Abstract
Kaposi's sarcoma-associated herpesvirus is associated with three human tumors: Kaposi's sarcoma, and the B cell lymphomas, plasmablastic lymphoma associated with multicentric Castleman's disease, and primary effusion lymphoma (PEL). Epstein-Barr virus, the closest human relative of Kaposi's sarcoma-associated herpesvirus, mimics host B cell signaling pathways to direct B cell development toward a memory B cell phenotype. Epstein-Barr virus-associated B cell tumors are presumed to arise as a consequence of this virus-mediated B cell activation. The stage of B cell development represented by PEL, how this stage relates to tumor pathology, and how this information may be used to treat the disease are largely unknown. In this study we used gene expression profiling to order a range of B cell tumors by stage of development. PEL gene expression closely resembles that of malignant plasma cells, including the low expression of mature B cell genes. The unfolded protein response is partially activated in PEL, but is fully activated in plasma cell tumors, linking endoplasmic reticulum stress to plasma cell development through XBP-1. PEL cells can be defined by the overexpression of genes involved in inflammation, cell adhesion, and invasion, which may be responsible for their presentation in body cavities. Similar to malignant plasma cells, all PEL samples tested express the vitamin D receptor and are sensitive to the vitamin D analogue drug EB 1089 (Seocalcitol).
Kaposi's sarcoma-associated herpesvirus (KSHV or HHV-8) was discovered in AIDS-associated Kaposi's sarcoma (1) and has since fulfilled criteria for causing all forms of this disease (2). KSHV is also present in all cases of two B cell lymphomas, primary effusion lymphoma (PEL) (3), and plasmablastic lymphoma associated with multicentric Castleman's disease (MCD) (4, 5). PEL usually presents as a lymphomatous effusion in the pleural, peritoneal or pericardial cavity without a contiguous tumor mass (6). It occurs predominantly in HIV-infected individuals and can be the primary presentation of KSHV infection.
Epstein-Barr virus (EBV), the other human γ-herpesvirus, is thought to control the development of its B cell host by mimicking cellular signaling pathways and guiding it toward a memory B cell in which latent virus can persist (7, 8). The EBV-associated lymphomas Burkitt's lymphoma (BL) and classical Hodgkin's lymphoma (HL) can arise when virus-induced B cell activation is not controlled. Although the majority of PEL cases are coinfected with EBV, evidence suggests that KSHV is directly responsible for transformation (6, 9). KSHV encodes a number of known oncogenes and constitutively active homologues of host genes involved in proliferation, signaling, and inhibition of apoptosis (9).
Tumors retain many of the characteristics of the cell type from which they arise. This property is especially true of B cell lymphomas, where distinctive morphology, immunophenotypes, and Ig gene sequences allow them to be classified by their cell of origin (10, 11). The advent of DNA microarray technology has shown that the global gene expression pattern (transcriptome) of tumors also mirrors that of the founding cell type (12). This finding is consistent with the model that malignant B cells are frozen at discrete developmental stages (13).
In the majority of instances, PEL resembles a transformed postgerminal center (GC) B cell (6). PEL cells express CD138 (syndecan-1) (14), and MUM1/IRF4 (multiple myeloma 1/IFN regulatory factor 4) (15), which is associated with late stages of B cell differentiation. The cells posses morphological features that bridge those of large-cell immunoblastic and anaplastic large-cell lymphoma (6). Most cases have mutated Ig genes whose sequences show evidence for selection by antigen (16, 17). However, some cases have unmutated Ig sequences, indicating a pre-GC origin and suggesting that PEL might represent transformation of B cells at different stages of ontogeny (16). Whereas most cases of PEL have functional Ig gene rearrangements and express Ig mRNA (16, 17), most do not express Ig protein (6).
In this study, we sought to define the stage of B cell development from which PEL is derived in order to understand the molecular basis of the PEL cell phenotype and tumor pathology. To achieve this end, we used DNA microarrays to compare the gene expression of PEL to B cell lines encompassing a range of different B cell malignancies.
Materials and Methods
Cells and RNA. All cell lines and primary samples were grown in RPMI medium 1640 (Invitrogen) with 10% FCS (Helena Biosciences, Sunderland, U.K.) and 100 units/ml penicillin/streptomycin (Invitrogen) in 5% CO2 at 37°C, except for DS-1 (supplemented with 10 units/ml IL-6; Sigma) and BEL (RPMI medium 1640 with Glutamax, 20% FCS, 1× nonessential amino acids, 1 mM sodium pyruvate, 50 μM 2-mercaptoethanol; Invitrogen). All cells were grown to a density of 1 × 106 cells per ml, split 1:4, and harvested at 1 × 106 cells per ml, except for BEL, which was split 1:2. Total RNA was purified with TRIzol (Invitrogen), DNase treated (Promega), repurified by phenol extraction and ethanol precipitation, and mRNA was purified using Oligotex (Qiagen, Valencia, CA). RNA quality was assessed by agarose gel electrophoresis and quantified by using UV spectrophotometry.
