Abstract
Changes in Bone Morphogenetic Protein (BMP) 2 gene expression and activity have been linked to many pathological conditions including cancer, osteoarthritis, and birth defects. BMP2 gene polymorphisms have been linked to osteoporosis and osteoarthritis. Sp1 and related proteins are widely expressed regulators of gene expression whose transcription activating abilities vary in different cells and on different genes. We present data indicating that the ratio of Sp1 and Sp3 isoforms varies in cells that express or do not express BMP2. Furthermore, the orientation of Sp1 sites conserved between four orders of mammals influences BMP2 expression. Together our data indicate that the stoichiometry and orientation of Sp1 and Sp3 complexes on the BMP2 promoter influences BMP2 expression.
Keywords: transcription, retinoids, evolution, embryonal carcinoma cells
1. Introduction
BMP2 is a key mediator of bone development and repair (Urist, 1965; Wozney et al., 1988; Lyons et al., 1989; Celeste et al., 1990; Lyons et al., 1990; Bostrom et al., 1995; Spector et al., 2001; Bouletreau et al., 2002; Cho et al., 2002; Gerstenfeld et al., 2003) and is uniquely required for fracture healing (Tsuji et al., 2006). The central role of BMP2 in initiating and regulating bone formation makes the finding that genetic variation in BMP2 is linked to osteoporosis logical (Styrkarsdottir et al., 2003; Ralston, 2005; Reneland et al., 2005; Xiong et al., 2006). The BMP2 gene also is linked to osteoarthritis, which has a complex genetic relationship with osteoporosis (Valdes et al., 2004; Valdes et al., 2006). Genetic manipulation of transgenic mice and human disorders highlight the importance of controlling the level of BMP signaling during bone and cartilage formation and adult bone homeostasis (Shafritz et al., 1996; Gong et al., 1999; Marcelino et al., 2001; Zhao et al., 2002; Devlin et al., 2003; Wu et al., 2003; Chen et al., 2004; Mishina et al., 2004; Usui et al., 2004).
Abnormal BMP2 gene expression also is specifically associated with osteoarthritis and with lung, breast, colon, pancreatic, and prostate cancers (Kleeff et al., 1999; Reinholz et al., 2002; Langenfeld et al., 2003; Hardwick et al., 2004; Horvath et al., 2004; Valdes et al., 2004). Loss of normal BMP2 expression occurs in prostate and breast cancers and in the microadenomas of familial adenomatous polyposis patients (Harris et al., 1994; Hardwick et al., 2004; Horvath et al., 2004). We and others have shown that recombinant BMP2 can inhibit cell proliferation and induce apoptosis in diverse cell types including cancer cells (e.g., (Rogers et al., 1992; Graham et al., 1994; Glozak and Rogers, 1996; Coucouvanis and Martin, 1999; Rodriguez-Leon et al., 1999; Kawamura et al., 2000; Hallahan et al., 2003; Zhang et al., 2003; Hardwick et al., 2004)). Thus, normal BMP2 protein levels may suppress tumors in some tissues and altering BMP2 expression may promote the transition from normality to malignancy.
In embryos, BMP2 is a morphogen produced in key signaling centers, such as the zone of polarizing activity (ZPA) in the limb bud and the enamel knot in tooth (Hogan, 1996; Vaahtokari et al., 1996; Martinez-Barbera et al., 1997; Vincent and Briscoe, 2001; Raftery and Sutherland, 2003). The level of a morphogen is critical, because cells measure the morphogen concentration and behave accordingly. For example, precisely regulated BMP2 levels specify the genes expressed in intermediate mesoderm and normal proepicardial identity (James and Schultheiss, 2005);(Schlueter et al., 2006).
The bottom line is that the concentration of BMP2 in specific tissues is critical because altered levels may cause developmental anomalies or influence the onset or progression of diverse human diseases. Thus identifying the molecular mechanisms that regulate the precise degree of BMP2 expression is of paramount importance.
The differentiation of F9 embryonal carcinoma cells is an excellent experimental model for elucidating how BMP2 expression is modulated. BMP2 is expressed at three distinct levels in F9 cells (Rogers et al., 1992; Rogers, 1996). Undetectable in undifferentiated stem cells, retinoic acid (RA) strongly induces the BMP2 transcript. Combined treatment with RA and drugs that elevate cAMP levels induces the BMP2 transcript 5 to 6 fold more than RA alone. Cyclic AMP elevation alone induces neither differentiation nor BMP2 expression. Our delineation of the elements that regulate BMP2 in F9 cells showed that both activating and repressing elements surround the promoter (Abrams et al., 2004). We previously used several approaches including chromatin immunoprecipitation (ChIP) analyses to show that these regulatory elements include a functional Sp1 consensus sequence.
Sp1 and other Sp and Krüpple-Like Factor (KLF) proteins regulate hundreds of genes (for review see (Bouwman and Philipsen, 2002; Li et al., 2004; Chu and Ferro, 2005)). The founding member, Sp1 and the highly similar Sp3 protein recognize a motif of GGGCGG or related GC-rich sequences (Gidoni et al., 1984). Sp1 and Sp3 can either enhance or repress gene activity. The exact function of these transcription factors depend on their relative ratios within cells, the nature of the binding site(s) within regulated promoters, and a variety of post-translational modifications, including proteolytic cleavage. Various transcription factors cooperate with Sp1 and Sp3 to modulate function (Li et al., 2004). For example, we found that the retinoid receptors potentiate Sp1 binding to the BMP2 site in vitro (Abrams et al., 2004). As in other RA-inducible genes that lack classical RAREs, but contain Sp1-binding sites, this retinoid receptor and Sp1 interaction may partly explain the absence of a classical retinoic acid response element (RARE) near the BMP2 promoter (Suzuki et al., 1999; Husmann et al., 2000; Shimada et al., 2001; Hao et al., 2003). We now present a more detailed analysis of how Sp1 and Sp3 modulate BMP2 expression.
2. Materials and methods
2.1. Luciferase Reporter Constructs
All nucleotide positions are indicated with respect to the murine distal BMP2 promoter that is 2,201 nt upstream of the initiator codon (ATG). All clones are of mouse (strain 129) origin. Constructs Bmp-LUC (nt -1,237 to 471, pGL1.7XX) and BmpΔSp1-LUC (nt -1,237 to -193 and -160 to 471, pGL1.7XXΔNot) were described in (Abrams et al., 2004). Construct BmpMutSp1-LUC (nt -1,237 to 471, with the CC at position nt -180 and -179 mutated to AA) was created by inserting annealed and blunted oligos into pBlueScript KSII cut with EcoRV. The sequence of the mutating oligo was: CCTTAGCGGCCGCGCGCTCGCCCCGAACCGCTCCACCGCGGCCGCCCCGT AGGGCGC. The resulting plasmid (pBlueMutSp1) was digested with Not I and a 32 bp fragment containing the mutated Sp1 site was purified and inserted into pGL1.7XX (construct A) cut with Not I to generate pGL1.7XXΔNotI+MutSp1. Construct BmpRevSp1-LUC (nt -1,237 to 471, except nt -193 to -161 in reverse orientation) was creating by digesting pGL1.7XX (construct A) with NotI and religating to select a Not I fragment in reverse orientation. All plasmids were sequenced to confirm that one copy of the sequence was in the expected orientation.
2.2. F9 Cell Culture and Differentiation
F9 embryonal carcinoma cells were plated on dishes pre-coated with 1% gelatin and incubated at 37 °C with 10% CO2. The culture media consisted of Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated calf serum and 2 mM glutamine. The cells were induced to differentiate into parietal endoderm by adding 1 μM all-trans-retinoic acid, 250 μM Bt2cAMP, and 500 μM theophylline (RACT). Undifferentiated control cells were treated with 250 μM Bt2cAMP and 500 μM theophylline (CT).
