Abstract
Transcriptional control by β-catenin and lymphoid enhancer-binding factor 1 (LEF1)/T cell factor regulates proliferation in stem cells and tumorigenesis. Here we provide evidence that transcriptional co repressor homeodomain interacting protein kinase 2 (HIPK2) controls the number of stem and progenitor cells in the skin and the susceptibility to develop squamous cell carcinoma. Loss of HIPK2 leads to increased proliferative potential, more rapid G1–S transition in cell cycle, and expansion of the epidermal stem cell compartment. Among the critical regulators of G1–S transition in the cell cycle, only cyclin D1 is selectively up-regulated in cells lacking HIPK2. Conversely, overexpression of HIPK2 suppresses LEF1/β-catenin-mediated transcriptional activation of cyclin D1 expression. However, deletion of the C-terminal YH domain of HIPK2 completely abolishes its ability to recruit another transcriptional corepressor CtBP and suppress LEF1/β-catenin-mediated transcription. To determine whether loss of HIPK2 leads to increased susceptibility to tumorigenesis, we treat wild-type, Hipk2+/−, andHipk2−/− mice with the two-stage carcinogenesis protocol. Our results indicate that more skin tumors are induced in Hipk2+/− and Hipk2−/− mutants, with most of the tumors showing shortened incubation time and malignant progression. Together, our results indicate that HIPK2 is a tumor suppressor that controls proliferation by antagonizing LEF1/β-catenin-mediated transcription. Loss of HIPK2 synergizes with activation of H-ras to induce tumorigenesis.
Keywords: cell cycle, proliferation, Wnt, corepressor
The process of skin carcinogenesis involves a series of transitional events, ranging from hyperplasia, dysplasia, and papilloma to invasive squamous cell carcinoma. The current model indicates that the progression of benign lesions to malignancy depends on the cell type targeted by these mutations (1, 2). Indeed, recent evidence has indicated that the bulge region of the hair follicles contains self-renewing, slow-cycling stem cells, which give rise to transient amplifying cells and several differentiated cell lineages in the interfollicular epidermis (3, 4). The progression of skin carcinogenesis can be modified by mutations involving Tgfβ1, Smad3, ubiquitin ligase Fbxw7/Cdc4, or Pten (5–9), suggesting that a genetic network synergizes with activated Ras mutation to promote the initiation or malignant transformation of skin tumors (10). Another important regulator of skin carcinogenesis is cyclin D1. Overexpression of cyclin D1 increases the propensity of skin carcinogenesis, whereas loss of cyclin D1 reduces tumor formation (11, 12). One mechanism to control cyclin D1 expression is through transcriptional regulation via the Wnt/β-catenin signaling pathway (13, 14), which consists of a core set of highly evolutionarily conserved proteins that have wide-ranging effects on gene expression affecting proliferation, migration, pluripotency, morphogenesis, and tumorigenesis (15–17). In the epidermis, targeted deletion of β-catenin during embryogenesis results in severe abnormalities in cell fate determination and maintenance of hair follicle formation (18). In contrast, mutations that activate β-catenin function lead to skin tumorigenesis in both humans and mice (19, 20).
Homeodomain interacting protein kinase (HIPK) contains three distinct members that regulate apoptosis, cell growth, and proliferation. HIPK2 has been identified as a transcriptional corepressor for homeodomain proteins NK3 and Brn3a (21, 22). Previous results have indicated that Brn3a and HIPK2 regulate a delicate balance of gene expression that controls programmed cell death in sensory and dopamine neurons (22–24). Both HIPK1 and HIPK2 can interact with human p53 and, under DNA damage conditions, activate p53 by promoting phosphorylation and acetylation (25–27). Contrary to the predicted function, HIPK1-deficient mice show a higher resistance to the development of DMBA-induced skin tumors (27). Recent evidence, however, has demonstrated a functional redundancy between HIPK1 and HIPK2 during development, which could compensate for the loss of HIPK1 (28). Indeed, HIPK2 has a wide range of functions, including promoting the degradation of CtBP in response to UV-induced DNA damage (29) and regulating cell proliferation through the Wnt signaling pathway (30). Furthermore, HIPK2 can interact with several proteins containing the high-mobility group I (HMGI), a domain highly conserved in transcription factors in the lymphoid enhancer-binding factor 1/T cell factor (LEF1-TCF) family (31).
In this work, we show that loss of HIPK2 leads to an expansion of epidermal stem cells due to increased proliferation. Our results reveal a mechanism of HIPK2 as a transcriptional corepressor of LEF1/β-catenin-mediated cyclin D1 expression. As a consequence, cells derived from Hipk2−/− mutants show increased cyclin D1 and an accelerated G1–S cell cycle transition. The mechanism by which HIPK2 achieves this suppressive effect is by forming a protein complex with β-catenin and LEF1 and by recruiting another corepressor, CtBP. Deletion of the CtBP-interacting domain abolishes the suppressive effect of HIPK2 on β-catenin. Consistent with these findings, both Hipk2+/− and Hipk2−/− mutants show increased propensity for tumor formation and malignant progression under the two-stage chemical carcinogenesis paradigm. Together these results provide evidence that HIPK2-mediated transcriptional programs restrict cell growth in epidermal stem and progenitor cells by suppressing cyclin D1 expression. Removal of such mechanisms leads to an expansion in the stem cell pool and increases the propensity for tumorigenesis.