Labeling and Microarray Hybridization. mRNA (1 μg) was used as a template for production of cDNA incorporating Cy5-dCTP (NEN) by using SuperScript II (Invitrogen) and random primers. Remaining mRNA was denatured and unincorporated nucleotides were removed by using Microcon YM-30 columns (Millipore). Cy5-labeled cDNA was mixed 1:1 with Cy3-labeled cDNA synthesized from a common reference. This mixture of mRNA was from eight different B cell lines in the following percentages: BC-3 + TPA (12-O-tetradecoylphorbol 13-acetate, 20 ng/ml; Sigma) for 72 h, 25%; JSC-1 + TPA for 48 h, 25%; Raji + TPA for 72 h, 20%; DoHH-2, 7.5%, Karpas-422, 7.5%, Nalm-6, 7.5%, and RPMI-8226, 7.5% (TPA was used to induce the expression of virus transcripts). The Cy5/Cy3 mix was hybridized to custom 5,808-element host-pathogen (comprising ∼5,700 human genes) cDNA microarrays under glass coverslips in custom-made chambers at 65°C for 16 h. The slides were washed in 2× saline phosphate/EDTA (0.15 M NaCl/10 mM phosphate, pH 7.4/1 mM EDTA) (SSPE) at 50°C, then 1× SSPE and 0.1× SPPE, both at room temperature. The arrays were scanned with a GenePix 4000B scanner (Axon Instruments, Union City, CA).
Cluster and Statistical Analysis of Array Expression Data. Data were extracted from microarray image files by using GENEPIX PRO 3.0 software (Axon Instruments). The log2 median of ratios were filtered to remove all flagged data and all data for which the signal was <1.5× background in the Cy3 channel or <2× background in the Cy5 channel. The data for each array were assembled and filtered for genes present in 80% of the arrays and were then median centered for both genes and arrays in Cluster (18). Data were grouped by average linkage hierarchical clustering by using the uncentered Pearson correlation coefficient as the similarity metric. The ordering of the nodes produced by clustering data from 26 arrays was first determined in Cluster by using a one-dimensional self-organizing map with the number of nodes set to √n. The clustered data were visualized by using TREEVIEW (18). Genes whose expression was significantly associated with PEL were found from a 36-array dataset that was processed as described. P values were generated by using the Mann-Whitney U test. Genes were sorted in ascending order by their P value and visualized in TREEVIEW. All datasets used in the analysis are available at www.biochem.ucl.ac.uk/bsm/virus_database/PEL.html.
Western Blotting. Cells (9 × 106) were lysed in 1 ml RIPA buffer with Complete protease inhibitor mixture (Roche Diagnostics, Lewes, U.K.) and benzonase (0.06 units/ml; Merck). Cell lysates (10 μl) were run on a 10% denaturing SDS/polyacrylamide gel. Equal loading was verified by Coomassie blue staining. Protein was transferred to Hybond-P membrane (Amersham Pharmacia Biotech, Amersham, U.K.) and blocked with milk (5%) and Tween 20 (0.1%; Sigma). Blots were incubated with mouse anti-ATF6 (Imgenex, San Diego) or anti-1,25-dihydroxyvitamin D3 receptor (VDR) (Santa Cruz Biotechnology) at 1:250 and 1:500 respectively, washed four times, incubated with rabbit anti-mouse (DAKO) at 1:2,000, washed four times, and stained with ECL plus (Amersham Pharmacia Biotech). Chemiluminescence was visualized by using Hyperfilm (Amersham Pharmacia Biotech) and a Compact X4 film processor (Xograph, Tetbury, U.K.).
Cell Proliferation Assay. Cells were seeded in 96-well plates at 1.5 × 105 cells per ml (3 × 104 cells per well) in normal medium. EB 1089 (Seocalcitol) (4 mM in isopropyl alcohol; Leo Pharmaceutical Products, Ballerup, Denmark) was added at halflog10 serial dilutions ranging from 0.1 to 100 nM in RPMI medium 1640 with 10% FCS. After 72 h at 37°C with 5% CO2, [methyl-3H]thymidine (Amersham Pharmacia Biotech) was added at 10 μCi/ml (1 Ci = 37 GBq) and left for an additional 5 h. Cells were harvested (Tomtec, Hamden, CT) onto Filtermat A filters (Wallac, Gaithersburg), washed, and 3H incorporation was measured by using MeltiLex A and a 1450 MicroBeta scintillation counter (Wallac).
Results
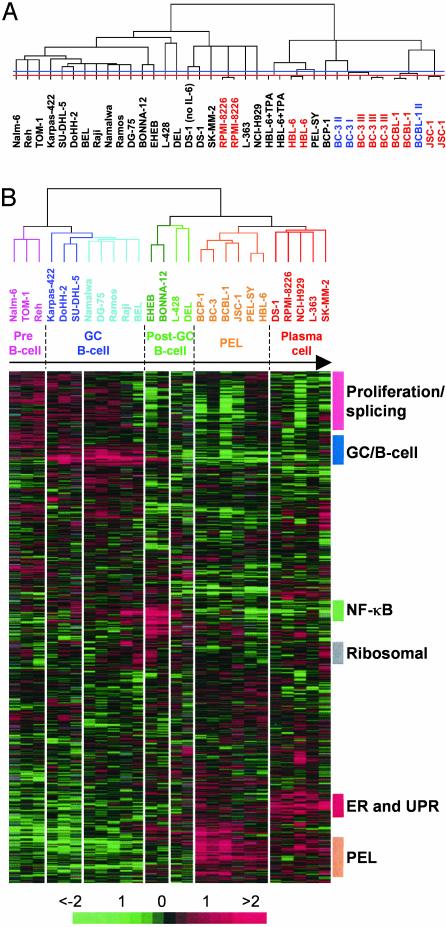
PEL Has a Plasma Cell Expression Profile. We assembled a collection of 24 different cell lines and two primary samples derived from different tumor types and stages of B cell development. Cy5-labeled cDNA from each sample was hybridized to a custom cDNA microarray containing probes for ∼5,700 human genes. Hierarchical clustering of 38 array experiments groups samples representing both technical replicates (same mRNA) and biological replicates (same cells regrown) together (Fig. 1A). The mean correlation coefficient for technical replicates is 0.96 (n = 7), and for biological replicates it is 0.90 (n = 9). In all cases, the branch lengths (representing correlation) are shorter between replicates than between different samples.
