Abstract
The mammalian gut represents a complex and diverse ecosystem, consisting of unique interactions between the host and microbial residents. Bacterial surfaces serve as an interface that promotes and responds to this dynamic exchange, a process essential to the biology of both symbionts. The human intestinal microorganism, Bacteroides fragilis, is able to extensively modulate its surface. Analysis of the B. fragilis genomic sequence, together with genetic conservation analyses, cross-species cloning experiments, and mutational studies, revealed that this organism utilizes an endogenous DNA inversion factor to globally modulate the expression of its surface structures. This DNA invertase is necessary for the inversion of at least 13 regions located throughout the genome, including the promoter regions for seven of the capsular polysaccharide biosynthesis loci, an accessory polysaccharide biosynthesis locus, and five other regions containing consensus promoter sequences. Bacterial DNA invertases of the serine site-specific recombinase family are typically encoded by imported elements such as phage and plasmids, and act locally on a single region of the imported element. In contrast, the conservation and unique global regulatory nature of the process in B. fragilis suggest an evolutionarily ancient mechanism for surface adaptation to the changing intestinal milieu during commensalism.
Considerable progress has been made in elucidating the mechanisms used by pathogenic bacteria to cause disease. In contrast, relatively little is known about how the multitude of bacteria that inhabit the human intestinal tract establish successful commensal and symbiotic relationships with the host. Bacteria that reside in a mammalian host over its lifetime must have mechanisms that allow them to interact with the host in a dynamic and responsive manner. Host mucosal surfaces have been shown to respond to the presence of their endogenous microflora (1). The surfaces of commensal bacteria also have a tremendous capacity for adapting to the changing environment of the host. Bacteroides thetaiotaomicron, a numerically abundant member of the intestinal microbiota, is able to extensively regulate the expression of surface proteins involved in nutrient utilization in response to environmental stimuli (2-5).
The ability of some Bacteroides species to vary their surface architecture also includes molecules not involved in nutrient acquisition. The human commensal microorganism Bacteroides fragilis is able to extensively change its surface by producing at least eight distinct capsular polysaccharides (PSA-H), each of which undergoes a reversible ON-OFF phenotype known as phase variation. Phase variation of seven of these polysaccharides is controlled by DNA inversions of the promoter regions of their biosynthesis loci, placing them in the correct or incorrect orientation for transcription of the downstream polysaccharide biosynthesis genes (6). An analogous system for the generation of surface diversity may also exist in B. thetaiotaomicron, whose genome was recently shown to contain seven polysaccharide biosynthesis loci, many of which are likely regulated by DNA inversions (5). In the intestinal ecosystem, the ability of these organisms to vary the expression of such a large array of surface polysaccharides may allow them to camouflage themselves from the host and other members of the intestinal microbiota. Because B. fragilis is one of the most clinically important anaerobes in extraintestinal sites, and its virulence is linked to production of its capsular polysaccharides (7, 8), elucidation of the factor(s) that governs these DNA inversions is paramount to understanding the complex interplay between the host and this microbe during both health and disease.
Factors that mediate DNA inversions, namely the DNA invertases, segregate into two distinct families: the tyrosine site-specific recombinases (Tsrs) (also known as the lambda integrase family) and the serine site-specific recombinases (Ssr) (recently reviewed in ref. 9). These two families are evolutionarily and mechanistically distinct, and both contain members with diverse functions including transposases, integrases, and DNA invertases. DNA invertases of the Ssr family are typically encoded by elements imported to bacteria from a phage (10-12) or plasmid (13) and act locally on the imported element. Many DNA invertases of the Tsr family have been shown to regulate phase variation of fimbriae by inverting a single area of the chromosome adjacent to the Tsr gene (14-16).
In this study, we demonstrate that the inversions of all seven invertible polysaccharide promoter regions of B. fragilis are mediated by a single member of the Ssr family, which we have designated Mpi. We show that in addition to these seven promoters, Mpi is also involved in inversion of six other promoter regions that control the transcription of uncharacterized products. To our knowledge, this is the first report of a DNA invertase acting globally to modulate the expression of multiple surface molecules encoded by regions distributed throughout a bacterial genome. The unique global regulatory nature and conservation of Mpi in the species suggest that its regulation of bacterial surface variability is ancient, conferring a selective advantage to the microbe during its coevolution with the mammalian host.
Materials and Methods
Analysis of Gene Conservation. Southern blot analysis was used to determine the conservation of the tsr and ssr genes within the species B. fragilis. An internal portion of each of the 30 site-specific recombinase genes was PCR amplified by using the primers listed in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. The purified PCR products were labeled by using the ECL Direct Labeling kit (Amersham Biosciences). Chromosomal DNA from 44 different B. fragilis strains was digested with EcoRI, and fragments were separated by agarose gel electrophoresis and transferred to nylon. Blots containing chromosomal DNA from at least 12 strains were probed with each of the labeled probes. If the probe hybridized under high stringency conditions with at least 95% of the B. fragilis strains tested, the gene was deemed conserved.