2.3. F9 Cell Transfection by Calcium Phosphate Precipitation
Transfections were performed essentially as described by Vasios et al. (Vasios et al., 1989). Briefly, F9 cells were plated at 1 × 106 or 0.3 × 106 (CT only) cells per 100-cm dish (Nunc) for 12 h, drugged for 48 h with CT or RACT, transfected by overnight calcium phosphate precipitation, and then cultured for an additional 24–48 h with drugs. Each 100-cm dish was co-transfected with 10 μg of reporter plasmid and 3 μg of pβAclacZ (Vasios et al., 1989) containing the lacZ coding region driven by the constitutive β-actin promoter.
2.4. Luciferase Assays
Cells were extracted, and luciferase activity was determined using the Promega Luciferase Assay System and a Monolight 2010 luminometer (Analytic Luminescence Laboratory). Luciferase activity was normalized for transfection efficiency by dividing the raw luciferase value by the units of β-galactosidase activity (1 unit = A420·μl−1·h−).
2.5. Nuclear Extracts
Nuclear extracts were prepared from F9 cells treated with CT or RACT (Masson et al., 1993). Briefly, confluent cells were washed two times with PBS, scraped, and collected by centrifugation for 10 min at 1,700 g at 4 °C. One ml of ice-cold Buffer A (50 mM Hepes-KOH (pH 7.8), 420 mM KCl, 0.1 mM EDTA (pH 8.0), 5 mM MgCl2, 20% glycerol, 1 mM phenylmethlsulfony fluoride (PMSF), 1 mM dithiothreitol (DTT), 2 μg/ml of aprotinin, and leupeptin) was added to resuspend the pellet. After a second centrifugation and resuspension, the pellet was incubated on ice for 10 min to swell and lyse the cells. After vortexing for 30 sec, lysis was confirmed by visual inspection. After centrifugation, the pelleted nuclei were resuspended in Buffer C (50 mM Hepes-KOH (pH 7.8), 25% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA (pH 8.0), 1 mM PMSF, 1mM DTT, 2μg/ml of aprotinin, and leupeptin) and incubated on ice for 15 min. After a 5 min centrifugation at 4°C, the supernatant was removed and stored at −80°C. The protein concentration was determined using the Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, CA).
2.7. Electrophoretic Mobility Shift Assays (EMSAs)
EMSAs were performed essentially as described in Glozak et al. (Glozak et al., 2003) except poly(dA-dT) was used to block nonspecific binding. Complementary oligonucleotides were annealed then purified by passage over Sephadex G50 spin columns. Dr. F. Kashanchi provided human recombinant Sp1 from Promega Corp. Supershifts were performed by adding antibodies (Santa Cruz Biotechnology Inc.) specific to Sp1 (sc-59), Sp3 (sc-13018), RARα (C-20), or RARβ (C-19) for 10 minutes after the normal incubation of nuclear extracts and probes.
2.8. Western blot analysis
Western blot analysis was performed as described in (Yatani et al., 2006). Four μg of protein were dissolved in 1% SDS, 100 mmol/liter Tris-HCl (pH 6.5), 10% glycerol, 0.05% bromphenol blue, 5% 2-mercaptoethanol, boiled, separated by 12.5% SDS-PAGE (30% Acryl:0.8%bis-Acryl) and electroblotted onto polyvinylidene difluoride membranes (Invitrogen, Carlsbad, CA). Blots were incubated for 30 min at room temperature with a 1:200 dilution of rabbit anti-Sp1 polyclonal antibody (sc-59, Santa Cruz Biotechnology Inc.) or 1:200 rabbit anti-Sp3 polyclonal antibody (sc-13018, Santa Cruz Biotechnology Inc.) in Tris-buffered saline containing 0.1% Tween 20 and 5% nonfat milk. Goat anti-rabbit IgG (H+L)-HRP conjugate (Bio-Rad 170–6515) secondary antibodies were used to visualize the protein bands using ECL reagent (Perkin Elmer Life Sciences, Inc). The bands were scanned utilizing a densitometer (Molecular Dynamics, Piscataway, NJ) and normalized for protein loading. Coomassie blue staining was used to verify equal protein loading of the blots.
3. Results and Discussion
3.1. F9 cells contain Sp1 and Sp3 proteins that bind a GC-rich region in the Bmp2 promoter
We previously showed that deletion of a 32 bp Not I fragment upstream of the distal Bmp2 promoter reduced Bmp2 reporter gene activity by nearly half (Abrams et al., 2004). A consensus Sp1 site (GGGCGG) in the reverse orientation relative to the promoter is located in this fragment. However, the 32 bp fragment is within a highly GC-rich region that may include other sites that Sp1 proteins could bind. Consequently, we used several annealed oligonucleotides and EMSAs to scan the GC-rich region shown in Fig. 1A for the ability to bind endogenous proteins in nuclear extracts from undifferentiated (dibutyryl cAMP and theophylline (CT)-treated) and differentiated (RACT-treated) F9 cells.
Fig. 1. Oligonucleotide mapping of a GC-rich region with several Sp1 sites.
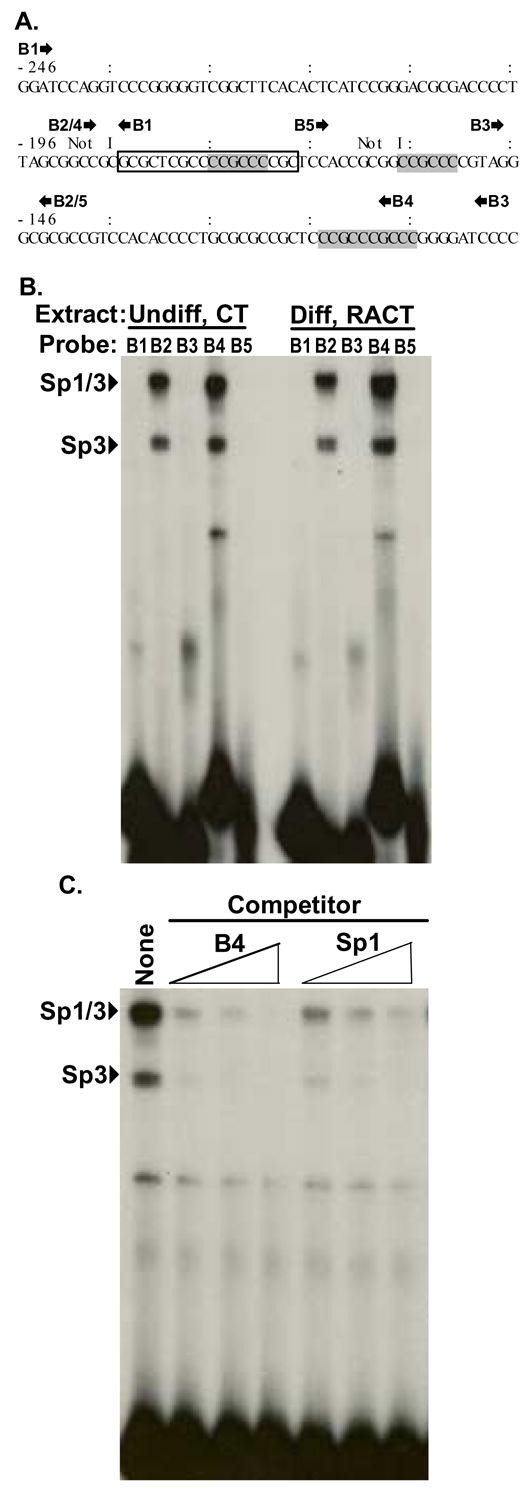
A. Bmp2 sequence from nt -246 to -97 relative to the distal promoter. Classical Sp1 sites (GGGCGG) in the reverse orientation are boldfaced and shaded. Two Not I restriction endonuclease sites (GCGGCCGC) used for the reporter gene constructs shown in Fig. 4 are marked. The approximate locations of the annealed oligonucleotide pairs used for EMSA in B are marked by forward (
 ) and reverse (
) and reverse (
 ) arrows. The box marks sequence present in oligos B2 and B4, but not oligos B1, B3, and B5. Because only B2 and B4 are gel shifted by Sp1/3 proteins, this must be the minimal sequence required for binding. B. Nuclear protein extracted from undifferentiated (Undiff) F9 cells treated with CT or differentiated (Diff) cells treated with RACT for 3 days were incubated with various annealed and radio-labeled oligonucleotide pairs: B1 (-246 nt to -187 nt), B2 (-192 nt to -143 nt), B3 (-148 nt to -98 nt), B4 (-190 nt to -107 nt), B5 (-168 nt to -143 nt). The autoradiograph shown is representative of 3 independent experiments. C. EMSA was performed with 32P-labeled B4 oligonucleotides and nuclear extract from CT-treated F9 cells. Competition assays were performed with 100-, 200-, and 500- fold molar excess of unlabeled B4 (lanes 2, 3, 4) or Sp1 (ATTCGATCGGGGCGGGGCGAGC, lanes 5, 6, 7) oligonucleotide.