Fig. 1.
Assembly of B cell tumors according to developmental stage by gene expression profiling. (A) Hierarchical clustering of filtered data (1,842 genes) from 38 arrays, hybridized with the samples named. Technical replicates, which are red, and biological replicates, which are blue, always cluster together. The dendogram relates samples by their gene expression pattern, with short branch lengths indicating similarity. The red and blue horizontal lines show the limit of technical and biological variation, respectively. (B) Hierarchical clustering of 26 samples and a filtered set of 1,987 genes. Each column represents one sample and each row one gene. Gene expression is shown as a pseudocolored representation of log2 expression ratio with red being above and green being below the row/column median level of expression as shown by the scale. Gray indicates data removed by filtering. Samples cluster by the tumor type from which they were derived: pink, acute lymphoblastic leukemia (ALL); dark blue, GC-like DLBCL and FL; light blue, BL; dark green, CLL and HCL; light green, HL and anaplastic DLBCL; orange, PEL; red, MM, PCL, and DS-1, which was derived from immunoblastic lymphoma. The ordering of the samples, which was determined by a one-dimensional self-organizing map, mirrors the order of their known normal B cell counterparts in B cell development (indicated by the arrow and names above). The colored bars to the right mark gene expression signatures, which vary in expression across the dataset. The bar labeled PEL indicates genes expressed strongly in PEL.
We reclustered the data without the replicates by using a set of 1,987 genes that passed our filtering criteria (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). Both the genes and samples were first ordered by using a self-organizing map algorithm and then grouped by hierarchical clustering. This ordering creates a spectrum of B cell neoplasia (Fig. 1B). Cell lines derived from each tumor type cluster together, indicating that they share a common gene expression pattern. The order of the samples parallels the respective stage of B cell development from which each tumor type is thought to originate. Cell lines established from precursor B (acute) lymphoblastic leukemia (ALL) all cluster together at the beginning, which is consistent with the transformation of one of the earliest stages of B cell development. This cluster is followed by the following tumors representing GC B cells: Follicular lymphoma (FL), GC-like diffuse large B cell lymphoma (DLBCL), and BL. Of these BL cell lines, Raji and Namalwa contain EBV. The major branch point separates pre-GC and GC B cells from post-GC B cells, with a few exceptions described later. The cell lines BONNA-12, derived from hairy cell leukemia (HCL), EHEB [chronic lymphocytic leukemia (CLL)], L-428 (HL) and DEL (anaplastic DLBCL) form the next branch. The position of these cell lines in this ordering of B cell development is consistent with their presumed derivation from post-GC B cells (10, 19). The two final major branches are composed of cell lines derived from PEL, and the plasma cell neoplasms multiple myeloma (MM) and plasma cell leukemia (PCL), respectively. Therefore, PEL is closely related to plasma cell tumors and may have a similar derivation. The cell line DS-1 (20), derived from a lymphoblastic leukemia resembling PEL, but which is KSHV-negative (see Fig. 6, which is published as supporting information on the PNAS web site), clusters with plasma cells.
To test whether primary PEL tumors also have a plasma cell expression pattern, cells taken from lymphomatous effusions of two AIDS patients were analyzed (PEL-SY (21) and BEL; Fig. 1B). PEL-SY clusters with PEL cell lines, whereas BEL clusters with cell lines derived from BL. BL-like effusions resembling PEL have previously been reported in AIDS patients (6). Fluorescence-activated cell sorter (FACS) analysis confirmed these classifications; PEL-SY is CD19- CD27-CD138+ as expected for PEL, whereas BEL is CD19+ CD27+ CD138- as expected for BL (data not shown). The microarrays also allowed simultaneous detection of virus transcripts; EBV was detected in both samples, whereas KSHV was only detected in PEL-SY (data not shown), as confirmed by virus-specific PCR (Fig. 6) and immunofluorescence assay (data not shown). Therefore, PEL cell lines maintain the global expression pattern of the primary tumor.
All PEL samples cluster together, regardless of EBV status (Fig. 6) or the mutation state of the Ig genes [HBL-6 and BCBL-1 are mutated (17); BC-3 is germ line (16)]. Therefore, PEL constitutes one disease, and cells do not require passage through the GC for its development.
Low Expression of B Cell Signatures in PEL and Plasma Cell Tumors. A number of B cell gene expression signatures were identified (Fig. 1B). The expression pattern of a number of genes contained within these expression signatures were confirmed by RT-PCR (see Fig. 7A, which is published as supporting information on the PNAS web site). Genes involved in cell proliferation show stronger expression in pre-B cell and GC B cell-derived tumors, as expected because of their dividing, undifferentiated nature (Proliferation signature, Fig. 1B, and Fig. 8, which is published as supporting information on the PNAS web site). This cluster also contains many genes involved in RNA maturation and splicing, which may represent the splicing of rearranged Ig V(D)J segments to the C region. The cluster of genes strongly expressed by GC B cell-derived cell lines (GC/B cell signature, Fig. 1B, and Fig. 9, which is published as supporting information on the PNAS web site) consists of known GC B cell markers and genes that act in B cell signaling pathways. This signature also includes genes involved in Ig class switching, DNA strand-break repair, and somatic hypermutation. Genes forming the NF-κB signature act both upstream and downstream of NF-κB during B cell activation (NF-κB; Fig. 1B, and Fig. 10, which is published as supporting information on the PNAS web site). The overexpression of this cluster in EHEB, BONNA-12, and L-428 is consistent with the expression of EBV LMP1 in EHEB and BONNA-12 (data not shown) and the constitutive activation of NF-κB in HL (22).