Plasmid Constructs. Plasmids containing regions encompassing each of the seven invertible polysaccharide promoter regions were constructed as follows: each of these regions and some flanking DNA was PCR amplified by using the primers listed in Table 2, which is published as supporting information on the PNAS web site. These PCR products were cloned into the PstI site of the Escherichia coli-Bacteroides shuttle vector pFD340 (17) and screened so that the forward primer (VarX-F) of each PCR produced a product with plasmid-borne primer C7. Subsequently, individual site-specific recombinase genes were amplified by using the primers listed in Table 2, cloned into the BamHI site of these plasmids, and screened for the proper orientation so that transcription was driven by the characterized plasmid-borne promoter (Figs. 1 A and 3A). Plasmids were introduced into B. fragilis 9343 (National Collection of Type Cultures) or Bacteroides vulgatus 8482 (American Type Culture Collection) by mobilization from E. coli with the conjugal helper plasmid RK231.
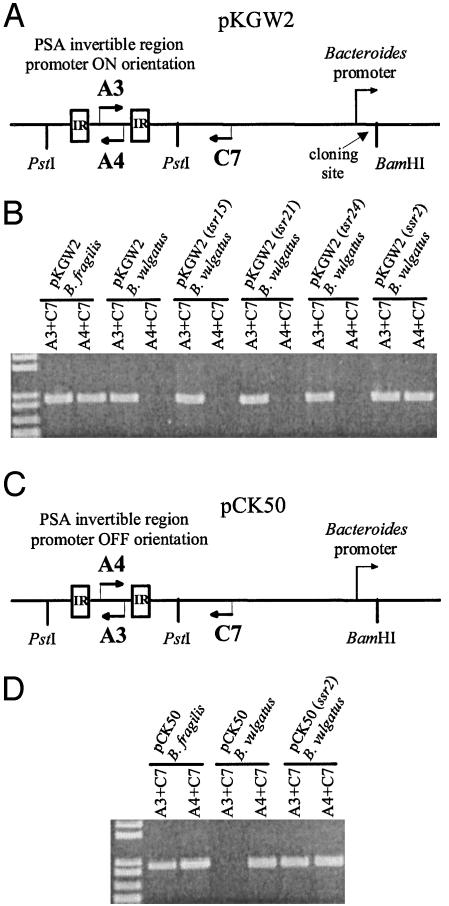
Fig. 1.
Inversion of the PSA promoter region by Ssr2. (A) Plasmid pKGW2 was constructed to monitor inversion of PSA promoter region from ON to OFF. The PSA promoter was cloned into the PstI site of pFD340, and its inversion to the OFF orientation was monitored by PCR using primers A4 and C7. Site-specific recombinases were cloned into the BamHI site, where their transcription is governed by a constitutive plasmid-borne promoter. (B) Ethidium bromide (EtBr)-stained agarose gel demonstrating the ability or inability of various candidate site-specific recombinases to invert the PSA promoter region. (C) Plasmid pCK50 was used to monitor inversion of the PSA promoter from the OFF orientation to the ON orientation. (D) EtBr-stained agarose gel demonstrating the ability of Ssr2 to invert the PSA promoter from the OFF orientation to the ON orientation.