) arrows. The box marks sequence present in oligos B2 and B4, but not oligos B1, B3, and B5. Because only B2 and B4 are gel shifted by Sp1/3 proteins, this must be the minimal sequence required for binding. B. Nuclear protein extracted from undifferentiated (Undiff) F9 cells treated with CT or differentiated (Diff) cells treated with RACT for 3 days were incubated with various annealed and radio-labeled oligonucleotide pairs: B1 (-246 nt to -187 nt), B2 (-192 nt to -143 nt), B3 (-148 nt to -98 nt), B4 (-190 nt to -107 nt), B5 (-168 nt to -143 nt). The autoradiograph shown is representative of 3 independent experiments. C. EMSA was performed with 32P-labeled B4 oligonucleotides and nuclear extract from CT-treated F9 cells. Competition assays were performed with 100-, 200-, and 500- fold molar excess of unlabeled B4 (lanes 2, 3, 4) or Sp1 (ATTCGATCGGGGCGGGGCGAGC, lanes 5, 6, 7) oligonucleotide.
Fig. 1B shows that oligos B2 and B4 bound two protein complexes strongly. The B2 and B4 oligos uniquely share the reversed Sp1 site previously noted in the 32 bp fragment that is required for full promoter activity. In contrast oligos B1, B3, and B5 failed to bind these two complexes. Oligos B3 and B5 contain consensus Sp1 sites that clearly cannot bind the same proteins as oligos B2 and B4. The 18 nucleotides (boxed in Fig. 1A) that are present in B2 and B4, but not in B1, B3, and B5, must be required for the ability of these oligos to effectively form these complexes in F9 cell extracts.
We next analyzed binding specificity by competition analyses using cold BMP2 and Sp1 annealed oligonucleotides. As expected unlabeled wild type B4 oligo competed efficiently with the radiolabeled probe for binding the two intense complexes (Fig. 1C). Similarly, a consensus Sp1 probe inhibited the binding of the BMP2-specific probe to the same two complexes. This indicated that the BMP2 sequence and the Sp1 sequence interacted with the same proteins in the complex nuclear extracts.
We then tested the hypotheses that the shifted protein:oligo complexes were Sp1 and/or the closely related protein, Sp3. The addition of antibodies directed against Sp1 and Sp3 caused distinct supershifted complexes (Fig. 2A, marked “Sp1 SS” and “Sp3 SS”). Running gels considerably longer than those shown in Fig. 1 revealed that the slower migrating, intensely-labeled complex, marked Sp1/3, had two components (Fig. 2A, B). Labeling in the top half of this complex was reduced by the addition of Sp1 antibody, but not Sp3 (compare lanes 1 and 6 to lanes 2 and 7). In contrast, Sp3 antibodies significantly reduced the intensities of the bottom half of the slower migrating complex and of the fastest migrating complex labeled Sp3 (Fig. 2A, B, compare lanes 1 and 6 to lanes 3 and 8). The identity of these complexes was confirmed by similar supershifts induced in HeLa cell extracts (Fig. 2B, compare lanes 11 to lanes 12 and 13). HeLa cells contain both Sp1 and Sp3 (Jackson and Tjian, 1989; Hagen et al., 1994). Thus the BMP2 Sp1 site can bind Sp1 and two isoforms of Sp3 proteins.
Fig. 2. Both Sp1 and Sp3 bind the Bmp2 sequence.
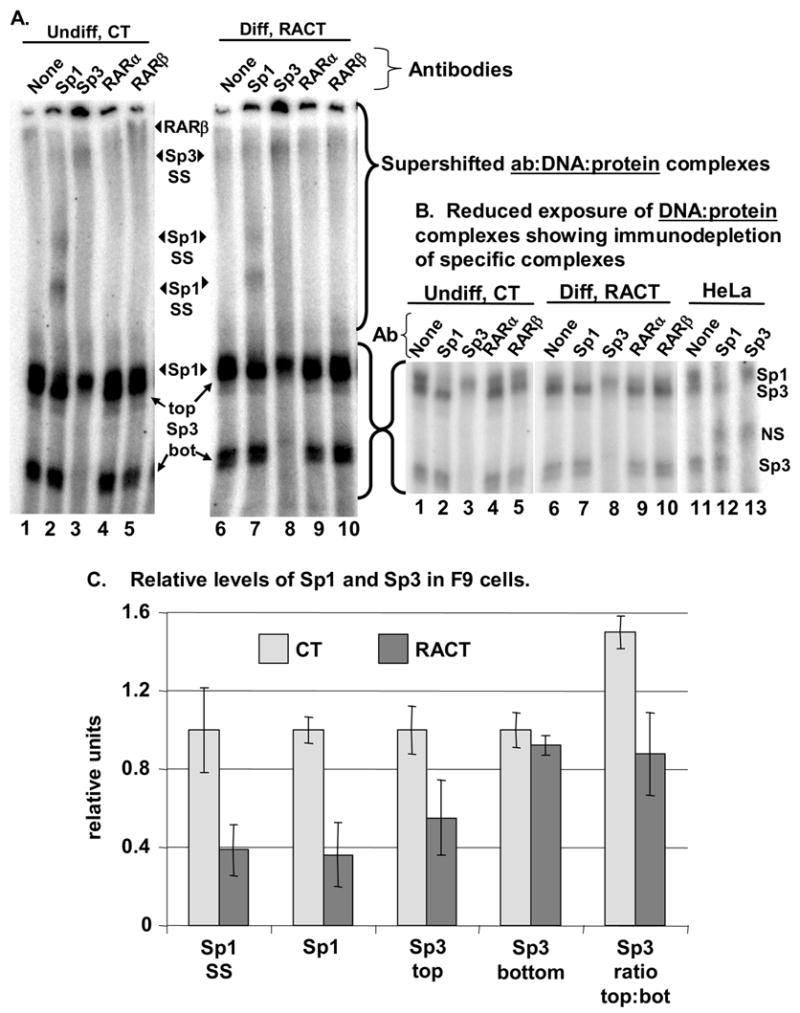
A, B. Gel mobility supershift experiments were carried out using nuclear extract from undifferentiated (CT) or differentiated (RACT) F9 cells (day 3) and B4 as probe. Supershift gels were electrophoresed longer than those shown in Fig. 1 to separate overlapping Sp1 and Sp3 complexes. A shows a longer exposure of the gel to emphasize the antibody:DNA:Sp1/3 complexes (supershifts). At this exposure the DNA:Sp1/3 complexes not bound by antibody are over-exposed. A shorter exposure of this region of the gel is shown in B. Anti-Sp1 antibody (Ab) caused the formation of two supershifted complexes (Sp1 SS) and reduced the amount of radiolabeled complex marked Sp1. This clearly revealed the slower migrating (top) form of Sp3. In contrast, anti-Sp3 antibody caused the formation of one supershifted complex (Sp3 SS) and reduced the amount of the two radiolabeled complexes marked Sp3 (top and bottom). Gel shifts using HeLa nuclear extracts, shown in B, illustrate the expected effect of complementary antibody depletion in a cell type where Sp1 and Sp3 have been extensively characterized (Jackson and Tjian, 1989; Hagen et al., 1994). The nonspecific (nonsp.) protein was shifted by both antibodies and has been observed previously (Hagen et al., 1994). Anti-RARβ, but not RARα, antibodies induced a greater supershift in extracts from CT-treated cells relative to RACT-treated cells. C. The counts in specific DNA:protein complexes that formed in extracts from CT- or RACT-treated cells were quantified. The first four pairs of bars indicate the relative levels of each complex with standard error measurements (SEM, n = 4–5): supershifted Sp1 (Sp1 SS), Sp1 bound to probe in the presence of Sp3 antibody (Sp1), the slower migrating Sp3 bound to probe in the presence of Sp1 antibody (Sp3 top), and the faster migrating Sp3 bound to probe (Sp3 bot). The 5th pair of bars shows the ratio of the slower to the faster migrating Sp3 forms (top:bot). Because this ratio was determined by dividing the counts in the top band by the counts in the lower band within each lane, variation between binding reactions is eliminated (n = 4).