Results
Expansion of Label-Retaining Cells and Transient Amplifying Cells in Hipk2−/− Mutants.
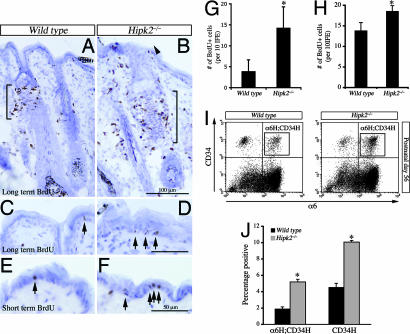
Using β-galactosidase inserted into the first coding exon of hipk2 (HIPK2LacZ) as a marker for HIPK2 expression, we found that HIPK2 was present diffusely in hair follicles [supporting information (SI) Fig. 7 A–E]. HIPK2, detected either by β-galactosidase or HIPK2 antibodies, was present in keratinocytes, transient amplifying (TA) cells (see SI Fig. 7 F and G) marked by a short pulse of BrdU, and label-retaining cells (LRCs) (see SI Fig. 7 H–K) marked by BrdU injected from postnatal day 1 (P1) to P3 twice a day followed by an 8-week chase period (32). To determine whether loss of HIPK2 affected cell growth and differentiation, we compared the number of LRCs and TA cells in the epidermis of wild-type (WT) and Hipk2−/− mutants. Our results showed that there were more LRCs in the bulge and infundibular regions of Hipk2−/− mutants (Fig. 1 A and B). Whereas there were very few BrdU+ LRCs in the basal layers of the interfollicular epidermis (IFE) in WT mice (<5 per 10 IFE), the number increased by at least 3-fold in the Hipk2−/− mutants (Fig. 1 C and D, arrows, and G). Similarly, TA cells in the epidermis, marked by a short-term BrdU labeling scheme, also showed a 40% increase in the IFE (Fig. 1 E, F, and H). To provide additional quantitative analyses of the stem and progenitor cells in the epidermis, we measured the number of integrin-α6- and CD34-positive cells at P56 by using FACS analyses. Interestingly, the number of α6high;CD34high double-positive cells (α6H;CD34H) and α6low;CD34high single-positive cells (CD34H) showed a consistent increase at P56 (Fig. 1 I and J). Collectively, the results indicated that loss of HIPK2 led to an expansion in the stem and progenitor cells in the epidermis.
Fig. 1.
Expansion of LRC and TA cells in the skin of adult Hipk2−/− mutant mice. (A–D) Loss of HIPK2 leads to a significant increase in LRC in the bulge region and the infundibulum of hair follicles (A and B, brackets and arrowhead). Expansion of LRC is also present in the IFE of Hipk2−/− mutants (C and D, arrows). (Scale bars: B, 100 μm; D, 50 μm.) (E and F) Short-term BrdU labeling scheme shows a significant increase in the number of TA cells in the IFE of Hipk2−/− mutants. (Scale bar: 50 μm.) (G and H) LRC in the IFE shows a 3-fold increase in Hipk2−/− mutant mice (G). A 40% increase in the TA cells within the IFE is also detected in the Hipk2−/− mutants (H). *, P = 0.005, Student's t test (n = 3 for WT and n = 4 for Hipk2−/−). (I and J) FACS analyses show that the numbers of α-6H;CD34H and CD34H cells in P56 Hipk2−/− mutants are increased by 2.7- and 2.2-fold, respectively. *, P = 0.02, Student's t test (n = 5 for WT and n = 4 for Hipk2−/−).
Loss of HIPK2 Results in Hyperproliferation in Keratinocytes and Mouse Embryonic Fibroblasts (MEFs).
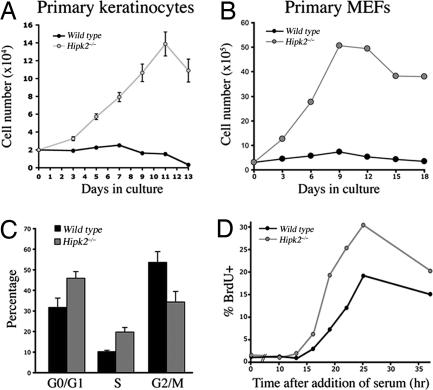
To determine the mechanism of HIPK2 in cell proliferation in the epidermis, we cultured primary keratinocytes from perinatal mice at a low density to determine their growth potential (Fig. 2A). Keratinocytes derived from Hipk2−/− mutants showed increased proliferative potential up to 2 weeks in culture, whereas those from WT littermates never showed a definitive increase in cell numbers (Fig. 2A). Consistent with the results from keratinocytes, MEFs from Hipk2−/− mutants also showed increased proliferative potential under 3T3 protocol, although they eventually entered senescence after three to four passages (Fig. 2B). To further investigate the mechanism of increased proliferation in Hipk2−/− keratinocytes and MEFs, we analyzed their cell cycle by using flow cytometry. Under standard culture conditions with 10% FBS, both Hipk2−/− MEFs and keratinocytes showed significantly more cells in the G0/G1 and S phases (Fig. 2C), suggesting potential defects in G1–S checkpoints. Indeed, under serum deprivation and cell cycle reinitiation, Hipk2−/− MEFs showed more BrdU uptake and a much faster transition from the G0 to the S phase (Fig. 2D).