None of these clusters is highly expressed in PEL, MM, or PCL, due to the action of the plasma cell transcription factor Blimp-1 (Fig. 7B and ref. 23). PEL and plasma cell tumors express increased levels of a number of genes implicated in oncogenesis, such as jun B, cyclin D2, FLICE inhibitory protein (FLIP), testis-enhanced gene transcript (TEGT), IRF4, tumor susceptibility gene (TSG) 101, PBX2, myeloid cell leukemia sequence (Mcl)-1, H-ras and raf1, and other downstream components of Ras signaling pathways, such as ERK3 and ELK1. Unlike MM and PCL, PEL does not exhibit disregulated c-myc expression (see Fig. 11, which is published as supporting information on the PNAS web site).
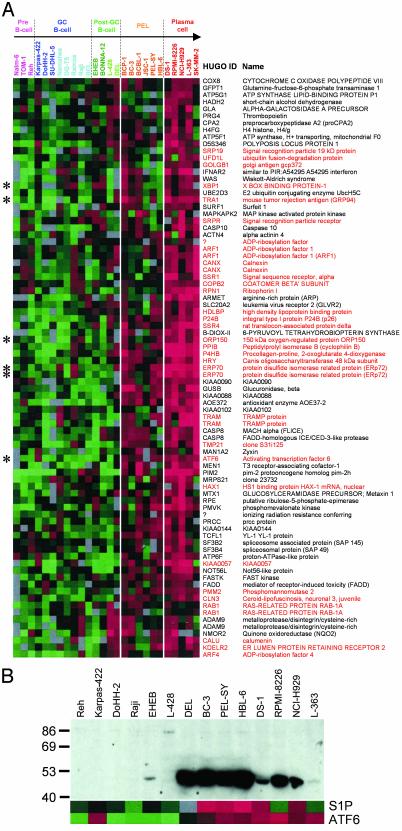
The Unfolded Protein Response (UPR) and Other Endoplasmic Reticulum (ER) Stress Pathways Are Activated in Plasma Cell Tumors. The ER and UPR cluster (Fig. 1B) is weakly up-regulated in PEL cells and is most strongly expressed in plasma cell tumors. This cluster consists mainly of genes whose protein products are localized to the ER or Golgi, or which are responsible for trafficking between the two compartments (red, Fig. 2A). Known components of the UPR are to be found within this cluster, such as GRP94, GRP170 (ORP150), and ERp72. These genes are activated by ATF6 during ER stress (24, 25), which is also present in this cluster. ATF6 induces expression of another gene in this cluster, XBP-1 (26), which is necessary for Ig secretion and plasma cell development (27). Thus, the UPR is an expression signature that defines plasma cells and links ER signaling to plasma cell development.
Fig. 2.
Activation of the unfolded protein response in PEL and plasma cell tumors. (A) Detail of the ER and UPR cluster (Fig. 1B), which has the highest expression in plasma cell derived tumors (red), and is also increased in PEL cells (orange). The Human Genome Organisation (HUGO) gene identifiers and gene names are indicated to the right of each row. Genes whose products have a role in ER or Golgi function or vesicle trafficking are named in red. Genes marked by asterisks (*) are discussed in the text. (B) Expression of ATF6 protein measured by Western blotting. The positions of molecular weight markers are shown on the left. The active 50-kDa form of ATF6, which induces the UPR, is expressed strongly in the anaplastic DLBCL-derived cell line DEL, PEL cells, and MM-derived cell lines. Except for a small number of cases, protein levels correlate with ATF6 and S1P mRNA expression measured by using microarrays (shown below).
On induction of ER stress, ATF6 (90 kDa) is cleaved by S1P and S2P, giving a 50-kDa form (28), which translocates to the nucleus to activate the UPR (25). Western blotting confirms the presence of the active 50-kDa form of ATF6 in PEL and MM cells (Fig. 2B). The amount of active ATF6 correlates with S1P and ATF6 mRNA levels as determined by cDNA arrays (Fig. 2B). These data indicate that the UPR, which leads to upregulation of the plasma cell transcription factor XBP-1, is activated in PEL.
The PEL expression signature contains genes that are overexpressed in PEL cells, or in both PEL and plasma cell tumors (see Fig. 12, which is published as supporting information on the PNAS web site). Many of the genes form part of pathways other than the UPR that are also triggered by ER stress. Sterol regulatory genes such as S1P, SCAP, NPC1, and downstream genes (phosphatidylserine synthase, LDL receptor, lipoprotein lipase, and caveolin) are all found in the PEL cluster. Genes involved in ER-associated degradation (ERAD) of misfolded proteins, such as components of the ER translocon, ubiquitination machinery, and the 26S proteasome, also show increased expression in PEL, MM, and PCL cells. The PEL cluster contains genes involved in inhibition of translation, such as eIF-2B and GCN1, and calpain and caspase 3, which are involved in the induction of apoptosis in cells with chronic ER stress.