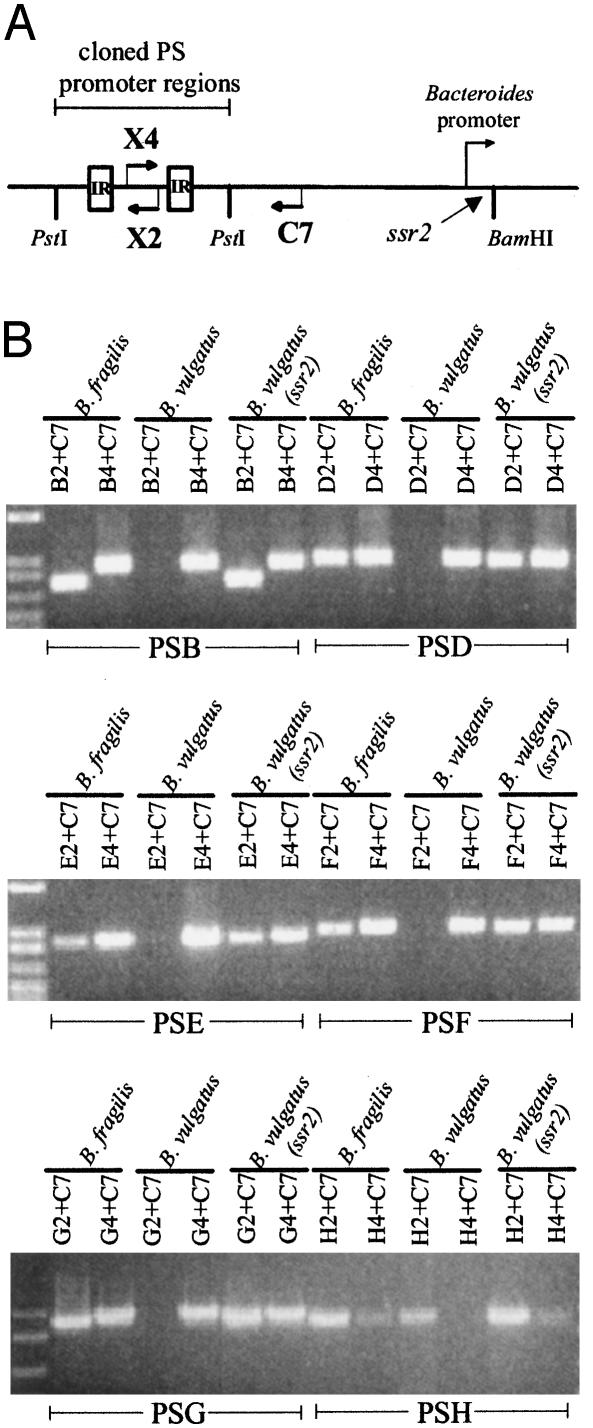
Fig. 3.
Direct role of Ssr2 (Mpi) in inversion of the PSB, PSD, PSE, PSF, PSG, and PSH promoter regions. (A) Diagrammatic representation of the PCR protocol that was used to detect inversion of each of six polysaccharide promoter regions when cloned into pFD340 and transferred to B. fragilis or B. vulgatus, with or without ssr2. The sequences of the primers used for each PCR are listed in Table 2. (B) EtBr-stained agarose gels demonstrating that each of the six promoter regions is able to invert in B. vulgatus only when ssr2 is present.
Construction of Δmpi (Δssr2) Mutants. Δmpi mutants were constructed so that 534 bp of the 591-bp mpi gene was removed. DNA segments upstream and downstream of the region to be deleted were PCR amplified by using primers listed in Table 2. PCR products were digested with BamHI and ligated into the Bacteroides conjugal suicide vector pJST55 (18). E. coli DH5α was transformed with the ligation mixture and resistant colonies were screened for the proper orientation of the left and right flanking DNA. The resulting plasmid was conjugally transferred into B. fragilis 9343, and cointegrates were selected by erythromycin resistance encoded by pJST55. The cointegrate strain was passaged, plated on nonselective medium, and replica-plated to medium containing erythromycin. Erythromycin-sensitive colonies were screened by PCR to select those acquiring the mutant genotype. Mutants were confirmed to be deletants in mpi by Southern blot analysis.
Western Blot Analysis. SDS/PAGE and Western blotting were performed essentially as described (19). Whole bacterial extracts were subjected to separation with 4-12% gradient SDS-polyacrylamide gels. Contents of the gels were transferred to poly(vinylidene difluoride) (Millipore) and probed with antisera specific to each of the eight polysaccharides of B. fragilis 9343, prepared as described (6).
Promoter Analysis. Reporter plasmid pLEC23 was used to assay for functional promoter activity. pLEC23 was created by modifying plasmid pFD340. The insertion sequence element of pFD340 containing the Bacteroides promoter was removed by digestion with PstI and BamHI. A 900-bp BamHI-PstI fragment of pXYLE10 (20) containing the promoterless xylE was inserted in place of the insertion sequence element. Bacteroides promoters cloned into the BamHI site of pLEC23 in the proper orientation will transcribe the downstream xylE, resulting in catechol 2,3-dioxygenase (XylE) activity. A 211-bp region between and including all but 11 bp of each of the MCR1 (Mpi-controlled region 1) inverted repeats (IRs) was PCR amplified by using primers MCR1-prom1 and MCR1-prom2 (Table 2). This DNA was cloned into pLEC23 in both orientations. XylE activity was measured as described (21). XylE assays were performed in triplicate and are reported as mean ± SEM.
Results
Identification of Candidate DNA Invertases. To identify candidate products that may be involved in inverting the polysaccharide promoter regions of B. fragilis, we used the B. fragilis 9343 genomic sequence made publicly available by the Sanger Centre (www.sanger.ac.uk/Projects/B_fragilis). To identify Tsr members, we searched for homologs of FimB, which controls a similar inversion system influencing the synthesis of type 1 fimbriae of E. coli (14). B. fragilis orthologs detected by this initial search were used to further query the B. fragilis database. In all, 28 distinct members of the Tsr family were detected and designated Tsr1-Tsr28 (Fig. 5, which is published as supporting information on the PNAS web site).