Fig. 2.
Loss of HIPK2 leads to increased proliferative potential in primary keratinocytes and MEFs. (A) Primary keratinocytes from Hipk2−/− mutants show increased proliferative potential. (B) Similarly, Hipk2−/− MEFs also showed increased proliferation under the standard 3T3 culture condition, although they undergo senescence after three to four passages. (C) FACS analyses show that Hipk2−/− MEFs contain more cells in the S phase. Student's t test (n = 3) and data are presented as mean ± SEM. (D) Uptake of BrdU after serum deprivation indicates the duration for the G1–S transition. As predicted from C and D, a higher percentage of MEFs from Hipk2−/− mutants show a more rapid entrance into the S phase upon addition of serum.
HIPK2 Suppresses β-Catenin-Mediated Transcriptional Activation of Cyclin D1.
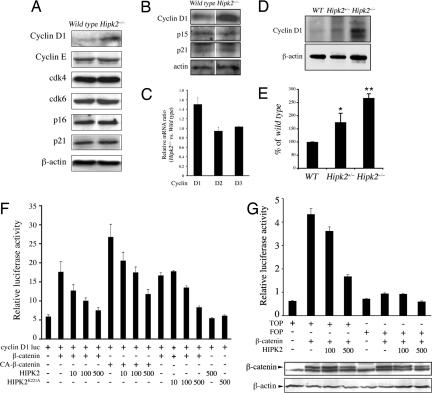
To characterize the cell cycle checkpoint defect in Hipk2−/− keratinocytes and MEFs, we examined the expression level of proteins that regulate G1–S phase transition, including cyclin D, cyclin E, G1-specific cyclin-dependent kinases, p16, and p21 (33). Although cyclin E, cdk4, cdk6, p16, and p21 showed no detectable difference between WT and Hipk2−/− MEFs, the level of cyclin D1 protein was up-regulated in Hipk2−/− MEFs and keratinocytes by 2.5- and 3-fold, respectively (Fig. 3 A and B). To exclude the possibility that loss of HIPK2 may affect the stability of cyclin D1, we determined cyclin D1 protein turnover and found that degradation of 35S-labeled cyclin D1 followed a similar kinetics in WT and Hipk2−/− MEFs (SI Fig. 8). Given the role of HIPK2 as a transcriptional corepressor, we reasoned that an increase in cyclin D1 protein may be due to an increase in transcription of cyclin D1 in Hipk2−/− MEFs. Consistent with this, quantitative RT-PCR (qRT-PCR) showed that cyclin D1 mRNA, but not D2 or D3, was up-regulated in Hipk2−/− MEFs (Fig. 3C). Similar to the results in MEFs and keratinocytes, protein extracts from skin of Hipk2+/− and Hipk2−/− mice also showed significant increases in cyclin D1 proteins (Fig. 3 D and E).
Fig. 3.
HIPK2 suppresses β-catenin-mediated transcription of cyclin D1. (A) Western blots showing increase of cyclin D1 in Hipk2−/− MEFs, whereas other regulators of G1 cell cycle, e.g., cyclin E, cdk4, cdk6, p16, and p21, are not affected. (B) Similarly, Hipk2−/− keratinocytes also show a 3-fold increase in cyclin D1 protein, whereas p15 and p21 show no detectable difference between WT and Hipk2−/− keratinocytes. (C) Selective increase of cyclin D1 mRNA in Hipk2−/− mutant MEFs. (D and E) Cyclin D1 protein also is increased in skin from Hipk2+/− and Hipk2−/− mice. *, P = 0.0189 between WT and Hipk2+/−; **, P = 0.00058 between WT and Hipk2−/−; Student's t test (n = 3). (F) HIPK2 suppresses the ability of β-catenin and constitutively active β-catenin (CA-β-catenin) to activate the cyclin D1 promoter. The kinase-inactive form of HIPK2, HIPK2K221A, continues to suppress cyclin D1 luciferase activity. (G) Similar to the cyclin D1 reporter, TOPFlash luciferase activity is inhibited by HIPK2 in a dose-dependent manner. HIPK2 has no effect on the stability of β-catenin protein.
