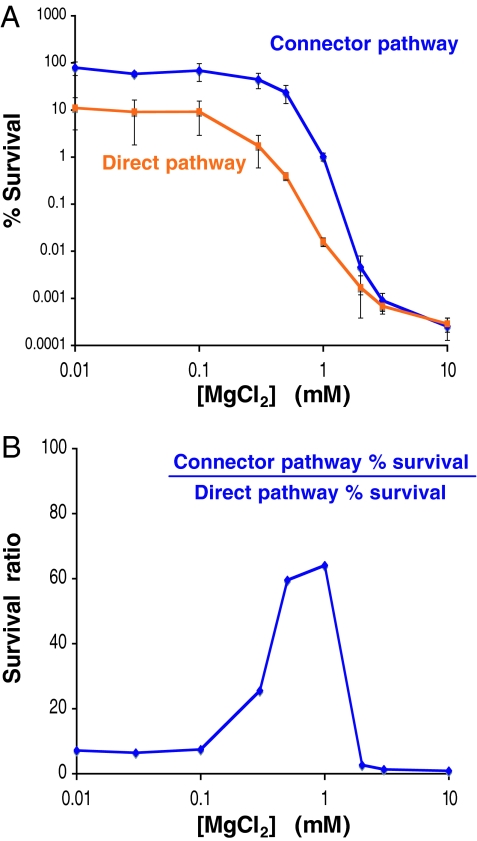
Fig. 5.
The connector-mediated pathway induces higher resistance to polymyxin B than the direct pathway. (A) Survival of ΔpmrC (EG14590) and an engineered strain (EG17352) that lacks the pmrC and pmrD genes and harbors synthetic PhoP boxes at the pbgP and ugd promoters after growth for 4 h in N-minimal medium with the indicated concentration of MgCl2, dilution in LB broth, and incubation in the presence of 5 μg/ml of polymyxin B for 1 h. Then, bacteria were diluted, plated on LB agar plates, and incubated overnight at 37°C. On the y axis, “% Survival” corresponds to the ratio of the number of colony-forming units in the presence of polymyxin B to the number of colony-forming units in the absence of polymyxin B × 100. (B) Survival ratio of ΔpmrC S. enterica (EG14590) relative to the engineered strain (EG17352) in the presence of polymyxin B after growth in N-minimal medium with the indicated concentration of MgCl2. The data set in A was used to generate this figure, which shows the most dramatic difference between the two strains in the 0.75–1 mM Mg2+ range.
Discussion
We have demonstrated that the connector-mediated pathway promoting expression of PmrA-regulated genes in response to the signal that activates the PhoP protein in wild-type S. enterica differs from the direct pathway, where PhoP acts at its target promoters, both in the expression levels (Fig. 2) and the dynamics (Figs. 3 and 4) of the transcriptional response. This finding indicates that connector proteins not only expand the spectrum of environments where genes can be expressed (by virtue of connecting systems that respond to different signals) but also affect the degree to which genes are regulated.
We determined that the connector-mediated pathway confers signal amplification. This was reflected on the mRNA levels achieved upon induction (Fig. 2B and SI Fig. 6) and on the degree of resistance to the antibiotic polymyxin B (Fig. 5), which is mediated by the products of these mRNAs. In addition to signal amplification, the connector-mediated pathway differed from the direct pathway in that it exhibited persistence of expression, that is, when S. enterica was switched from inducing to noninducing conditions, gene expression remained for a longer period in organisms harboring the connector-mediated-pathway architecture. The connector protein, PmrD, is essential for expression persistence because extending its half-life resulted in prolonged expression of the PmrA-activated pbgP gene (SI Fig. 7). An increase in the half-life of the PmrD protein also resulted in higher levels of pbgP expression (SI Fig. 7 and SI Table 1), which is in agreement with the model predictions regarding the significant role of PmrD levels in signal amplification (Fig. 3B). These findings suggest that signal amplification and expression persistence are closely connected and are likely caused by the same factors. Thus, we anticipate that increased affinity of PmrD for PmrA-P, which is predicted to promote increased output levels will also increase persistence of expression. From the point of view of evolution, it is possible that persistence of expression is a “side effect” of positive selection for signal amplification in the connector-mediated pathway. In any case, these properties may allow S. enterica to express PmrA-activated genes continuously in fluctuating environments.