A similar set of analyses was used to detect members of the Ssr family. The B. fragilis genome was searched by using the pfam00239 motif, a 139-aa consensus sequence present in members of the Ssr family (22). Three products similar to site-specific recombinases were retrieved and designated Ssr1-Ssr3. The sizes and protein sequences of Ssr2 and Ssr3 identified them as members of the resolvase/invertase branch of the Ssr family. In contrast, Ssr1 is larger (585 aa) and segregates into the large serine recombinase branch, whose members mediate integration (23), excision (24), and transposition (25) events, but not DNA inversion. Therefore, of these three Ssr products, only Ssr2 and Ssr3 were further considered as candidate DNA invertases.
To narrow the list of candidates that may be involved in the inversions of the polysaccharide promoter regions, we took advantage of the fact that these invertible regions are contained on conserved areas of the B. fragilis chromosome (8, 26). Therefore, the factor(s) that govern these DNA inversions should also be conserved in the species. Southern blot analysis using internal portions of each of the 30 candidate DNA invertase genes as probes revealed that ssr2 and nine of the 28 tsr genes (Fig. 5) are conserved in B. fragilis. The ssr3 gene was present in only 5 of the 17 B. fragilis strains probed, and was therefore not further considered. ssr3 has subsequently been designated finB and is plasmid-borne (27). ssr2 has alternatively been designated finA, although no function has been described for its product (27). These hybridization analyses reduced the number of potential candidates from 30 to 10.
Ssr2 Mediates Inversion of the PSA Promoter Region. To determine whether any of these 10 conserved products are involved in inverting the polysaccharide promoter regions, we constructed plasmid pKGW2 (Fig. 1A). pKGW2 contains a 420-bp region including the PSA invertible promoter region cloned into the E. coli-Bacteroides shuttle vector pFD340. A PCR product is generated from this plasmid by using the plasmid-borne primer C7 with primer A3 (within the PSA invertible region). When pKGW2 is conjugally transferred into B. fragilis, the PSA promoter region undergoes inversion from the ON-to-OFF orientation, as demonstrated by the generation of a PCR product with primers C7 and A4 (Fig. 1B). However, when pKGW2 is transferred into a different Bacteroides species, B. vulgatus, no inversion is detected. Candidate DNA invertase genes were cloned into the BamHI site of pKGW2, where their transcription is controlled by a plasmid-borne promoter. The resulting plasmids were conjugally transferred to B. vulgatus, and the ability of the candidate DNA invertases to mediate inversion of the PSA promoter was monitored by PCR using primers C7 and A4. As shown in Fig. 1B, products of three different conserved Tsr genes (tsr15, tsr21, and tsr24) did not cause inversion of the PSA promoter. In contrast, when ssr2 was cloned into pKGW2, DNA inversion was detected in the B. vulgatus background. PCR/digestion, a quantitative assay to determine orientations of invertible regions (6, 28), demonstrated that at least half of the PSA promoters of this construct had flipped to the OFF orientation (Fig. 6, which is published as supporting information on the PNAS web site). Therefore, Ssr2 mediates inversion of the PSA promoter region from the ON-to-OFF orientation in B. vulgatus. To determine whether Ssr2 could also bring about inversion of the PSA promoter from the OFF-to-ON orientation, plasmid pCK50, which is identical to pKGW2 except that the PSA promoter is initially OFF, was constructed (Fig. 1C). As shown in Fig. 1D, Ssr2 also mediates inversion of the PSA promoter from the OFF-to-ON orientation. ssr2 is present in the compiled B. fragilis 9343 genome sequence at base pairs 3237948-3238541 and is not transcriptionally coupled to other genes because both the upstream and downstream genes are transcribed in the opposite orientation.
All Seven Invertible Polysaccharide Promoter Regions Are Locked in ssr2 Deletion Mutants. To further confirm the involvement of Ssr2 in the inversion of the PSA promoter region and to determine whether this product is involved in inverting the other six polysaccharide promoter regions, ssr2 deletion mutants were constructed whereby an internal 534-bp portion of the 594-bp gene was removed by allelic exchange. Eight deletion mutants were obtained (Δssr2 mutants 2, 4, 8, 10, 42, 44, 48, and 62) and analyzed for inversions of the seven polysaccharide promoter regions by PCR (Fig. 2A). In each of these eight mutants, all seven of the invertible polysaccharide promoter regions are locked and unable to invert relative to wild type. These results demonstrate global control of multiple DNA inversions by a single site-specific recombinase. Fig. 2B shows the three different genotypes represented in the eight mutants in regard to polysaccharide promoter orientation. To ensure that the deletion of ssr2 was responsible for locking the polysaccharide promoter regions, ssr2 was cloned into expression vector pFD340, creating plasmid pKGW4R, and added in trans to Δssr2 mutant 44. As shown in the bottom panel of Fig. 2B, each of the polysaccharide promoter regions regained the ability to invert when ssr2 was provided in trans.