Our results indicated that HIPK2 suppressed β-catenin-mediated activation of cyclin D1 (Fig. 3F). The suppressive effect of HIPK2 did not require phosphorylation on the critical Ser/Thr residues of β-catenin because HIPK2 suppressed the transcriptional activity of a constitutively active form of β-catenin (CA-β-catenin) in which amino acids 29–48 were deleted (Fig. 3F). Interestingly, the kinase-inactive form of HIPK2 (HIPK2K221A) showed a similar efficacy in suppressing cyclin D1 expression, suggesting that the kinase activity of HIPK2 was dispensable. To determine whether the effect of HIPK2 was directly mediated through β-catenin and LEF1/TCF, we used the TOPFlash luciferase reporter that contained only LEF1/TCF binding sites as a highly specific reporter for Wnt/β-catenin signaling. Similar to the results with the cyclin D1 luciferase reporter, HIPK2 suppressed TOPFlash activity activated by β-catenin (Fig. 3G). The presence of HIPK2 did not alter the level of β-catenin, indicating that HIPK2 did not affect the stability or turnover of β-catenin (Fig. 3G and SI Fig. 9).
Transcriptional Repression of HIPK2 Requires Recruitment of Co repressor CtBP via the C-Terminal YH Domain.
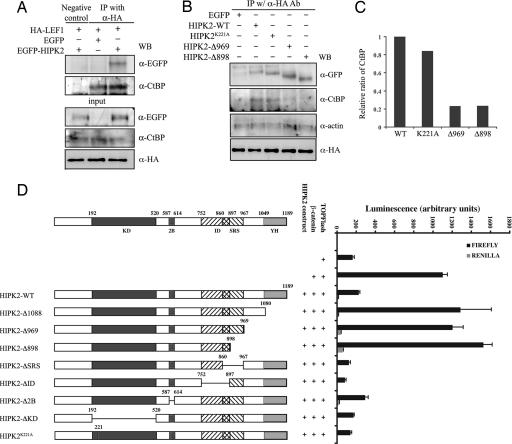
To determine whether suppression of β-catenin/LEF1-mediated gene expression by HIPK2 required the formation of a protein complex, we performed a series of coimmunoprecipitation (co-IP) assays to detect the presence of HIPK2 with LEF1 and other known transcriptional corepressors. Interestingly, classical corepressors, such as HDAC1, were not detected in the HIPK2–β-catenin–LEF1 complex. Furthermore, HIPK2 continued to suppress β-catenin luciferase activity even in the presence of trichostatin A (TSA), a specific inhibitor for HDACs (G.W. and E.J.H., unpublished data). In contrast, the transcriptional corepressor C-terminal binding protein (CtBP) was consistently detected in the complex with LEF1 and HIPK2 in the nuclear extracts, despite the overall reduction of CtBP level in the cytosol (Fig. 4A) (29). In the absence of HIPK2, CtBP can be detected in a protein complex with LEF1, albeit at a much lower affinity (Fig. 4A). The recruitment of CtBP to the HIPK2–LEF1 complex did not require the kinase activity of HIPK2. In contrast, HIPK2 mutants, HIPK2-Δ969 and HIPK2-Δ898, which lacked the C-terminus YH domain and the adjacent consensus CtBP binding motif PLNLS (amino acids 1031–1035), showed a marked reduction in their ability to form a protein complex with CtBP (Fig. 4 B and C). A series of luciferase assays confirmed that HIPK2 mutants with deletions involving the YH domain or adjacent regions, such as HIPK2-Δ1088, HIPK2-Δ969, and HIPK2-Δ898, failed to suppress the β-catenin-activated TOPFlash activity (Fig. 4D). In contrast, deletion of the kinase domain (HIPK2-ΔKD), the alternatively spliced exon 2B (HIPK2-Δ2B), Brn3a-interacting domain (HIPK2-ΔID), or the speckle-retention signal (HIPK2-ΔSRS), did not affect the suppressive activity of HIPK2 (Fig. 4D).
Fig. 4.
The YH domain in the C terminus of HIPK2 is required to recruit transcriptional corepressor CtBP and suppress β-catenin-mediated transcription. (A) Co-IP shows that HIPK2 can be detected in a protein complex with LEF1 and transcriptional corepressor CtBP. (B and C) The interaction between HIPK2 and CtBP requires the YH domain in the C terminus of HIPK2 because HIPK2 mutants lacking this domain, including HIPK2-Δ969 and HIPK2-Δ898, fail to coimmunoprecipitate with CtBP. Quantification of the interaction between HIPK2 and CtBP is shown in C. (D) Deletion of the YH domain of HIPK2 abolishes the ability of HIPK2 to suppress β-catenin. Truncation mutants in the HIPK2 C terminus (HIPK2-Δ1088, HIPK2-Δ969, and HIPK2-Δ898), the kinase domain (HIPK2-ΔKD), the Brn3a-interacting domain (HIPK2-ΔID), and the speckle-retention signal (HIPK2-ΔSRS) are generated to map the domain required for the suppressor activity of HIPK2. Deletion of the YH domain in the C terminus completely abolishes the suppressor effect of HIPK2, whereas loss of the kinase domain, the Brn3a-interacting domain, or speckle-retention signal has no detectable effect.
Two-Stage Skin Carcinogenesis Treatment Reveals Tumor Suppressor Function of HIPK2.