A number of genes associated with Alzheimer's disease, including presenilin 1, nicastrin, and Fe65, show an increase in their expression in PEL and plasma cell tumors (see Fig. 13, which is published as supporting information on the PNAS web site). Their coexpression therefore suggests that they may function in a single gene network, related to ER function, which shows greater activity in plasma cell tumors.
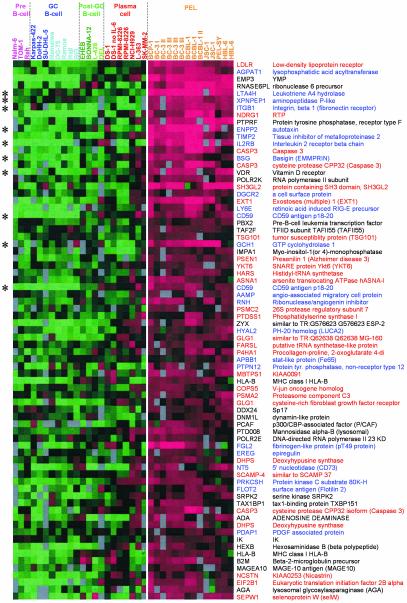
PEL Gene Expression Pattern Helps Explain Tumor Pathology. PEL presents in the serous body cavities and thus has quite a different pathology to tumors derived from plasma cells. To explain its unusual pathology we used a Mann-Whitney U test to find which genes are most significantly overexpressed in PEL compared with other B cell neoplasias (Fig. 3). Most of these genes are also present in the PEL gene expression signature (Fig. 12). Many of these genes are involved in inflammation, adhesion, and invasion (blue in Fig. 3). Inflammatory genes include leukotriene A4 hydrolase, IL-2 receptor β-subunit, GTP cyclohydrolase, aminopeptidase P, and CD59. A number of genes in the PEL cluster are specifically involved in leukocyte adhesion and extravasation during inflammation such as integrin α6 (CD49f) and integrin β1 (CD29), which together form the laminin-binding protein VLA-6. FACS analysis confirmed increased expression of integrin α6 on most PEL cells compared with cell lines derived from other B cell tumors (see Fig. 14, which is published as supporting information on the PNAS web site). All samples tested express surface integrin β1 (data not shown). Integrin α6 expression determined by microarrays predicts the intensity of cell-surface staining measured by FACS analysis (r = 0.73, P < 0.001). Genes implicated in matrix remodeling, invasion, and metastasis are also associated with PEL. These genes include pM5 protein, EMMPRIN (CD147), autotaxin, and metastasis-associated (mta)1.
Fig. 3.
Identification of genes that define PEL. The 75 genes most associated with PEL by using a Mann-Whitney U test (P values from 2.91 × 10-7 to 1.36 × 10-4). Each column represents one sample, and each row represents one gene (identified by HUGO ID and gene name). The majority of genes can be classified by their involvement in either inflammation, adhesion, and invasion (blue) or ER, Golgi, and vesicle trafficking (red).
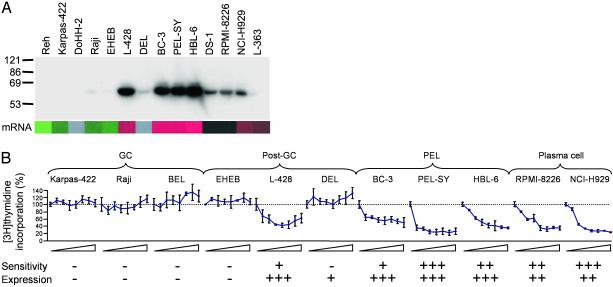
PEL Is Sensitive to the Vitamin D Analogue Drug EB 1089 (Seocalcitol). We identified a number of known drug targets and resistance markers in the dataset whose expression is a function of developmental stage. PEL, plasma cell tumors, and the HL cell line L-428 exhibit elevated expression of VDR mRNA (Fig. 12). The levels of VDR protein expression follow those of its transcript measured using microarrays (Fig. 4A). It is known that MM is sensitive to growth inhibition by the vitamin D analogue EB 1089 (29). From our microarray data, we therefore postulated that PEL cells would also be sensitive to vitamin D analogue drugs. To test this hypothesis, we measured the effect of EB 1089 on cell proliferation of a number of different B cell lines and primary tumor samples in vitro (Fig. 4B). EB 1089 inhibited the growth of all MM and PEL samples tested, albeit to varying extents. Of samples derived from post-GC B cells, only L-428 expresses high levels of VDR and shows sensitivity to EB 1089. Tumor cells representing earlier stages of B cell development, such as GC-like DLBCL and BL, do not express VDR and are refractory to EB 1089.
Fig. 4.
PEL is sensitive to the vitamin D analogue drug EB 1089 (Seocalcitol). (A) Expression of the VDR measured by Western blotting. The receptor is strongly expressed in PEL cells. Protein levels correlate with VDR mRNA expression measured by using microarrays (shown below). (B) In vitro [3H]thymidine incorporation of cell lines and primary tumor cells as percentage of no drug control after addition of EB 1089 at concentrations increasing at half-log10 intervals from 0.1 to 100 nM (means and SD shown, n = 3). Sensitivity to EB 1089 and expression of VDR are scored underneath.