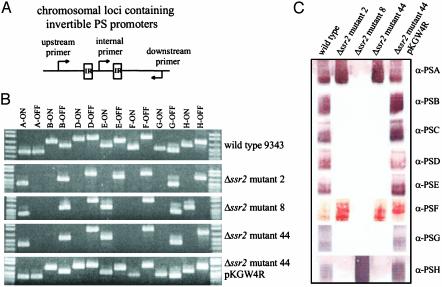
Fig. 2.
Ssr2 is necessary for inversion of all seven polysaccharide promoter regions. (A) Diagrammatic representation of the PCR method used to detect DNA inversions of each of the seven invertible promoter regions. Each of the primers contained within the IRs was used in a PCR with both the upstream and downstream primers individually. (B) EtBr-stained agarose gel demonstrating that the promoters of each of the seven polysaccharide biosynthesis loci is present in only a single orientation in the Δssr2 mutants relative to wild type. Three distinct locked polysaccharide promoter patterns were detected in the panel of eight mutants exemplified by Δssr2 mutants 2, 8, and 44. The bottom panel shows complementation of the locked genotypes when ssr2 is added in trans to Δssr2 mutant 44 (pKGW4R). (C) Western blots demonstrating the polysaccharide phenotypes of the three representative Δssr2 mutants.
Phenotypic Analysis of Polysaccharide Expression by Δssr2 Mutants. Phenotypic analyses demonstrated a high correlation between the locked orientation of a promoter region and the expression, or lack of expression, of that polysaccharide (Fig. 2C). The polysaccharides whose promoters are locked OFF are not produced in any of the eight mutants with the exception of PSF. The PSF promoter is locked OFF in all mutants; however, all mutants except mutant 8 produce PSF. These data suggest that there may be a secondary promoter downstream of the invertible PSF promoter. The PSC biosynthesis locus is the only one of the eight polysaccharide regions that does not contain an invertible promoter (6). Despite this fact, PSC is not produced in any of the Δssr2 mutants, demonstrating that its synthesis is also regulated in some manner by Ssr2. In addition, the PSE promoter is locked ON in all mutants except mutant 2; however, the PSE polysaccharide is not synthesized by any of the Δssr2 mutants. Δssr2 mutant 8 is the only mutant where a promoter other than those of PSA or PSE is locked ON, namely PSH. Although this mutant produces a large amount of PSH, it is the only mutant that does not produce PSA. These combined phenotypic data suggest that the expression of each of the eight polysaccharides of B. fragilis is complex, interdependent, and regulated by Ssr2 either directly or indirectly. Phenotypic analysis of Δssr2 mutant 44 containing ssr2 in trans (pKGW4R) revealed that expression of each of the polysaccharides is restored, correlating with the ability of the promoters to undergo DNA inversion.
Inversion of Six Other PS Promoter Regions by Ssr2. The ssr2 mutational data presented above demonstrate that Ssr2 is necessary for the inversion of each of the seven polysaccharide promoter regions. However, these data do not demonstrate the direct role for Ssr2 in the inversion of these regions previously shown for the PSA locus. For these analyses, we created plasmid constructs similar to pKGW2 containing one of the six promoter regions of PSB, PSD, PSE, PSF, PSG, or PSH (Fig. 3A). We first confirmed that each of these constructs contained sufficient DNA in cis for the inversions to occur by demonstrating that each region undergoes inversion when placed in B. fragilis (Fig. 3B). Similarly, we showed that B. vulgatus does not synthesize a product that inverts these regions. When ssr2 was cloned into the BamHI site of these plasmids, each of the six promoter regions was able to invert in B. vulgatus (Fig. 3B). These data demonstrate a direct role for Ssr2 in the inversion of each of the seven invertible polysaccharide promoter regions. Based on these data, we changed the designation of ssr2 to mpi for multiple promoter invertase.