To further demonstrate that our nuclear extracts are biologically relevant, we also show the results of supershifts with antibodies to RARα and β. The antibody specific to RARβ, but not RARα, supershifted a BMP2 probe:Sp1/3 complex specifically in extracts from CT-treated cells, but not RACT-treated cells (Fig. 2A, lanes 5 and 10). Thus, as predicted from our previous ChIP results (Abrams et al., 2004), Sp1 and RARβ appear to simultaneously bind the BMP2 promoter region in nuclear extracts from undifferentiated cells.
Multiple experiments revealed that the relative levels of Sp1 and Sp3 differed in extracts from differentiated or undifferentiated cells (Fig. 2A–C). To quantify the levels of probe-bound Sp1 or Sp3, we used several approaches. As shown in the first pair of bars in Fig. 2C, the amount of Sp1 supershifted by the Sp1 antibody was significantly higher in extracts from CT-treated cells relative to RACT-treated cells (n=4). Directly measuring the probe:Sp1 complex was complicated by the migration of a probe:Sp3 complex (Sp3 top) immediately below the Sp1 complex. We abolished the Sp3 complex at this position by immunodepletion (supershift with a specific Sp3 antibody), leaving only the Sp1 complex (Fig. 2B, lanes 3, 8, and 13). Measurements of the amount of Sp1-bound probe in 4 independent reactions containing Sp3 antibody replicated the Sp1 supershift results (Fig. 2C, 2nd pair of bars). Thus differentiation reduced the level of endogenous Sp1 bound to BMP2 probe.
We also quantified the two forms of Sp3 by measuring the amount of Sp3 bound by probe in the presence of Sp1 antibody. As shown in Fig. 2C, the slower migrating form of Sp3 (top) was clearly more abundant in CT-treated cells relative to RACT-treated cells (n=4). In contrast, the level of the faster migrating form of Sp3 (bottom) was not changed between CT- and RACT-treated cells (n=5). To confirm that differentiation influenced the relative levels of these proteins, we also directly measured the ratio of the top form to the bottom form in 4 independent reactions (Fig. 2C, 5th pair of bars). The advantage of measuring a ratio within one lane is that variation between binding assays is eliminated. This analysis proved that the relative level of Sp1 and Sp3 isoforms was altered during differentiation.
Because resolving specific proteins on non-denaturing gels is challenging, we also used western blot analysis to quantify the levels of the Sp1 and Sp3 proteins. We made nuclear extracts from F9 cells treated with CT or RACT for 3 and 4 days. The RACT-treated cells, but not the the CT-treated cells, are fully differentiated and express high BMP2 levels. As shown in Fig. 3, both undifferentiated and differentiated cells synthesize Sp1 and Sp3. However, in differentiated cells, the level of Sp1 and the 115 kDa isoform of Sp3 are reduced relative to the 78 and 80 kDa Sp3 isoforms. This result is consistent with the quantitative EMSA results (Fig. 2) and our previous demonstration by ChIP that Sp1 occupies the Bmp2 promoter in undifferentiated, CT-treated cells, but not in RACT-treated cells (Abrams et al., 2004).
Fig. 3. Western blot analysis of Sp1 and Sp3 levels in nuclear extracts from undifferentiated or differentiated F9 cells.
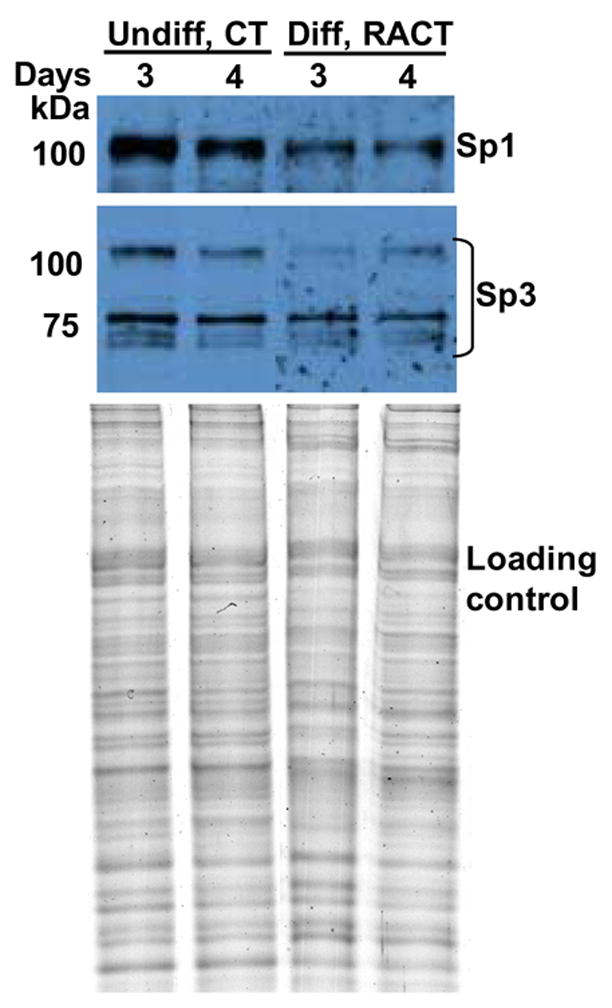
Nuclear extracts (4 μg protein) from cells treated with CT or RACT for 3 and 4 days as indicated were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and incubated with polyclonal antibodies to Sp1 and Sp3. The bottom panel shows a Coomassie Blue-stained gel of the same amount of each extract indicating that similar amounts of protein were loaded for each sample.
The nature of the Sp1 or Sp3 proteins that bind in vivo can be modulated by interactions with other transcription factors. Indeed, we demonstrated here that an RARβ antibody can supershift a complex containing Sp1/3 and BMP2 sequence (Fig. 2A, lane 5). This is consistent with our previous demonstration that RARs potentiated the binding of Sp1 to the Bmp2 sequence in vitro and that RARβ co-occupied the Bmp2 sequence with Sp1 only in undifferentiated cells. The relative levels of Sp1 and Sp3 have been shown to influence the expression of other genes (Li et al., 2004; Safe and Abdelrahim, 2005). In F9 cells, it appears that the stoichiometry of complexes containing retinoid receptors, Sp1, and Sp3 on the Bmp2 promoter differs between undifferentiated cells and differentiated cells. This may contribute to the effect of retinoid treatment on Bmp2 expression.