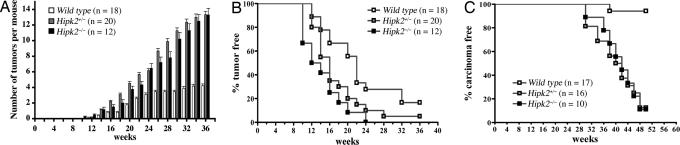
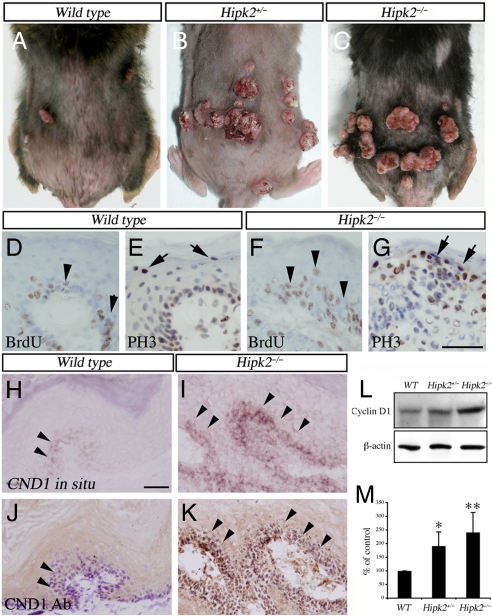
Given the important role of cyclin D1 as an oncogene, we reasoned that an increase in cyclin D1 might predispose Hipk2−/− mutants to tumor formation. Consistent with this prediction, Hipk2−/− MEFs formed more colonies than WT MEFs under clonogenic culture conditions (SI Fig. 10). Transfection with an activated form of H-ras (H-rasG12V) further induced enlarged colonies in Hipk2−/− MEFs (SI Fig. 10), suggesting that Ras activation synergized with a loss of HIPK2 to increase the proliferative potentials. Although neither Hipk2+/− nor Hipk2−/− mutant mice developed spontaneous tumors, when challenged with the two-stage skin carcinogenesis protocol (34), both showed an ≈3-fold increase in tumor number compared with that in WT (Figs. 5A and 6 A–C). Consistent with previous studies, all tumors, regardless of genotype, contained the common CTA mutation in H-ras (SI Fig. 11). The tumor-free latency was significantly shorter in Hipk2+/− and Hipk2−/− mutants. By 24 weeks, essentially every Hipk2−/− mouse had developed skin tumors (Fig. 5B). Whereas tumors in all WT mice were benign squamous papillomas, many Hipk2+/− and Hipk2−/− mutants showed features of carcinoma in situ or invasive squamous cell carcinoma (Fig. 5C). Similar to the Hipk2−/− MEFs and keratinocytes in culture, tumor cells in Hipk2−/− mutants showed a higher labeling index for BrdU and phosphohistone 3 (PH3) in the suprabasal layers of tumors (Fig. 6 D–G), a feature associated with a high probability of malignant progression (35). Consistent with the results from keratinocytes and MEFs (Fig. 3 A–C), tumors from Hipk2+/− and Hipk2−/− showed a higher level of cyclin D1 (Fig. 6 I and K–M). Although cyclin D1+ cells in WT tumors were restricted to the basal layer, many such cells in Hipk2−/− tumors extended to the suprabasal layers (Fig. 6 H and J, arrowheads). Together, these results supported the notion that loss of HIPK2 and DMBA-induced H-ras activation acted synergistically to promote skin tumorigenesis in Hipk2−/− mutants.
Fig. 5.
Increased propensity of skin carcinogenesis in Hipk2+/− and Hipk2−/− mutant mice. (A) Two-stage skin carcinogenesis paradigm induces more tumors in Hipk2+/− and Hipk2−/− mutants compared with WT littermates (P < 0.0001). (B) Incubation time for tumor formation is much shorter in Hipk2+/− and Hipk2−/− mutants (P = 0.0151 for WT and Hipk2+/−; P = 0.004 for WT and Hipk2−/−). (C) A higher percentage of the skin tumors in Hipk2+/− and Hipk2−/− mutants develop into carcinoma (P < 0.0001). Data in B and C are presented by using the Kaplan–Meier method, and statistical analyses are performed by using Student's t test.
Fig. 6.
Increased cell proliferation and cyclin D1 expression in tumors from Hipk2+/− and Hipk2−/− mutants. (A–C) Gross images of skin tumors in WT, Hipk2+/−, and Hipk2−/− mice. (D–I) Histopathological analyses of the squamous neoplasms show increased BrdU (arrowheads) and phosphohistone 3 (PH3) (arrows) labeling indices in Hipk2−/− mutants. (H–K) In situ hybridization and immunohistochemistry in adjacent sections show that cyclin D1 mRNA and protein levels (arrowheads) are significantly increased in the tumors from Hipk2−/− mutants. (Scale bars: 50 μm.) (L and M) Western blot analyses confirm increased cyclin D1 protein in tumors from Hipk2+/− and Hipk2−/− mice. *, P = 0.027 between WT and Hipk2+/−; **, P = 0.00358 between WT and Hipk2−/−; Student's t test (n = 3).