DNA Inversion of Other Chromosomal Loci by Mpi. DNA invertases require the presence of IRs flanking the invertible DNA segment. The seven invertible polysaccharide promoter regions are flanked by IRs of 19-21 bp that contain a consensus core sequence (6). The upstream IR core consensus sequence is ARACGTTCGT, followed by 168-194 bp of intervening DNA, followed by the downstream core consensus IR sequence ACGAACGTYT. We exploited this commonality to search for other regions of the B. fragilis genome that might be subject to DNA inversion by Mpi, searching for additional regions matching the pattern ARACGTTCGTN{90,250}ACGAACGTYT by using the findpatterns program (wisconsin package, Version 10.2 for UNIX, Accelrys, Burlington, MA). This initial search retrieved 92 unique segments of the B. fragilis genome when three mismatches were allowed. Because the core IR consensus sequence is part of a larger IR in each of the seven polysaccharide regions, each of these 92 segments was examined for the presence of larger IRs by using the program einverted [distributed as part of the emboss suite of programs (29)] with a threshold detection score of 30. Thirteen segments remained after this search: seven of these were the previously identified polysaccharide promoter regions (1), and the remaining six were uncharacterized.
These six new regions each contain a set of IRs 19-23 bp in length and separated by 119-187 bp of intervening DNA (Fig. 4A). Two pairs of these newly found regions are closely linked on the B. fragilis chromosome; thus, the six regions encompass four new chromosomal loci. These regions were designated MCR1, MCR2, MCR3a, MCR3b, MCR4a, and MCR4b. ORF maps of these regions are shown in Fig. 4B.
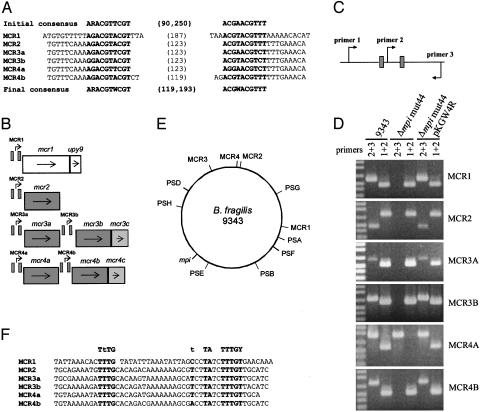
Fig. 4.
Characterization of six additional loci whose DNA inversions are controlled by Mpi. (A) The top line represents the upstream and downstream core consensus Mpi recognition sequence (ARACGTTCGTN{90,250}ACGAACGTYT) used to query the B. fragilis genome database. The IR sequences of the six new Mpi-controlled regions are shown. The bottom line shows the revised Mpi consensus recognition sequence based on these new additions. (B) Diagrammatic representation of the four new genomic regions containing six invertible regions and the immediate downstream ORFs. Genes highlighted in similar shades of gray are homologous to each other. IRs are shown as small boxes. (C) Design of the PCR used to examine each of the MCRs for inversion. Small boxes represent the upstream and downstream IR of each of the six regions. (D) EtBr-stained agarose gel demonstrating that each MCR undergoes inversion regulated by Mpi. (E) A scaled representation of the 5,205,140-bp B. fragilis 9343 genome demonstrating the relative locations of mpi and each of the Mpi-controlled regions. (F) Consensus promoter sequence present in each of the MCR invertible regions. The top line shows the B. fragilis consensus promoter sequence. The last nucleotide reported for each regions is adjacent to the first nucleotide of the downstream IR.
The results of the PCR analyses outlined in Fig. 4C demonstrate that the DNA regions between each of these six new IRs do in fact undergo inversion in wild-type bacteria (Fig. 4D). To determine whether these inversions are controlled by Mpi, we analyzed the ability of each of these regions to invert in the Δmpi (Δssr2) mutants (Fig. 4D and Table 3, which is published as supporting information on the PNAS web site). The DNA regions between each of these six new IRs are locked in all Δmpi mutants relative to wild type. When mpi is provided in trans, each of these six regions regains the ability to invert (Fig. 4D).
Using the compiled B. fragilis genomic sequence, we determined that the 11 Mpi-controlled regions (encompassing 13 distinct inversion events) are scattered throughout the ≈5.2-Mb genome (Fig. 4E). In addition, none of these 11 regions are adjacent to mpi; the closest is the PSE promoter region, which is ≈206 kb from mpi. Analysis of the partially sequenced B. fragilis 638R genome demonstrated that, like the polysaccharide promoter regions (8, 26), the MCRs are also conserved.
The two genes downstream of the MCR1 invertible region (Fig. 4B) are homologous to genes contained in polysaccharide biosynthesis loci. The protein designated Mcr1 is homologous to Wza, an outer membrane lipoprotein of E. coli involved in the secretion of surface polysaccharides (30). The gene immediately downstream of mcr1, designated upy9, is a member of the upxY gene family, whose members are present just downstream of each of the promoter regions of the eight polysaccharide biosynthesis loci of B. fragilis (6). Each of the remaining five MCRs is extremely similar. All contain identical IRs with 117-119 bp of intervening DNA, and each downstream IR is 146-151 bp upstream of the gene(s) putatively regulated by the DNA inversion. The mcr genes encode products that comprise two families of proteins. The first gene downstream of each of the five IRs encodes products that are 48.5-84.7% identical over their entire length (Fig. 7A, which is published as supporting information on the PNAS web site). These products demonstrate no significant homology to any sequences in the database and, as such, represent an unknown family of proteins. The MCR3b and MCR4b regions each contain an additional gene (mcr3c and mcr4c, respectively) whose products are 92.1% identical to each other (Fig. 7B) and have a close homolog of unknown function in the B. thetaiotaomicron genome (5).