Discussion
Analysis of PEL Gene Expression in Relation to Other B Cell Tumor Types Provides Insights Into Its Pathology. Our data show that PEL has a gene expression profile similar to transformed plasma cells. This finding is consistent with expression of CD138 and IRF4 in the majority of cases (14, 15). The low expression of mature B cell genes demonstrates that PEL has completed the critical part of plasma cell development controlled by Blimp-1. However, the tumor cells appear to be arrested during up-regulation of XBP-1 and other genes of the UPR. It has been suggested that PEL constitutes two diseases (16), because some cases have mutated Ig genes, whereas others are unmutated. Here we show that all cases examined share an expression profile, regardless of whether the cells have passed through the GC. Notably, CLL cases have also been shown to share a common general expression pattern that is independent of Ig hypermutation (19, 30). Taken together, these studies suggest that B cells activated in extrafollicular sites pass along the same pathway of B cell differentiation as B cells leaving the GC, and thus give rise to the same tumor types.
EBV-infected tumors are presumed to arise when virus-induced B cell development toward memory B cells is not controlled (7, 8). KSHV-associated tumors may therefore arise in an analogous manner. The expression pattern of PEL cells suggests that KSHV may act to direct B cells toward a plasma cell fate. The KSHV-infected B cells seen in MCD resemble extrafollicular plasmablasts (all express unmutated IgM; refs. 5 and 31). Both KSHV-associated B cell tumors may therefore be blocked at stages along the plasma cell developmental pathway. This finding may point to long-lived plasma cells being the reservoir of latent KSHV infection in vivo. Our data show that a number of cellular homologues of KSHV genes are increased in post-GC cells (cyclin D2, FLIP, IRF4, Mcl-1, and CD59). KSHV also encodes a functional IL-6 homologue (vIL-6) (9). Therefore, we hypothesize that KSHV pirated some host genes to help the virus direct B cell development toward plasma cells. It will be interesting to see whether vIL-6 is able to induce the expression of XBP-1, as is the case for its human counterpart (32). The block in the UPR (this article) and Ig production (6) seen in PEL cells could be because of KSHV infection. Interestingly, the latent KSHV protein K15 interacts with the ER-localized HAX-1 (33), which is present in the ER and UPR cluster.
Genes that are most able to define PEL by their overexpression are involved in inflammation, adhesion, and invasion. FACS analysis confirms the high expression of VLA-6 on PEL cells. Integrin α6 contributes to the invasive properties of certain tumor types (34). Interaction of VLA-6 with laminin may contribute to the localization of eosinophils (35) and TH1 cells (36) to inflammatory sites. Galectin-3, which is overexpressed by PEL and other tumors of late-stage B cells, is involved in neutrophil extravasation (37), and may be a positive regulator of inflammatory responses in the peritoneal cavity (38). The expression of these adhesion molecules, together with the absence of others responsible for homotypic adhesion and homing to lymphoid tissue (39), may be responsible for the presentation of PEL as a body cavity-based effusion. If PEL cells posses a normal cellular counterpart, it may be a type of plasma cell or plasmablast that is targeted to sites of inflammation, and could be defined by CD138 and VLA-6 expression.
Activation of ER Stress Pathways in PEL and Plasma Cell Tumors. By comparing tumors from all stages of B cell development, we have revealed the changes in expression pattern associated with a plasma cell phenotype. Plasma cell tumors overexpress genes of the UPR. These tumors may therefore connect ER stress to plasma cell differentiation and Ig secretion through XBP-1. The clustering of numerous ER, Golgi, and trafficking genes with those known to be induced by the UPR indicates that the UPR affects the entire secretory pathway in plasma cells. A number of genes involved in ATP synthesis also show increased expression in plasma cells (Fig. 2A), which is consistent with the increased amounts of energy required for secretory protein biogenesis. The UPR may be triggered in plasma cells because of the production of secreted Ig. This finding suggests that the relatively limited activation of the UPR in PEL is related to the lack of Ig production and the low expression of Ig λ-light chain and J chain (Fig. 11). However, these data infer that the production of the active form of ATF6 does not depend on Ig synthesis.
The expression pattern of genes involved in Alzheimer's disease (Fig. 13) suggests they may function in a single gene network in plasma cells. Alzheimer's-associated genes are involved in regulation of ER calcium levels through AICD and Fe65 (40, 41). A number of other genes involved in ER calcium regulation, such as calnexin, calumenin, calmodulin, calreticulin, calpain, and annexins I, V, and VII, also show increased expression in PEL and plasma cell tumors. A function for Alzheimer's genes in B cell calcium regulation is supported by the description of perturbed calcium homeostasis in peripheral lymphocytes from Alzheimer's patients (42).
The Plasma Cell Phenotype of PEL May Lead to Novel Therapies. The presence of VDR generally correlates with a sensitivity to the vitamin D analogue drug EB 1089 (Fig. 4). The expression of VDR in PEL and its sensitivity to growth inhibition by EB 1089 are consistent with a plasma cell derivation. Therefore, PEL may respond to other agents that are active against plasma cell tumors. However, the high expression of lung resistance protein in PEL (Fig. 12) could render the tumor resistant to melphalan (43). Interestingly, Kaposi's sarcoma cells also express high levels of VDR and 1α,25-dihydroxyvitamin D3 inhibits tumor growth in vitro and in vivo (44). Vitamin D analogue drugs may therefore prove to be potential therapeutics for all KSHV-associated diseases.