3.2. The orientation of Sp1 sites are conserved among BMP2 genes from four mammalian orders
We used newly emerging genome sequences to assess the relative importance of the Sp1 site in the context of evolution. The degree of conservation of specific sequences may reveal regions where variation exerts significant positive or negative effects on survival over the millennia. As shown previously (Abrams et al., 2004) and in Fig. 4A, the reversed Sp1 site is conserved in humans. Sites with one G substitution are present in the Bmp2 promoters of another primate, squirrel monkey, dog, and cow. Immediately upstream of this Sp1 site, dog, cow, and both primates, but not rodents, also have another Sp1 sequence. Interestingly, all sites in all species are reversed. These species represent four orders of mammals (Rodentia, Primates, Carnivores, and Artiodactyls) and an evolutionary separation of 92 million years (Ureta-Vidal et al., 2003). As we show for the mouse gene (Fig. 4D), the orientation of these sites may be functionally relevant.
Fig. 4. A conserved Sp1 site is the functional element mediating RA response of Bmp2 gene.
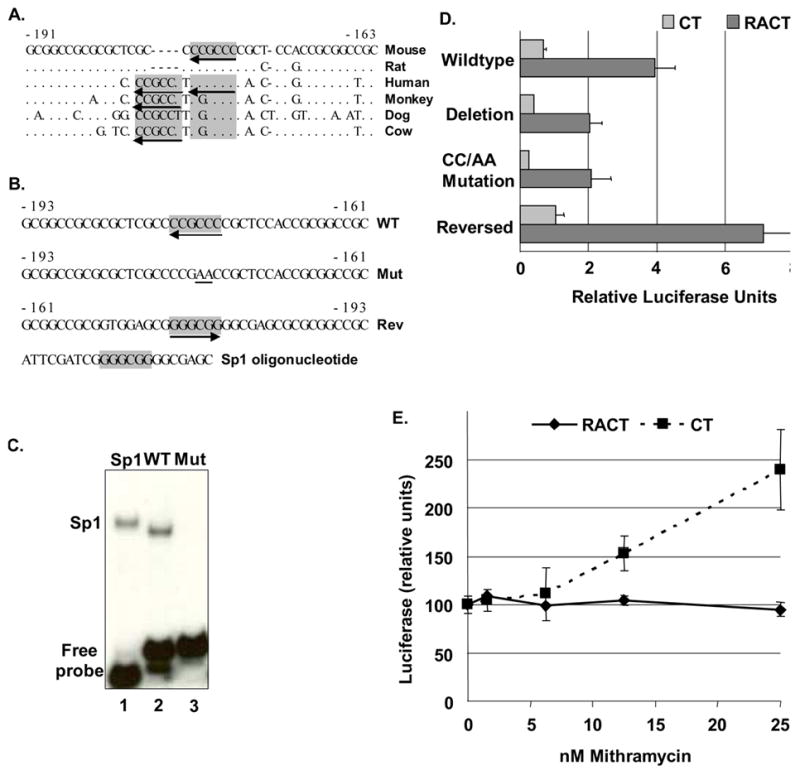
A. The Bmp2 upstream region (-191 nt to -163 nt relative to the murine distal promoter, M. musculus, NW_000178) and the corresponding sequences from rat (R. norvegicus, AABR02024700), human (AF040249), squirrel monkey (S. sciureus, AY494188), dog (C. familiaris, NC_006606.2) and cow (B. taurus, NC_007311) were aligned with MultiPipMaker (bio.cse.psu.edu/pipmaker). Identical nucleotides are indicated by “.”; gaps by “-”. The Sp1 sites are shaded. Arrows indicate orientation. Additional perfect sites in the same reverse orientation are present in the primate, dog, and cow promoters. B. Wild-type and modified Not I fragments from the mouse Bmp2 gene from -193 nt to -161 nt relative to the distal promoter that were used for EMSA (C) and reporter gene analysis (D). The Sp1 sites (GGGCGG) are boldfaced and shaded. The orientation of the naturally-reversed wild-type (WT) Sp1 site is indicated by an arrow. The wild-type Sp1 site was mutated (Mut) by converted a “CC” to “AA” as shown. The sequence of the Not I fragment in the reverse orientation is shown with the Sp1 site indicated. C. EMSA analysis was performed with annealed WT or Mut Bmp2 oligos or the Sp1 consensus oligos labeled with 32P and recombinant human Sp1 protein. D. Reporter genes with wild type or altered Bmp2 promoters (-1237 nt to 471 nt) were co-transfected into F9 cells treated with CT or RACT. In BmpΔSp1-Luc, the -193 nt to -161 nt fragment was deleted. The Sp1 site in BmpMutSp1-Luc was mutated as shown in B. In BmpRevSp1-Luc, the -193 nt to -161 nt Not I fragment was reversed as shown in B. Cells were co-transfected with a plasmid that constitutively expresses lacZ to normalize transfection efficiency. According to a Student’s t test (two-sample assuming equal variances) the luciferase activity generated from each modified reporter construct differed significantly from the wild type construct in RACT-treated cells (p < 0.05), but not in CT-treated cells. E. F9 cells were drug treated and transfected as above except that cells were exposed to mithramycin (MTM) at the time of transfection. The relative luciferase activity +/− SEM is plotted relative to MTM concentration (n = 2–4). The values showing the effect of MTM in CT-treated cells (Abrams et al., 2004) are shown to illustrate the different effect of MTM on BMP2 reporter gene expression in undifferentiated and differentiated cells.
3.3. The specific sequence and orientation of the Sp1 site modulates Bmp2 reporter gene expression
EMSAs and western blots determined that F9 cells contained Sp1 and Sp3 proteins that bind immediately upstream of the BMP2 promoter. To determine the role of the reversed Sp1 site, we generated specific mutations in the context of a luciferase reporter gene containing the Bmp2 promoter. We previously showed that deletion of the 32 bp Not I fragment containing the Sp1 site reduced the activity of the reporter gene by half (Fig. 4D, (Abrams et al., 2004)). We used site-directed mutagenesis to disrupt the Sp1 site as shown in Fig. 4B (Mut). Recombinant Sp1 protein bound oligonucleotides containing the wild type 32 bp fragment as well as the Sp1 consensus sequence (Fig. 4C, compare lanes 1 and 2). In contrast, oligonucleotides with the mutated site were unable to interact with the recombinant Sp1 protein (Fig. 4C, lane 3). Furthermore, the activity of the reporter gene with this mutation was indistinguishable from the activity of the reporter gene with the 32 bp deletion (Fig. 4D). This confirmed that the reversed Sp1 site accounted for the activating influence of the 32 bp Not I fragment.
We previously showed that mithramycin (MTM), an inhibitor of Sp1:DNA binding, induced the BMP2 reporter gene in specifically in CT-treated cells (Abrams et al., 2004). We now provide our previously unpublished data demonstrating that MTM did not alter reporter gene expression in RACT-treated cells. This suggests that Sp1-like proteins primarily influence BMP2 expression in undifferentiated cells.
We also tested a reporter gene with the 32 bp Not I fragment in an orientation that was flipped relative to the promoter. This change placed the Sp1 site in a forward orientation (“Rev” in Fig. 4B), but did not change its distance from the transcription start site. The activity of this reporter gene was significantly greater than that of the wild type construct (Fig. 4D). This suggests that an, as yet unidentified, activator binds this site. Inhibiting Sp1 binding, e.g., with mithramycin, may enable this activator to bind and induce BMP2 expression in undifferentiated cells. Altering the context of the site as in the 32 bp reversal may improve the affinity of the activator for the site or the relationship of the activator with other proteins in the region.