It is noteworthy that some of the genes identified as being activated via the PhoP–PmrD–PmrA connector-mediated pathway in S. enterica (i.e., pbgP and ugd) are under direct transcriptional control of the PhoP protein in the related enteric pathogen Yersinia pestis, which lacks a pmrD gene (22). The Y. pestis pbgP and ugd promoters harbor binding sites for both the PhoP and PmrA proteins, which allows Y. pestis to express these genes in response to the same signals that activate their transcription in S. enterica. The 4-aminoarabinose modification brought about by the PbgP and Ugd proteins is required not only for resistance to polymyxin B (18, 19, 23), which is enhanced in the connector-mediated pathway present in S. enterica (Fig. 5), but also to metal ions present in soil (24). Moreover, it has been implicated in S. enterica infection of chicken macrophages (25) and in Y. pestis survival within murine macrophages (26). Because the connector-mediated and direct pathways differ in their output, it is possible that the distinct regulatory designs used by S. enterica and Y. pestis to express the 4-aminoarabinose-modifying genes contribute to the distinct niches that these bacterial species occupy.
The connector-mediated pathway can be viewed as a regulatory architecture consisting of two separate single-input modules (27) joined by a connector protein. Our analysis demonstrates that this architecture has properties that are distinct from those of the individual single-input modules, but are in common with other multistage expression control circuits. The regulatory logic of the connector-mediated pathway is analogous to that of the coherent OR-gate version of the feedforward loop (FFL), one of the most frequently encountered network motifs (28). Similarly to this FFL, the connector-mediated pathway is characterized by persistence of expression upon deactivation (Fig. 4); yet, the connector-mediated pathway exhibits delayed induction (Fig. 3A) and signal amplification (Fig. 2), which have not been reported for this FFL type (28, 29). On the other hand, there exist striking and unexpected similarities between the dynamical behavior of the connector-mediated pathway, which consists of two transcriptional activators connected by a protein at a posttranslational level, and that corresponding to a synthetic transcriptional cascade consisting of three repressors (30). Both pathways are qualitatively equivalent with respect to induction lags and expression persistence (30–32). Thus, it is likely that other multistage regulatory circuits with linear structure will exhibit similar properties.
Materials and Methods
We describe the bacterial strains, plasmids, growth conditions, construction of chromosomal mutants, and degradation rate analysis by quantitative Western blotting in the SI Text. Bacterial strains and plasmids used in this study are listed in SI Table 2. Primers used in the construction of chromosomal mutants are listed in SI Table 3.
Quantitative Analysis of Transcription.
Bacterial cells from an overnight culture grown in N-minimal medium (pH 7.7, with 10 mM MgCl2) were added to 25 ml of fresh medium with 50 times dilution and shaken at 37°C for 4 h, which constitutes noninducing conditions for the PhoP/PhoQ and PmrA/PmrB systems. After the cells had been spun down, the pellet was resuspended in 175 μl of fresh medium. This fresh cell suspension was used as noninducing cells for all quantitative transcriptional analysis.
For activation analysis, the PhoP/PhoQ and PmrA/PmrB systems were induced by adding the cell suspension of the wild-type 14028s strain or the engineered strain (EG17344) to 25 ml of fresh medium containing no MgCl2 with 500 times dilution, which results in an approximate final concentration of Mg2+ of 20 μM, and shaken at 37°C. For steady-state analysis, a cell suspension of the wild-type strain 14028s or the engineered strain (EG17344) was added to 2 ml of fresh medium containing different concentrations of MgCl2 with 1,000 times dilution and shaken at 37°C for 2 h. For repression analysis, the PhoP/PhoQ and PmrA/PmrB systems were preinduced by adding the cell suspension of wild-type 14028s strain or the engineered strain (EG17344) to 25 ml of fresh medium containing no MgCl2 with 500 times dilution, which results in an approximate final Mg2+ concentration of 20 μM, and shaken at 37°C for 2 h. To preinduce the PmrA/PmrB system in high-Fe3+ condition, a cell suspension of a pmrD mutant (strain EG11491) was added to 25 ml of fresh medium containing no MgCl2 and 100 μM FeCl3 and shaken at 37°C for 1 h. Two hundred fifty microliters of 1 M MgCl2 or 750 μl of 100 mM deferoxamine mesylate was added to the culture to repress the PhoP/PhoQ and/or PmrA/PmrB systems.
An aliquot (1 ml for activation analysis, 2 ml for steady-state analysis, and 0.5 and 1 ml for Mg2+- and Fe3+ chelator-mediated repression analysis, respectively) of bacterial cell culture was collected at the indicated time points, mixed immediately with 1/5 vol of 5% phenol, pH 4.3 (Sigma, St. Louis, MO)/95% ethanol to inactivate cellular RNases, and kept on ice for at least 30 min. After the cells had been spun down at 3,220 × g for 5 min at 4°C, the supernatant was carefully removed. The cell pellet was kept at −80°C until the following day.