MCR Invertible Regions Contain Promoter Sequences. Analysis of the MCRs depicted in Fig. 4B suggested that the regions that undergo inversion contain promoters that control transcription of the downstream gene(s). We analyzed the DNA between each of these new IRs and found that in one orientation, each aligned perfectly with the described B. fragilis consensus promoter sequence (Fig. 4F) (31). Promoter activity of the MCR1 region was confirmed by using reporter plasmid pLEC23, which utilizes the enzymatic activity encoded by xylE to detect promoters. The DNA between the MCR1 IRs yielded 1.11 ± 0.03 milliunits of XylE/mg protein when cloned into pLEC23 in the putatively ON orientation. In comparison, absolutely no XylE activity was detected when this same DNA segment was cloned into pLEC23 in the putatively OFF orientation. The demonstration of this functional promoter in the MCR1 region, coupled with the presence of identical promoter sequences between the IRs of the other MCRs, allowed us to designate ON and OFF orientations for each region. PCR analysis was performed to determine the orientation in which each of the Mpi-controlled regions was locked in each of the eight Δmpi mutants (Table 3). Except for Δmpi mutants 4, 10, and 44, which are identical in regard to the ON-OFF orientations of all 13 Mpi-controlled promoters, the other five mutants all differ in their locked orientations. These data suggest that the deletion of mpi locked these 13 promoter regions in the orientation in which they were at the time of deletion. Because some promoters are locked ON in some mutants and OFF in other mutants, Mpi is necessary for the inversion of these regions in both orientations.
Discussion
The unique characteristics of the predominant commensal and symbiotic microorganisms that allow them to thrive in the mammalian gut are just beginning to be appreciated. The surfaces of these microorganisms are crucial to their adaptive response to, and dynamic interaction with, the host. The ability of these microorganisms to modulate their surfaces is likely essential to their long-term survival in the host.
In this study, we have identified a factor that mediates extensive surface variability of the numerically abundant commensal microorganism B. fragilis. This factor, Mpi, is a Ssr with unique characteristics: it is conserved in the species, it is involved in the inversions of multiple conserved promoter regions (including all seven invertible polysaccharide promoters), and the regions controlled by it are distributed throughout the B. fragilis genome.
Phenotypic analysis of the mpi mutants reveals that ultimate expression of the eight capsular polysaccharides of B. fragilis relies on a complex and interdependent system. The orientation of an Mpi-controlled promoter is insufficient to predict whether a particular polysaccharide will be expressed. Moreover, the pattern of promoter orientations observed in the mpi mutants suggests that whether a particular promoter is ON or OFF at any given time is not influenced to any obvious degree by the status of the other Mpi-controlled promoters. This system affords B. fragilis the flexibility to globally modulate its surface components in a way that efficiently provides for a great degree of antigenic variability. Furthermore, the conserved nature of this system among the species, coupled with the observation that the regions controlled by Mpi are distributed throughout the genome, argues strongly for this being an evolutionarily ancient system, and thus an integral part of the organism's adaptation to its ecological niche.
The genome of the numerically abundant gut symbiont B. thetaiotaomicron was recently reported to contain seven polysaccharide biosynthesis loci, many of which are likely to be regulated by DNA inversions (5). It is intriguing to note that this genome encodes the closest homolog of Mpi. Therefore, the unique global mechanism of surface modulation described in this study may have broad implications for the complex interplay between the host and other abundant members of its microbiota.
Supplementary Material
Acknowledgments
We are grateful to the Pathogen Sequencing Group at the Sanger Institute for producing the Bacteroides fragilis 9343 and 638R genomic sequence data, S. Mazmanian for valuable discussions, A. Nichols for Southern blotting, C. J. Smith for pFD340, and M. Malamy for pJST55. This work was supported by National Institutes of Health-National Institute of Allergy and Infectious Diseases Grant AI44193.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: EtBr, ethidium bromide; IR, inverted repeat; MCR, Mpi-controlled region; Ssr, serine site-specific recombinase; Tsr, tyrosine site-specific recombinase.