The precise expression level of BMP2 must be tightly regulated, because elevated or reduced levels of this essential morphogen will influence cell behavior. Sp1 and Sp3 are nearly ubiquitously expressed transcription factors whose activities are controlled by both cell-specific and gene-specific parameters. A human single nucleotide polymorphism that is one of the best predictors of susceptibility to osteoporotic fracture slightly increases the affinity of Sp1 for a binding site in the COL1A1 gene (Mann et al., 2001; Mann and Ralston, 2003). The effect of the modest elevation in collagen alpha 1(I) protein relative to alpha 2(I) caused by this increase in Sp1 binding affinity illustrates how subtle mutations influence complex biological processes. Despite the relatively few mutations that would be required to convert sequences in the GC-rich region to forward orientation Sp1 sites, these have not evolved over a lengthy evolutionary time frame. Thus, the presence and orientation of the BMP2 Sp1 sites may confer a selective advantage.
3.4. Conclusions
BMP2 controls essential processes including cell survival, growth rate, differentiation, and apoptosis. Because BMP2 is such a potent growth factor, many mechanisms have evolved to precisely regulated expression and function. Indeed, the mammalian BMP2 promoter and 3′ untranslated regions are functionally conserved between four orders of mammals (Abrams et al., 2004; Fritz et al., 2004; Fritz et al., 2006). Negative regulation of BMP2 expression and function is a common theme. Numerous highly conserved antagonists such as Noggin repress BMP2 regulation at the extracellular level (Canalis et al., 2003). Repressors have been identified that block every intracellular signaling transduction step (Massague et al., 2005; Hill, 2006). Our observations that BMP2 RNAs are less stable in cells that do not express BMP2 suggest that negative post-transcriptional regulatory mechanisms influence BMP2 expression (Fritz et al., 2004). At the transcriptional level, we showed that RARβ and Sp1 proteins assemble on the inactive BMP2 promoter and disassemble after retinoids induce BMP2 expression (Fig. 2, (Abrams et al., 2004)). We propose the hypothesis that relief of repression at multiple regulatory levels is a primary means by which BMP2 gene expression is induced.
Acknowledgments
We appreciate the technical assistance of Liping Zhang and Dr. Minzhen He. This work was supported in part by the Molecular Resource Facility at the UMDNJ – NJ Medical School, by National Institute of Child Health and Human Development Grant #HD31117, and by grants from the Foundation of UMDNJ.
Abbreviations
- BMP
Bone Morphogenetic Protein
- RA
retinoic acid
- RACT
RA and dibutyryl cAMP and theophylline
- EMSAs
electrophoretic mobility shift assays
- ChIP
chromatin immunoprecipitation
- RAR
retinoic acid receptor
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- PCR
polymerase chain reaction
- EDTA
ethylenediamine tetraacetic acid
- bp
base pairs
- nt
nucleotides
- UTR
untranslated region
- PBS
phosphate buffered saline
- DTT
dithiothreitol
- PMSF
phenylmethylsulfony fluoride
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrams KL, Xu J, Nativelle-Serpentini C, Dabirshahsahebi S, Rogers MB. An evolutionary and molecular analysis of Bmp2 expression. J Biol Chem. 2004;279:15916–28. doi: 10.1074/jbc.M313531200. [DOI] [PubMed] [Google Scholar]
- Bostrom MP, Lane JM, Berberian WS, Missri AA, Tomin E, Weiland A, Doty SB, Glaser D, Rosen VM. Immunolocalization and expression of bone morphogenetic proteins 2 and 4 in fracture healing. J Orthop Res. 1995;13:357–67. doi: 10.1002/jor.1100130309. [DOI] [PubMed] [Google Scholar]
- Bouletreau PJ, Warren SM, Spector JA, Peled ZM, Gerrets RP, Greenwald JA, Longaker MT. Hypoxia and VEGF up-regulate BMP-2 mRNA and protein expression in microvascular endothelial cells: implications for fracture healing. Plast Reconstr Surg. 2002;109:2384–97. doi: 10.1097/00006534-200206000-00033. [DOI] [PubMed] [Google Scholar]
- Bouwman P, Philipsen S. Regulation of the activity of Sp1-related transcription factors. Mol Cell Endocrinol. 2002;195:27–38. doi: 10.1016/s0303-7207(02)00221-6. [DOI] [PubMed] [Google Scholar]
- Canalis E, Economides AN, Gazzerro E. Bone morphogenetic proteins, their antagonists, and the skeleton. Endocr Rev. 2003;24:218–35. doi: 10.1210/er.2002-0023. [DOI] [PubMed] [Google Scholar]
- Celeste AJ, Iannazi JA, Taylor RC, Hewick RM, Rosen V, Wang EA, Wozney JM. Identification of transforming growth factor β family members present in bone-inductive protein purified from bovine bone. Proc Natl Acad Sci USA. 1990;87:9843–9847. doi: 10.1073/pnas.87.24.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–41. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- Cho TJ, Gerstenfeld LC, Einhorn TA. Differential temporal expression of members of the transforming growth factor beta superfamily during murine fracture healing. J Bone Miner Res. 2002;17:513–20. doi: 10.1359/jbmr.2002.17.3.513. [DOI] [PubMed] [Google Scholar]
- Chu S, Ferro TJ. Sp1: regulation of gene expression by phosphorylation. Gene. 2005;348:1–11. doi: 10.1016/j.gene.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Coucouvanis E, Martin GR. BMP signaling plays a role in visceral endoderm differentiation and cavitation in the early mouse embryo. Development. 1999;126:535–46. doi: 10.1242/dev.126.3.535. [DOI] [PubMed] [Google Scholar]
- Devlin RD, Du Z, Pereira RC, Kimble RB, Economides AN, Jorgetti V, Canalis E. Skeletal overexpression of noggin results in osteopenia and reduced bone formation. Endocrinology. 2003;144:1972–8. doi: 10.1210/en.2002-220918. [DOI] [PubMed] [Google Scholar]
- Fritz DT, Jiang S, Xu J, Rogers MB. A polymorphism in a conserved posttranscriptional regulatory motif alters bone morphogenetic protein 2 (BMP2) RNA:protein interactions. Mol Endocrinol. 2006;20:1574–86. doi: 10.1210/me.2005-0469. [DOI] [PubMed] [Google Scholar]
- Fritz DT, Liu D, Xu J, Jiang S, Rogers MB. Conservation of Bmp2 post-transcriptional regulatory mechanisms. J Biol Chem. 2004;279:48950–8. doi: 10.1074/jbc.M409620200. [DOI] [PubMed] [Google Scholar]
- Gerstenfeld LC, Cullinane DM, Barnes GL, Graves DT, Einhorn TA. Fracture healing as a post-natal developmental process: molecular, spatial, and temporal aspects of its regulation. J Cell Biochem. 2003;88:873–84. doi: 10.1002/jcb.10435. [DOI] [PubMed] [Google Scholar]
- Gidoni D, Dynan WS, Tjian R. Multiple specific contacts between a mammalian transcription factor and its cognate promoters. Nature. 1984;312:409–13. doi: 10.1038/312409a0. [DOI] [PubMed] [Google Scholar]
- Glozak MA, Li Y, Reuille R, Kim KH, Vo MN, Rogers MB. Trapping and characterization of novel retinoid response elements. Mol Endocrinol. 2003;17:27–41. doi: 10.1210/me.2002-0192. [DOI] [PubMed] [Google Scholar]