Total RNA isolation was performed by using an SV Total RNA Isolation System (Promega, Madison, WI) according to the manufacturer's instructions with the following modifications: cells were resuspended in 10 μl of 10 mM Tris/1 mM EDTA, pH 8 (TE buffer) containing lysozyme (50 mg/ml), and incubated at room temperature for 5 min, and lysed by adding the mixture of 75 μl of SV RNA Lysis buffer (Promega, Madison, WI) and 90 μl of 10 mM Tris/1 mM EDTA, pH 8. The reverse transcription reaction (10 μl) consisted of ≈8 ng total RNA, 2.5 μM random hexamers, and TaqMan Reverse Transcription Reagents (Applied Biosystems, Foster City, CA). Quantitative real-time PCR analysis was carried out by using an appropriate primer set, cDNAs, and an SYBR Green PCR Master Mix (Applied Biosystems) by an ABI PRISM 7000 Sequence Detection System (Applied Biosystems) according to the manufacturer's instructions. Data were normalized with the values corresponding to 16S RNA. For analysis of gene expression at steady state, we obtained the induction ratio by dividing the mRNA levels after growth at the different concentrations of MgCl2 by those corresponding to growth in 10 mM MgCl2. For analysis of gene expression when organisms were switched from inducing to repressing conditions, data were further normalized with the mRNA values of target genes in inducing and noninducing conditions (at time 0) as 1 and 0, respectively. The data correspond to mean values of two independent experiments performed in duplicate. Error bars correspond to the standard deviation. Primers used in quantitative real-time PCR, which were designed by using Primer Express (Applied Biosystems), are listed in SI Table 4.
Polymyxin B Sensitivity Assay.
A 1-h polymyxin B sensitivity assay was performed as described in ref. 8. The data correspond to mean values of two independent experiments performed in duplicate. The error bars in Fig. 5A correspond to the standard deviation.
Mathematical Models: Construction and Analysis.
The mathematical models of the direct and connector-mediated pathways are ordinary differential equation systems describing the temporal changes in the concentrations of the main chemical components of the two pathways. The direct regulation model consists of one equation for the concentration of the pbgP mRNA. The connector-mediated regulation model comprises four equations that describe changes in the concentrations of the pbgP mRNA, PmrD, PmrA-P, and PmrA-P/PmrD. In both pathways, the concentration of PhoP-P is the independent variable that represents the input signal, whereas the concentration of the pbgP mRNA is the output signal. The chemical interactions are modeled by using mass action kinetics; transcriptional control is described by using sigmoidal control functions. The explicit form of the equations, derivation details, and descriptions of mathematical analyses are given in the SI Text.
All computations (including model fitting) were performed in MATLAB R2007a (MathWorks, Natick, MA). The experimental data describing PhoP-P activation dynamics (10) were approximated by a fifth-degree polynomial by using the functions pchip and polyfit. Model-fitting to the data in Fig. 3A was performed with MATLAB's SimBiology Toolbox (MathWorks) by using the optimization functions ga (for the connector-mediated pathway model) and fmincon (for the direct regulation model). In our model-fitting experiments, we used the following general strategy. The initial values for the concentrations of PhoP-P and pbgP mRNA were selected based on experimental data; for other variables (connector-mediated pathway), the initial values were arbitrary. To adjust the parameter (rate/equilibrium constant) values, the model-fitting function sbioparamestim was used. The fitting procedure was initialized with randomized sets of parameter values, which resulted in different fits; the set giving the best fit was selected as the final set. At this stage, the initial values for the reactant concentrations remained unchanged. Next, the steady-state concentration values corresponding to the concentration of PhoP-P under repressing conditions were calculated. We set the initial values for the reactant concentrations equal to the steady-state values, ran the fitting procedure (without randomization) on the model, and calculated the steady state for the new fit; the final fit was obtained after several iterations of this procedure.
Supplementary Material
Acknowledgments
We thank T. Latifi and M. Cromie for technical assistance; M. Goulian, O. A. Igoshin, K. Nishino, D. Shin, J. C. Perez, C. Mouslim, H. Huang, and I. Zwir for discussions; and B. L. Bassler, B. Cohen, L. Harvey, H. Huang and I. Zwir for comments on the manuscript. This work was supported, in part, by National Institutes of Health Grant 42336 to E.A.G., who is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704462104/DC1.