Supplementary Material
Acknowledgments
We thank Leo Pharmaceutical Products for supplying EB 1089; Lyn Healy and Mel Greaves for the cell lines Nalm-6, TOM-1, Reh, and SU-DHL-5; Ursula Ayliffe and Rob Miller for providing PEL-SY and BEL patient samples; and Roger Buxton at the National Institute for Medical Research for the use of the Axon array scanner. This work was supported by the Medical Research Council (R.G.J., P.K., and R.A.W.) and Cancer Research UK (K.M., N.C., C.B., and R.W.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PEL, primary effusion lymphoma; KSHV, Kaposi's sarcoma-associated herpesvirus; EBV, Epstein-Barr virus; ER, endoplasmic reticulum; BL, Burkitt's lymphoma; HL, Hodgkin's lymphoma; GC, germinal center; DLBCL, GC-like diffuse large B cell lymphoma; MM, multiple myeloma; PCL, plasma cell leukemia; UPR, unfolded protein response; VDR, 1,25-dihydroxyvitamin D3 receptor.
References
- 1.Chang, Y., Cesarman, E., Pessin, M. S., Lee, F., Culpepper, J., Knowles, D. M. & Moore, P. S. (1994) Science 266, 1865-1869. [DOI] [PubMed] [Google Scholar]
- 2.Boshoff, C. & Weiss, R. A. (2001) Philos. Trans. R. Soc. London B 356, 517-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cesarman, E., Chang, Y., Moore, P. S., Said, J. W. & Knowles, D. M. (1995) N. Engl. J. Med. 332, 1186-1191. [DOI] [PubMed] [Google Scholar]
- 4.Soulier, J., Grollet, L., Oksenhendler, E., Cacoub, P., Cazals-Hatem, D., Babinet, P., d'Agay, M. F., Clauvel, J. P., Raphael, M., Degos, L., et al. (1995) Blood 86, 1276-1280. [PubMed] [Google Scholar]
- 5.Dupin, N., Diss, T. L., Kellam, P., Tulliez, M., Du, M. Q., Sicard, D., Weiss, R. A., Isaacson, P. G. & Boshoff, C. (2000) Blood 95, 1406-1412. [PubMed] [Google Scholar]
- 6.Cesarman, E. & Knowles, D. M. (1999) Semin. Cancer Biol. 9, 165-174. [DOI] [PubMed] [Google Scholar]
- 7.Thorley-Lawson, D. A. & Babcock, G. J. (1999) Life Sci. 65, 1433-1453. [DOI] [PubMed] [Google Scholar]
- 8.Kieff, E. & Rickinson, A. B. (2001) in Fields' Virology, eds. Knipe, D. K. & Howley, P. M. (Lippincott, Williams & Wilkins, Philadelphia), Vol. 2, pp. 2511-2574. [Google Scholar]
- 9.Jenner, R. G. & Boshoff, C. (2002) Biochim. Biophys. Acta 1602, 1-22. [DOI] [PubMed] [Google Scholar]
- 10.Kuppers, R., Klein, U., Hansmann, M. L. & Rajewsky, K. (1999) N. Engl. J. Med. 341, 1520-1529. [DOI] [PubMed] [Google Scholar]
- 11.Jaffe, E. S., Harris, N. L., Stein, H. & Vardiman, J. W. (2001) in World Health Organization Classification of Tumours, eds. Kleihues, P. & Sobin, L. H. (IARC Press, Lyon, France).
- 12.Shaffer, A. L., Rosenwald, A., Hurt, E. M., Giltnane, J. M., Lam, L. T., Pickeral, O. K. & Staudt, L. M. (2001) Immunity 15, 375-385. [DOI] [PubMed] [Google Scholar]
- 13.Greaves, M. F. (1986) Science 234, 697-704. [DOI] [PubMed] [Google Scholar]
- 14.Gaidano, G., Gloghini, A., Gattei, V., Rossi, M. F., Cilia, A. M., Godeas, C., Degan, M., Perin, T., Canzonieri, V., Aldinucci, D., et al. (1997) Blood 90, 4894-4900. [PubMed] [Google Scholar]
- 15.Carbone, A., Gloghini, A., Cozzi, M. R., Capello, D., Steffan, A., Monini, P., De Marco, L. & Gaidano, G. (2000) Br. J. Haematol. 111, 247-257. [DOI] [PubMed] [Google Scholar]
- 16.Matolcsy, A., Nador, R. G., Cesarman, E. & Knowles, D. M. (1998) Am. J. Pathol. 153, 1609-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fais, F., Gaidano, G., Capello, D., Gloghini, A., Ghiotto, F., Roncella, S., Carbone, A., Chiorazzi, N. & Ferrarini, M. (1999) Leukemia 13, 1093-1099. [DOI] [PubMed] [Google Scholar]
- 18.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein, U., Tu, Y., Stolovitzky, G. A., Mattioli, M., Cattoretti, G., Husson, H., Freedman, A., Inghirami, G., Cro, L., Baldini, L., et al. (2001) J. Exp. Med. 194, 1625-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bock, G. H., Long, C. A., Riley, M. L., White, J. D., Kurman, C. C., Fleisher, T. A., Tsokos, M., Brown, M., Serbousek, D., Schwietermann, W. D., et al. (1993) Cytokine 5, 480-489. [DOI] [PubMed] [Google Scholar]
- 21.Lankester, K. J., Lishman, S., Ayliffe, U., Kocjan, G., Spittle, M. F. & Miller, R. F. (1998) Int. J. STD AIDS 9, 616-618. [DOI] [PubMed] [Google Scholar]