Discussion
Transcriptional Regulation of Cyclin D1 by β-Catenin and HIPK2.
Overexpression of cyclin D1 is commonly detected in colon, breast, and skin cancers (36). Several important regulatory elements have been identified in the promoter sequence of the cyclin D1 gene, suggesting that transcriptional control of cyclin D1 may harbor potential tumorigenic targets (13, 14). Results in this study indicate that HIPK2 is an important component of an intrinsic transcriptional mechanism that negatively regulates cyclin D1 expression (Figs. 2, 4, and 6). Our results indicate that the C terminus of HIPK2 is required to interact with the corepressor, CtBP, which in turn promotes the suppressor function of HIPK2 (Fig. 4) (37). Deletion of the YH domain (HIPK2-Δ1088) and the putative CtBP binding motif (HIPK2-Δ969 and HIPK2-Δ898) completely abolishes the repressor effect of HIPK2 in TOPFlash reporter assays (Fig. 4). Results obtained by using construct HIPK2-Δ1088 are particularly intriguing because they suggest that, at least in the context of β-catenin-dependent activation of TOPFlash, the integrity of the C-terminal region of HIPK2 is important for the recruitment of additional transcriptional corepressors, such as CtBP.
Transcriptional Control of Proliferation in Epidermal Stem Cells.
Lineage tracing by using genetic targeting, pulse chase with labeled nucleotides, and transplantation experiments have provided solid evidence that epidermal stem cells contribute to the formation and homeostasis of epidermis and its adnexal structures (3, 4, 38, 39). Within its niche, epidermal stem cells are subjected to a variety of exogenous factors that control its growth, proliferation, migration, and differentiation (40). One prominent mechanism that controls the development and maintenance of epidermal stem cells is the Wnt/β-catenin signaling pathway. Conditional deletion of β-catenin during embryogenesis leads to a blockage of initial placode and hair follicle formation during morphogenesis and a complete loss of hair cycling during the first catagen phase in postnatal life (18). These results underscore the dual roles of β-catenin in cell fate determination and the maintenance of hair follicle formation, suggesting that, in the absence of β-catenin, epidermal stem cells favor a fate toward epidermis. Interestingly, a high level of LEF1 is expressed in the hair follicles during morphogenesis, and targeted deletion of Lef1 also leads to phenotypes similar to β-catenin-conditional mutants with a severe reduction of whiskers and hair follicles (41). Together these results indicate a cooperative role of β-catenin and LEF1 in the morphogenesis and fate determination in hair follicles.
Based on the characteristics of HIPK2 as a transcriptional corepressor of the β-catenin–LEF1 complex, one would predict that a common set of target genes may be either up- or down-regulated in the epidermal stem cells of Hipk2−/− mutants and transgenic mice that express a stabilized form of β-catenin (ΔN/ΔN). Indeed, both mice show a similar up-regulation in cyclin D1. However, several prominent differences are present. First, the number of the two distinct populations of stem cells is increased by 2- to 3-fold in Hipk2−/− mutants (Fig. 1), but not in ΔN/ΔN mice at comparable ages (42). It is possible that loss of HIPK2 may perturb the expression of additional target genes, which contributes to such differences. Alternatively, it is possible that the K14 promoter, used to generate the ΔN/ΔN mice, selectively targets the constitutively active form of β-catenin to a more restricted population of epidermal progenitor cells and preserves the microenvironment in the stem cell niche. In contrast, loss of HIPK2 may affect a broader population of cells, including dermal fibroblasts, infiltrating T cells, and macrophages.
Tumor Suppressor Effects of HIPK2 in Skin Carcinogenesis.
Although HIPK2 has been implicated in regulating p53-dependent cell growth and apoptosis (25, 26), several lines of evidence indicate that this mechanism may not be conserved across different species. First, the patterns of cell growth in primary and E1A-transformed Hipk2−/− MEFs are substantially different from that of p53−/− mutants (SI Fig. 12). Second, the induction of total p53 protein and phosphorylation on several critical Ser and Thr residues in response to various cell injury conditions remains intact in Hipk2−/− MEFs (SI Fig. 12). Finally, the pattern of tumor growth and the spectrum of differentiation in Hipk2+/− and Hipk2−/− mutants, such as increased skin tumor formation and multilineage features, is very different from the paradoxical reduction of papilloma number in p53−/− mutants (43). Together these results argue that HIPK2 and p53 have different, nonoverlapping functions in the control of cell growth and tumorigenesis. Indeed, preliminary results indicate that partial loss of both HIPK2 and p53 synergistically enhances radiation-induced thymic lymphoma (J.-H.M. and A.B., unpublished data), suggesting that HIPK2 and p53 act in trans, rather than in cis, to regulate cell growth and tumorigenesis.