References
- 1.Bry, L., Falk, P. G., Midtvedt, T. & Gordon, J. I. (1996) Science 273, 1380-1383. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, K. L. & Salyers, A. A. (1989) J. Bacteriol. 171, 3199-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hwa, V. & Salyers, A. A. (1992) J. Bacteriol. 174, 342-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reeves, A. R., D'Elia, J. N., Frias, J. & Salyers, A. A. (1996) J. Bacteriol. 178, 823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu, J., Bjursell, M. K., Himrod, J., Deng, S., Carmichael, L. K., Chiang, H. C., Hooper, L. V. & Gordon, J. I. (2003) Science 299, 2074-2076. [DOI] [PubMed] [Google Scholar]
- 6.Krinos, C. M., Coyne, M. J., Weinacht, K. G., Tzianabos, A. O., Kasper, D. L. & Comstock, L. E. (2001) Nature 414, 555-558. [DOI] [PubMed] [Google Scholar]
- 7.Onderdonk, A. B., Kasper, D. L., Cisneros, R. L. & Bartlett, J. G. (1977) J. Infect. Dis. 136, 82-89. [DOI] [PubMed] [Google Scholar]
- 8.Coyne, M. J., Tzianabos, A. O., Mallory, B., Kasper, D. L. & Comstock, L. E. (2001) Infect. Immun. 69, 4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith, M. C. M. & Thorpe, H. M. (2002) Mol. Microbiol. 44, 299-307. [DOI] [PubMed] [Google Scholar]
- 10.Kwoh, D. Y. & Zipser, D. (1981) Virology 114, 291-296. [DOI] [PubMed] [Google Scholar]
- 11.Iida, S., Meyer, J., Kennedy, K. E. & Arber, W. (1982) EMBO J. 1, 1445-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iida, S., Sandmeier, H., Hubner, P., Hiestand-Nauer, R., Schneitz, K. & Arber, W. (1990) Mol. Microbiol. 4, 991-997. [DOI] [PubMed] [Google Scholar]
- 13.Chen, J., Leblanc, D. J. & Galli, D. M. (2002) J. Bacteriol. 184, 5926-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klemm, P. (1986) EMBO J. 5, 1389-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, X., Lockatel, C. V., Johnson, D. E. & Mobley, H. L. (2002) Mol. Microbiol. 45, 865-874. [DOI] [PubMed] [Google Scholar]
- 16.Honarvar, S., Choi, B. K. & Schifferli, D. M. (2003) Mol. Microbiol. 48, 157-171. [DOI] [PubMed] [Google Scholar]
- 17.Smith, C. J., Rogers, M. B. & McKee, M. L. (1992) Plasmid 27, 141-154. [DOI] [PubMed] [Google Scholar]
- 18.Thompson, J. S. & Malamy, M. H. (1990) J. Bacteriol. 172, 2584-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coyne, M. J., Kalka-Moll, W., Tzianabos, A. O., Kasper, D. L. & Comstock, L. E. (2000) Infect. Immun. 68, 6176-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein, D. C. (1992) Gene 117, 157-158. [DOI] [PubMed] [Google Scholar]
- 21.Zukowski, M. M., Gaffney, D. F., Speck, D., Kauffmann, M., Findeli, A., Wisecup, A. & Lecocq, J. P. (1983) Proc. Natl. Acad. Sci. USA 80, 1101-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S. R., Griffiths-Jones, S., Howe, K. L., Marshall, M. & Sonnhammer, E. L. (2002) Nucleic Acids Res. 30, 276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuura, M., Noguchi, T., Yamaguchi, D., Aida, T., Asayama, M., Takahashi, H. & Shirai, M. (1996) J. Bacteriol. 178, 3374-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrasco, C. D., Ramaswamy, K. S., Ramasubramanian, T. S. & Golden, J. W. (1994) Genes Dev. 8, 74-83. [DOI] [PubMed] [Google Scholar]
- 25.Wang, H. & Mullany, P. (2000) J. Bacteriol. 182, 6577-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Comstock, L. E., Pantosti, A. & Kasper, D. L. (2000) Infect. Immun. 68, 6182-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patrick, S., Parkhill, J., McCoy, L. J., Lennard, N., Larkin, M. J., Collins, M., Sczaniecka, M. & Blakely, G. (2003) Microbiology 149, 915-924. [DOI] [PubMed] [Google Scholar]
- 28.Kim, J. K., Gunther, N. W., Zhao, H., Johnson, D. E., Keay, S. K. & Mobley, H. L. T. (1998) Infect. Immun. 66, 3303-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rice, P., Longden, I. & Bleasby, A. (2000) Trends Genet. 16, 276-277. [DOI] [PubMed] [Google Scholar]
- 30.Drummelsmith, J. & Whitfield, C. (2000) EMBO J. 4, 57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bayley, D. P., Rocha, E. R. & Smith, C. J. (2000) FEMS Microbiol. Lett. 193, 149-154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.