References
- 1.Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton, FL: Chapman & Hall/CRC; 2006. [Google Scholar]
- 2.Kato A, Groisman EA. Genes Dev. 2004;18:2302–2313. doi: 10.1101/gad.1230804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eguchi Y, Utsumi R. Trends Biochem Sci. 2005;30:70–72. doi: 10.1016/j.tibs.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Bougdour A, Wickner S, Gottesman S. Genes Dev. 2006;20:884–897. doi: 10.1101/gad.1400306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tu X, Latifi T, Bougdour A, Gottesman S, Groisman EA. Proc Natl Acad Sci USA. 2006;103:13503–13508. doi: 10.1073/pnas.0606026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoch JA, Silhavy TJ. Two-Component Signal Transduction. Washington, DC: Am Soc Microbiol; 1995. [Google Scholar]
- 7.Inouye M, Dutta R. Histidine Kinases in Signal Transduction. San Diego: Academic; 2003. [Google Scholar]
- 8.Kox LF, Wosten MM, Groisman EA. EMBO J. 2000;19:1861–1872. doi: 10.1093/emboj/19.8.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin D, Groisman EA. J Biol Chem. 2005;280:4089–4094. doi: 10.1074/jbc.M412741200. [DOI] [PubMed] [Google Scholar]
- 10.Shin D, Lee EJ, Huang H, Groisman EA. Science. 2006;314:1607–1609. doi: 10.1126/science.1134930. [DOI] [PubMed] [Google Scholar]
- 11.Wösten MM, Kox LF, Chamnongpol S, Soncini FC, Groisman EA. Cell. 2000;103:113–125. doi: 10.1016/s0092-8674(00)00092-1. [DOI] [PubMed] [Google Scholar]
- 12.García Véscovi E, Soncini FC, Groisman EA. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 13.Kato A, Tanabe H, Utsumi R. J Bacteriol. 1999;181:5516–5520. doi: 10.1128/jb.181.17.5516-5520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minagawa S, Ogasawara H, Kato A, Yamamoto K, Eguchi Y, Oshima T, Mori H, Ishihama A, Utsumi R. J Bacteriol. 2003;185:3696–3702. doi: 10.1128/JB.185.13.3696-3702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lejona S, Aguirre A, Cabeza ML, Garcia Vescovi E, Soncini FC. J Bacteriol. 2003;185:6287–6294. doi: 10.1128/JB.185.21.6287-6294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zwir I, Shin D, Kato A, Nishino K, Latifi T, Solomon F, Hare JM, Huang H, Groisman EA. Proc Natl Acad Sci USA. 2005;102:2862–2867. doi: 10.1073/pnas.0408238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto K, Ogasawara H, Fujita N, Utsumi R, Ishihama A. Mol Microbiol. 2002;45:423–438. doi: 10.1046/j.1365-2958.2002.03017.x. [DOI] [PubMed] [Google Scholar]
- 18.Groisman EA, Kayser J, Soncini FC. J Bacteriol. 1997;179:7040–7045. doi: 10.1128/jb.179.22.7040-7045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunn JS, Lim KB, Krueger J, Kim K, Guo L, Hackett M, Miller SI. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 20.Lee H, Hsu FF, Turk J, Groisman EA. J Bacteriol. 2004;186:4124–4133. doi: 10.1128/JB.186.13.4124-4133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raetz CRH, Whitfield C. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winfield MD, Groisman EA. Appl Environ Microbiol. 2003;69:3687–3694. doi: 10.1128/AEM.69.7.3687-3694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CRH. J Biol Chem. 2001;276:43122–43131. doi: 10.1074/jbc.M106961200. [DOI] [PubMed] [Google Scholar]
- 24.Nishino K, Hsu FF, Turk J, Cromie MJ, Wosten MM, Groisman EA. Mol Microbiol. 2006;61:645–654. doi: 10.1111/j.1365-2958.2006.05273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Jansen R, Gaastra W, Arkesteijn G, van der Zeijst BA, van Putten JP. Infect Immun. 2002;70:5319–5321. doi: 10.1128/IAI.70.9.5319-5321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grabenstein JP, Fukuto HS, Palmer LE, Bliska JB. Infect Immun. 2006;74:3727–3741. doi: 10.1128/IAI.00255-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen-Orr SS, Milo R, Mangan S, Alon U. Nat Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 28.Mangan S, Alon U. Proc Natl Acad Sci USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalir S, Mangan S, Alon U. Mol Syst Biol. 2005;1 doi: 10.1038/msb4100010. 2005.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hooshangi S, Thiberge S, Weiss R. Proc Natl Acad Sci USA. 2005;102:3581–3586. doi: 10.1073/pnas.0408507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDaniel R, Weiss R. Curr Opin Biotechnol. 2005;16:476–483. doi: 10.1016/j.copbio.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 32.Hooshangi S, Weiss R. Chaos. 2006;16 doi: 10.1063/1.2208927. 026108. [DOI] [PubMed] [Google Scholar]
- 33.Kato A, Latifi T, Groisman EA. Proc Natl Acad Sci USA. 2003;100:4706–4711. doi: 10.1073/pnas.0836837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.