- 22.Bargou, R. C., Emmerich, F., Krappmann, D., Bommert, K., Mapara, M. Y., Arnold, W., Royer, H. D., Grinstein, E., Greiner, A., Scheidereit, C. & Dorken, B. (1997) J. Clin. Invest. 100, 2961-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaffer, A. L., Lin, K. I., Kuo, T. C., Yu, X., Hurt, E. M., Rosenwald, A., Giltnane, J. M., Yang, L., Zhao, H., Calame, K. & Staudt, L. M. (2002) Immunity 17, 51-62. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida, H., Haze, K., Yanagi, H., Yura, T. & Mori, K. (1998) J. Biol. Chem. 273, 33741-33749. [DOI] [PubMed] [Google Scholar]
- 25.Haze, K., Yoshida, H., Yanagi, H., Yura, T. & Mori, K. (1999) Mol. Biol. Cell 10, 3787-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida, H., Okada, T., Haze, K., Yanagi, H., Yura, T., Negishi, M. & Mori, K. (2000) Mol. Cell. Biol. 20, 6755-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reimold, A. M., Iwakoshi, N. N., Manis, J., Vallabhajosyula, P., Szomolanyi-Tsuda, E., Gravallese, E. M., Friend, D., Grusby, M. J., Alt, F. & Glimcher, L. H. (2001) Nature 412, 300-307. [DOI] [PubMed] [Google Scholar]
- 28.Ye, J., Rawson, R. B., Komuro, R., Chen, X., Dave, U. P., Prywes, R., Brown, M. S. & Goldstein, J. L. (2000) Mol. Cell 6, 1355-1364. [DOI] [PubMed] [Google Scholar]
- 29.Puthier, D., Bataille, R., Barille, S., Mellerin, M. P., Harousseau, J. L., Ponzio, A., Robillard, N., Wijdenes, J. & Amiot, M. (1996) Blood 88, 4659-4666. [PubMed] [Google Scholar]
- 30.Rosenwald, A., Alizadeh, A. A., Widhopf, G., Simon, R., Davis, R. E., Yu, X., Yang, L., Pickeral, O. K., Rassenti, L. Z., Powell, J., et al. (2001) J. Exp. Med. 194, 1639-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du, M. Q., Liu, H., Diss, T. C., Ye, H., Hamoudi, R. A., Dupin, N., Meignin, V., Oksenhendler, E., Boshoff, C. & Isaacson, P. G. (2001) Blood 97, 2130-2136. [DOI] [PubMed] [Google Scholar]
- 32.Wen, X. Y., Stewart, A. K., Sooknanan, R. R., Henderson, G., Hawley, T. S., Reimold, A. M., Glimcher, L. H., Baumann, H., Malek, L. T. & Hawley, R. G. (1999) Int. J. Oncol. 15, 173-178. [DOI] [PubMed] [Google Scholar]
- 33.Sharp, T. V., Wang, H. W., Koumi, A., Hollyman, D., Endo, Y., Ye, H., Du, M. Q. & Boshoff, C. (2002) J. Virol. 76, 802-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cress, A. E., Rabinovitz, I., Zhu, W. & Nagle, R. B. (1995) Cancer Metastasis Rev. 14, 219-228. [DOI] [PubMed] [Google Scholar]
- 35.Georas, S. N., McIntyre, B. W., Ebisawa, M., Bednarczyk, J. L., Sterbinsky, S. A., Schleimer, R. P. & Bochner, B. S. (1993) Blood 82, 2872-2879. [PubMed] [Google Scholar]
- 36.Colantonio, L., Iellem, A., Clissi, B., Pardi, R., Rogge, L., Sinigaglia, F. & D'Ambrosio, D. (1999) Blood 94, 2981-2989. [PubMed] [Google Scholar]
- 37.Sato, S., Ouellet, N., Pelletier, I., Simard, M., Rancourt, A. & Bergeron, M. G. (2002) J. Immunol. 168, 1813-1822. [DOI] [PubMed] [Google Scholar]
- 38.Hsu, D. K., Yang, R. Y., Pan, Z., Yu, L., Salomon, D. R., Fung-Leung, W. P. & Liu, F. T. (2000) Am. J. Pathol. 156, 1073-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boshoff, C., Gao, S. J., Healy, L. E., Matthews, S., Thomas, A. J., Coignet, L., Warnke, R. A., Strauchen, J. A., Matutes, E., Kamel, O. W., et al. (1998) Blood 91, 1671-1679. [PubMed] [Google Scholar]
- 40.Cao, X. & Sudhof, T. C. (2001) Science 293, 115-120. [DOI] [PubMed] [Google Scholar]
- 41.Leissring, M. A., Murphy, M. P., Mead, T. R., Akbari, Y., Sugarman, M. C., Jannatipour, M., Anliker, B., Muller, U., Saftig, P., De Strooper, B., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 4697-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eckert, A., Forstl, H., Zerfass, R., Hennerici, M. & Muller, W. E. (1997) Neurobiol. Aging 18, 281-284. [DOI] [PubMed] [Google Scholar]
- 43.Filipits, M., Drach, J., Pohl, G., Schuster, J., Stranzl, T., Ackermann, J., Konigsberg, R., Kaufmann, H., Gisslinger, H., Huber, H., et al. (1999) Clin. Cancer Res. 5, 2426-2430. [PubMed] [Google Scholar]
- 44.Masood, R., Nagpal, S., Zheng, T., Cai, J., Tulpule, A., Smith, D. L. & Gill, P. S. (2000) Blood 96, 3188-3194. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.