- Glozak MA, Rogers MB. Specific induction of apoptosis in P19 embryonal carcinoma cells by retinoic acid and BMP2 or BMP4. Devl Biol. 1996;179:458–470. doi: 10.1006/dbio.1996.0275. [DOI] [PubMed] [Google Scholar]
- Gong Y, et al. Heterozygous mutations in the gene encoding noggin affect human joint morphogenesis. Nat Genet. 1999;21:302–4. doi: 10.1038/6821. [DOI] [PubMed] [Google Scholar]
- Graham A, Francis-West P, Brickell P, Lumsden A. The signalling molecule BMP4 mediates apoptosis in the rhombencephalic neural crest. Nature. 1994;372:684–686. doi: 10.1038/372684a0. [DOI] [PubMed] [Google Scholar]
- Hagen G, Muller S, Beato M, Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. Embo J. 1994;13:3843–51. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallahan AR, Pritchard JI, Chandraratna RA, Ellenbogen RG, Geyer JR, Overland RP, Strand AD, Tapscott SJ, Olson JM. BMP-2 mediates retinoid-induced apoptosis in medulloblastoma cells through a paracrine effect. Nat Med. 2003;9:1033–8. doi: 10.1038/nm904. [DOI] [PubMed] [Google Scholar]
- Hao H, Qi H, Ratnam M. Modulation of the folate receptor type beta gene by coordinate actions of retinoic acid receptors at activator Sp1/ets and repressor AP-1 sites. Blood. 2003;101:4551–60. doi: 10.1182/blood-2002-10-3174. [DOI] [PubMed] [Google Scholar]
- Hardwick JC, Van Den Brink GR, Bleuming SA, Ballester I, Van Den Brande JM, Keller JJ, Offerhaus GJ, Van Deventer SJ, Peppelenbosch MP. Bone morphogenetic protein 2 is expressed by, and acts upon, mature epithelial cells in the colon. Gastroenterology. 2004;126:111–21. doi: 10.1053/j.gastro.2003.10.067. [DOI] [PubMed] [Google Scholar]
- Harris SE, Harris MA, Mahy P, Wozney J, Feng JQ, Mundy GR. Expression of bone morphogenetic protein messenger RNAs by normal rat and human prostate and prostate cancer cells. Prostate. 1994;24:204–11. doi: 10.1002/pros.2990240406. [DOI] [PubMed] [Google Scholar]
- Hill CS. Turning off Smads: identification of a Smad phosphatase. Dev Cell. 2006;10:412–3. doi: 10.1016/j.devcel.2006.03.008. [DOI] [PubMed] [Google Scholar]
- Hogan B. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- Horvath LG, Henshall SM, Kench JG, Turner JJ, Golovsky D, Brenner PC, O’Neill GF, Kooner R, Stricker PD, Grygiel JJ, Sutherland RL. Loss of BMP2, Smad8, and Smad4 expression in prostate cancer progression. Prostate. 2004;59:234–42. doi: 10.1002/pros.10361. [DOI] [PubMed] [Google Scholar]
- Husmann M, Dragneva Y, Romahn E, Jehnichen P. Nuclear receptors modulate the interaction of Sp1 and GC-rich DNA via ternary complex formation. Biochem J. 2000;352(Pt 3):763–72. [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Tjian R. Purification and analysis of RNA polymerase II transcription factors by using wheat germ agglutinin affinity chromatography. Proc Natl Acad Sci U S A. 1989;86:1781–5. doi: 10.1073/pnas.86.6.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James RG, Schultheiss TM. Bmp signaling promotes intermediate mesoderm gene expression in a dose-dependent, cell-autonomous and translation-dependent manner. Dev Biol. 2005;288:113–25. doi: 10.1016/j.ydbio.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Kawamura C, Kizaki M, Yamato K, Uchida H, Fukuchi Y, Hattori Y, Koseki T, Nishihara T, Ikeda Y. Bone morphogenetic protein-2 induces apoptosis in human myeloma cells with modulation of STAT3. Blood. 2000;96:2005–11. [PubMed] [Google Scholar]
- Kleeff J, Maruyama H, Ishiwata T, Sawhney H, Friess H, Buchler MW, Korc M. Bone morphogenetic protein 2 exerts diverse effects on cell growth in vitro and is expressed in human pancreatic cancer in vivo. Gastroenterology. 1999;116:1202–16. doi: 10.1016/s0016-5085(99)70024-7. [DOI] [PubMed] [Google Scholar]
- Langenfeld EM, Calvano SE, Abou-Nukta F, Lowry SF, Amenta P, Langenfeld J. The mature bone morphogenetic protein-2 is aberrantly expressed in non-small cell lung carcinomas and stimulates tumor growth of A549 cells. Carcinogenesis. 2003;24:1445–54. doi: 10.1093/carcin/bgg100. [DOI] [PubMed] [Google Scholar]
- Li L, He S, Sun JM, Davie JR. Gene regulation by Sp1 and Sp3. Biochem Cell Biol. 2004;82:460–71. doi: 10.1139/o04-045. [DOI] [PubMed] [Google Scholar]
- Lyons K, Pelton R, Hogan B. Patterns of expression of murine Vgr-1 and BMP-2a RNA suggest that transforming growth factor-β-like genes coordinately regulate aspects of embryonic development. Genes & Devl. 1989;3:1657–1668. doi: 10.1101/gad.3.11.1657. [DOI] [PubMed] [Google Scholar]
- Lyons KM, Pelton RW, Hogan BLM. Organogenesis and pattern formation in the mouse: RNA distribution patterns suggest a role for Bone Morphogenetic Protein-2A (BMP-2A) Development. 1990;109:833–844. doi: 10.1242/dev.109.4.833. [DOI] [PubMed] [Google Scholar]
- Mann V, Hobson EE, Li B, Stewart TL, Grant SF, Robins SP, Aspden RM, Ralston SH. A COL1A1 Sp1 binding site polymorphism predisposes to osteoporotic fracture by affecting bone density and quality. J Clin Invest. 2001;107:899–907. doi: 10.1172/JCI10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann V, Ralston SH. Meta-analysis of COL1A1 Sp1 polymorphism in relation to bone mineral density and osteoporotic fracture. Bone. 2003;32:711–7. doi: 10.1016/s8756-3282(03)00087-5. [DOI] [PubMed] [Google Scholar]
- Marcelino J, Sciortino CM, Romero MF, Ulatowski LM, Ballock RT, Economides AN, Eimon PM, Harland RM, Warman ML. Human disease-causing NOG missense mutations: effects on noggin secretion, dimer formation, and bone morphogenetic protein binding. Proc Natl Acad Sci U S A. 2001;98:11353–8. doi: 10.1073/pnas.201367598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Barbera JP, Toresson H, Da Rocha S, Krauss S. Cloning and expression of three members of the zebrafish Bmp family: Bmp2a, Bmp2b and Bmp4. Gene. 1997;198:53–9. doi: 10.1016/s0378-1119(97)00292-8. [DOI] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Masson N, Hurst HC, Lee KA. Identification of proteins that interact with CREB during differentiation of F9 embryonal carcinoma cells. Nucleic Acids Res. 1993;21:1163–9. doi: 10.1093/nar/21.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina Y, Starbuck MW, Gentile MA, Fukuda T, Kasparcova V, Seedor JG, Hanks MC, Amling M, Pinero GJ, Harada S, Behringer RR. Bone morphogenetic protein type IA receptor signaling regulates postnatal osteoblast function and bone remodeling. J Biol Chem. 2004;279:27560–6. doi: 10.1074/jbc.M404222200. [DOI] [PubMed] [Google Scholar]
- Raftery LA, Sutherland DJ. Gradients and thresholds: BMP response gradients unveiled in Drosophila embryos. Trends Genet. 2003;19:701–8. doi: 10.1016/j.tig.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Ralston SH. Genetic determinants of osteoporosis. Curr Opin Rheumatol. 2005;17:475–9. doi: 10.1097/01.bor.0000166385.62851.92. [DOI] [PubMed] [Google Scholar]