Regulators of the G1 cell cycle progression play an important role in skin carcinogenesis. For instance, conditional overexpression of cyclin D1 using the Cre-lox approach increases carcinogenesis, even in genetic backgrounds known to confer resistance to DMBA/TPA treatment (11). Conversely, loss of cyclin D1 results in a marked decrease in skin tumors in transplant recipients of keratinocytes infected with a retrovirus carrying v-H-ras or transgenic mice expressing v-H-ras (12). Our results indicate that loss of HIPK2 “de-represses” cyclin D1 transcription. It is most likely that the higher level of cyclin D1 in keratinocytes and epidermal progenitor cells of Hipk2−/− mutants predisposes these mice to more skin tumors with much shorter latency. Indeed, both Hipk2+/− and Hipk2−/− mice are more prone to tumorigenesis (Figs. 5 and 6 and SI Fig. 13). Consistent with this observation, MEFs and keratinocytes derived from Hipk2+/− mice showed faster growth rate, cell cycle abnormalities, and increase in cyclin D1, suggesting that haploinsufficiency could induce these phenotypes.
The tumorigenic potentials of another member in the HIPK family, HIPK1, have been investigated by Kondo et al. (27). Unlike Hipk2-null cells, Hipk1−/− MEFs show a reduced proliferative potential and increased sensitivity to cell death under high-dose γ-irradiation. Most intriguingly, mice lacking HIPK1 have reduced tumor formation and malignant transformation under the same carcinogenic conditions. Although these distinct differences raise the possibility that HIPK1 and HIPK2 may have opposite functions, it is possible that HIPK2 may compensate for the loss of HIPK1. One unique feature of the skin tumors in Hipk2−/− mutants is that the majority of tumors in Hipk2+/− and Hipk2−/− mutants progress into carcinoma in situ or invasive carcinoma (Figs. 5 and 6 and SI Fig. 13), raising the possibility that loss of HIPK2 may lead to dysregulation in these or other unidentified genetic pathways, e.g., Tgfβ1, Smad3, ubiquitin ligase Fbxw7/Cdc4, or Pten (5–9).
Materials and Methods
Characterization of Primary and Stable MEFs.
Primary MEFs were isolated from embryonic day (E) 13.5 embryos and maintained in DMEM and 10% FBS. All experiments were performed on MEFs at passage two to three. For establishment of stable MEF cell lines, primary MEFs were transfected with plasmid containing viral oncogene E1A. Positive clones were isolated, expanded, and maintained under puromycin (2.5 μg/ml; Invitrogen, San Diego, CA). Standard 3T3 cell culture protocol was used to characterize the proliferative potentials of WT and Hipk2−/− MEFs. For FACS analyses of cell cycle, MEFs were cultured in the presence of serum, trypsinized, washed twice with PBS, and fixed with ice-cold 70% ethanol for 3 h. After washing with PBS, cells were incubated in 0.25 mg/ml of RNaseA in PBS for 30 min at 37°C and stained with propidium iodide (Molecular Probes, Carlsbad, CA) at a final concentration of 20 μg/ml for 30 min at 4°C. DNA content was measured by using FACSCalibur and analyzed by CellQuest software (Becton Dickinson, San Jose, CA).
To determine the G1–S transition of the cell cycle, MEFs were plated on six-well plates at 2 × 105 cells per well. After 48 h, cells were washed twice with DMEM and then incubated in a low concentration of serum (0.1%) for 72 h. Cells were allow to reenter the cell cycle by replacing the culture medium with that containing 10% FCS. For each time point, cells were pulse-labeled with BrdU (50 mg/kg) for 1 h, harvested, and stained with BrdU antibody (Novocastra, Norwell, MA) and propidium iodide. BrdU-positive cells were counted under microscope and represented as the percentage of the total number of cells.
Mouse Primary Keratinocyte Culture and FACS Analysis of Epidermal Stem Cells.
For primary keratinocyte culture, full-thickness skin taken from P1–3 mice was treated with dispase overnight at 4°C, and the epidermis was peeled off from the dermis and trypsinized to create a single-cell suspension. Cells were plated in EMEM/EBSS medium (BioWhittaker, Walkersville, MD) supplemented with 10% chelexed FBS, 0.05 mM CaCl2, penicillin/streptomycin, and 0.2 μg/ml amphotericin B at 37°C under 5% CO2 in dishes precoated with fibronection and collagen solution (Sigma–Aldrich, St. Louis, MO) for 5 h. Unattached cells were removed by washing with PBS, and attached cells were further cultured in the presence of EGF at 50 ng/ml. For FACS analyses, single-cell suspension of primary mouse keratinocytes was isolated from P1–3, P42, or P56 mouse dorsal skin as described above. One million cells were labeled with anti-α6-integrin antibody conjugated to FITC and anti-CD34 conjugated to Phycoerithrin (PharMingen, San Diego, CA) for 30 min at room temperature. Cells were then analyzed by using FACSCalibur and analyzed by using CellQuest software (Becton Dickinson).
See SI Materials and Methods for detailed experimental procedures for histology, luciferase assays, qRT-PCR, and co-IP.