- Reinholz MM, Iturria SJ, Ingle JN, Roche PC. Differential gene expression of TGF-beta family members and osteopontin in breast tumor tissue: analysis by real-time quantitative PCR. Breast Cancer Res Treat. 2002;74:255–69. doi: 10.1023/a:1016339120506. [DOI] [PubMed] [Google Scholar]
- Reneland RH, Mah S, Kammerer S, Hoyal CR, Marnellos G, Wilson SG, Sambrook PN, Spector TD, Nelson MR, Braun A. Association between a variation in the phosphodiesterase 4D gene and bone mineral density. BMC Med Genet. 2005;6:9. doi: 10.1186/1471-2350-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Leon J, Merino R, Macias D, Ganan Y, Santesteban E, Hurle JM. Retinoic acid regulates programmed cell death through BMP signalling. Nat Cell Biol. 1999;1:125–6. doi: 10.1038/10098. [DOI] [PubMed] [Google Scholar]
- Rogers M. Receptor-selective retinoids implicate RAR alpha and gamma in the regulation of bmp-2 and bmp-4 in F9 embryonal carcinoma cells. Cell Growth Differ. 1996;7:115–122. [PubMed] [Google Scholar]
- Rogers MB, Rosen V, Wozney JM, Gudas LJ. Bone Morphogenetic Proteins-2 and 4 are Involved in the Retinoic Acid-induced Differentiation of Embryonal Carcinoma Cells. Molec Biol Cell. 1992;3:189–196. doi: 10.1091/mbc.3.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe S, Abdelrahim M. Sp transcription factor family and its role in cancer. Eur J Cancer. 2005;41:2438–48. doi: 10.1016/j.ejca.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Schlueter J, Manner J, Brand T. BMP is an important regulator of proepicardial identity in the chick embryo. Dev Biol. 2006;295:546–58. doi: 10.1016/j.ydbio.2006.03.036. [DOI] [PubMed] [Google Scholar]
- Shafritz A, Shore E, Gannon F, Zasloff M, Taub R, Muenke M, Kaplan F. Overexpression of an osteogeneic morphogen in fibrodysplasia ossificans progressiva. NE J Med. 1996;335:555–561. doi: 10.1056/NEJM199608223350804. [DOI] [PubMed] [Google Scholar]
- Shimada J, Suzuki Y, Kim SJ, Wang PC, Matsumura M, Kojima S. Transactivation via RAR/RXR-Sp1 interaction: characterization of binding between Sp1 and GC box motif. Mol Endocrinol. 2001;15:1677–92. doi: 10.1210/mend.15.10.0707. [DOI] [PubMed] [Google Scholar]
- Spector JA, Luchs JS, Mehrara BJ, Greenwald JA, Smith LP, Longaker MT. Expression of bone morphogenetic proteins during membranous bone healing. Plast Reconstr Surg. 2001;107:124–34. doi: 10.1097/00006534-200101000-00018. [DOI] [PubMed] [Google Scholar]
- Styrkarsdottir U, et al. Linkage of Osteoporosis to Chromosome 20p12 and Association to BMP2. PLoS Biol. 2003;1:E69. doi: 10.1371/journal.pbio.0000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Shimada J, Shudo K, Matsumura M, Crippa MP, Kojima S. Physical interaction between retinoic acid receptor and Sp1: mechanism for induction of urokinase by retinoic acid. Blood. 1999;93:4264–76. [PubMed] [Google Scholar]
- Tsuji K, Bandyopadhyay A, Harfe BD, Cox K, Kakar S, Gerstenfeld L, Einhorn T, Tabin CJ, Rosen V. BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat Genet. 2006;38:1424–9. doi: 10.1038/ng1916. [DOI] [PubMed] [Google Scholar]
- Ureta-Vidal A, Ettwiller L, Birney E. Comparative genomics: genome-wide analysis in metazoan eukaryotes. Nat Rev Genet. 2003;4:251–62. doi: 10.1038/nrg1043. [DOI] [PubMed] [Google Scholar]
- Urist M. Bone: formation by autoinduction. Science. 1965;150:893–899. doi: 10.1126/science.150.3698.893. [DOI] [PubMed] [Google Scholar]
- Usui M, Yoshida Y, Tsuji K, Oikawa K, Miyazono K, Ishikawa I, Yamamoto T, Nifuji A, Noda M. Tob deficiency superenhances osteoblastic activity after ovariectomy to block estrogen deficiency-induced osteoporosis. Proc Natl Acad Sci U S A. 2004;101:6653–8. doi: 10.1073/pnas.0303093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaahtokari A, Aberg T, Jernvall J, Keranen S, Thesleff I. The enamel knot as a signaling center in the developing mouse tooth. Mech Dev. 1996;54:39–43. doi: 10.1016/0925-4773(95)00459-9. [DOI] [PubMed] [Google Scholar]
- Valdes AM, Hart DJ, Jones KA, Surdulescu G, Swarbrick P, Doyle DV, Schafer AJ, Spector TD. Association study of candidate genes for the prevalence and progression of knee osteoarthritis. Arthritis Rheum. 2004;50:2497–507. doi: 10.1002/art.20443. [DOI] [PubMed] [Google Scholar]
- Valdes AM, Van Oene M, Hart DJ, Surdulescu GL, Loughlin J, Doherty M, Spector TD. Reproducible genetic associations between candidate genes and clinical knee osteoarthritis in men and women. Arthritis Rheum. 2006;54:533–9. doi: 10.1002/art.21621. [DOI] [PubMed] [Google Scholar]
- Vasios GW, Gold JD, Petkovich M, Chambon P, Gudas LJ. A Retinoic Acid-responsive Element is Present in the 5′ Flanking Region of the Laminin B1 Gene. Proc Natl Acad Sci USA. 1989;86:9099–9103. doi: 10.1073/pnas.86.23.9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JP, Briscoe J. Morphogens. Curr Biol. 2001;11:R851–4. doi: 10.1016/s0960-9822(01)00514-0. [DOI] [PubMed] [Google Scholar]
- Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, Hewick RM, Wang EA. Novel regulators of bone formation: Molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- Wu XB, Li Y, Schneider A, Yu W, Rajendren G, Iqbal J, Yamamoto M, Alam M, Brunet LJ, Blair HC, Zaidi M, Abe E. Impaired osteoblastic differentiation, reduced bone formation, and severe osteoporosis in noggin-overexpressing mice. J Clin Invest. 2003;112:924–34. doi: 10.1172/JCI15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong D, Shen H, Zhao L, Xiao1 P, Yang T, Guo Y, Wang W, Guo Y, Liu Y, Recker R, Deng H. A Robust and Comprehensive Analysis of 20 Osteoporosis Candidate Genes by Very High-Density Single-Nucleotide Polymorphism Screen among 405 Caucasian Nuclear Families Identified Significant Association and Gene-Gene Interaction. J Bone Miner Res. 2006 doi: 10.1359/JBMR.060808. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatani A, Shen YT, Yan L, Chen W, Kim SJ, Sano K, Irie K, Vatner SF, Vatner DE. Down regulation of the L-type Ca2+ channel, GRK2, and phosphorylated phospholamban: protective mechanisms for the denervated failing heart. J Mol Cell Cardiol. 2006;40:619–28. doi: 10.1016/j.yjmcc.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Zhang S, Fantozzi I, Tigno DD, Yi ES, Platoshyn O, Thistlethwaite PA, Kriett JM, Yung G, Rubin LJ, Yuan JX. Bone morphogenetic proteins induce apoptosis in human pulmonary vascular smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2003;285:L740–54. doi: 10.1152/ajplung.00284.2002. [DOI] [PubMed] [Google Scholar]
- Zhao M, Harris SE, Horn D, Geng Z, Nishimura R, Mundy GR, Chen D. Bone morphogenetic protein receptor signaling is necessary for normal murine postnatal bone formation. J Cell Biol. 2002;157:1049–60. doi: 10.1083/jcb.200109012. [DOI] [PMC free article] [PubMed] [Google Scholar]