Supplementary Material
Acknowledgments
We thank Drs. W. Holleran and W. Lowry for the protocols to isolate keratinocytes and epidermal stem cells; Drs. P. Leboit, B. Ruben, and S. Kakar for reviewing the skin tumor pathology; and Drs. R.T. Moon and O. Tetsu for valuable reagents. This work was supported by the University of California, San Francisco, Medical Student Research Program (S.K.); National Institutes of Health Grants NS48393 (to E.J.H.), NS44223 (to E.J.H.), and CA84244 (to A.B.); Department of Energy Grant DE-FG02-03ER63630 (to A.B.); and a Veterans Affairs Merit Review Award (to E.J.H.).
Abbreviations
- En
embryonic day n
- Pn
postnatal day n
- LRC
label-retaining cells
- TA cells
transient amplifying cells
- LEF1
lymphoid enhancer-binding factor 1
- TCF
T cell factor
- MEF
mouse embryonic fibroblast.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703213104/DC1.
References
- 1.Owens DM, Watt FM. Nat Rev Cancer. 2003;3:444–451. doi: 10.1038/nrc1096. [DOI] [PubMed] [Google Scholar]
- 2.Perez-Losada J, Balmain A. Nat Rev Cancer. 2003;3:434–443. doi: 10.1038/nrc1095. [DOI] [PubMed] [Google Scholar]
- 3.Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G. Nat Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- 4.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Cui W, Fowlis DJ, Bryson S, Duffie E, Ireland H, Balmain A, Akhurst RJ. Cell. 1996;86:531–542. doi: 10.1016/s0092-8674(00)80127-0. [DOI] [PubMed] [Google Scholar]
- 6.Glick AB, Lee MM, Darwiche N, Kulkarni AB, Karlsson S, Yuspa SH. Genes Dev. 1994;8:2429–2440. doi: 10.1101/gad.8.20.2429. [DOI] [PubMed] [Google Scholar]
- 7.Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, Brown K, Bryson S, Balmain A. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 8.Vijayachandra K, Lee J, Glick AB. Cancer Res. 2003;63:3447–3452. [PubMed] [Google Scholar]
- 9.Mao JH, To MD, Perez-Losada J, Wu D, Del Rosario R, Balmain A. Genes Dev. 2004;18:1800–1805. doi: 10.1101/gad.1213804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao JH, Balmain A. Curr Opin Genet Dev. 2003;13:14–19. doi: 10.1016/s0959-437x(03)00005-4. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto H, Ochiya T, Takeshita F, Toriyama-Baba H, Hirai K, Sasaki H, Sasaki H, Sakamoto H, Yoshida T, Saito I, Terada M. Cancer Res. 2002;62:1641–1647. [PubMed] [Google Scholar]
- 12.Robles AI, Rodriguez-Puebla ML, Glick AB, Trempus C, Hansen L, Sicinski P, Tennant RW, Weinberg RA, Yuspa SH, Conti CJ. Genes Dev. 1998;12:2469–2474. doi: 10.1101/gad.12.16.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tetsu O, McCormick F. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 14.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Logan CY, Nusse R. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 16.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. Nat Rev Genet. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 17.Reya T, Clevers H. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 18.Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 19.Chan EF, Gat U, McNiff JM, Fuchs E. Nat Genet. 1999;21:410–413. doi: 10.1038/7747. [DOI] [PubMed] [Google Scholar]
- 20.Gat U, DasGupta R, Degenstein L, Fuchs E. Cell. 1998;95:605–614. doi: 10.1016/s0092-8674(00)81631-1. [DOI] [PubMed] [Google Scholar]
- 21.Kim YH, Choi CY, Lee SJ, Conti MA, Kim Y. J Biol Chem. 1998;273:25875–25879. doi: 10.1074/jbc.273.40.25875. [DOI] [PubMed] [Google Scholar]
- 22.Wiggins AK, Wei G, Doxakis E, Wong C, Tang AA, Zang K, Luo EJ, Neve RL, Reichardt LF, Huang EJ. J Cell Biol. 2004;167:257–267. doi: 10.1083/jcb.200406131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doxakis E, Huang EJ, Davies AM. Curr Biol. 2004;14:1761–1765. doi: 10.1016/j.cub.2004.09.050. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Pho V, Bonasera SJ, Holzmann J, Tang AT, Hellmuth J, Tang S, Janak PH, Tecott LH, Huang EJ. Nat Neurosci. 2007;10:77–86. doi: 10.1038/nn1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G, et al. Nat Cell Biol. 2002;4:11–19. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- 26.Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W, Will H, Schmitz ML. Nat Cell Biol. 2002;4:1–10. doi: 10.1038/ncb715. [DOI] [PubMed] [Google Scholar]
- 27.Kondo S, Lu Y, Debbas M, Lin AW, Sarosi I, Itie A, Wakeham A, Tuan J, Saris C, Elliott G, et al. Proc Natl Acad Sci USA. 2003;100:5431–5436. doi: 10.1073/pnas.0530308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Isono K, Nemoto K, Li Y, Takada Y, Suzuki R, Katsuki M, Nakagawara A, Koseki H. Mol Cell Biol. 2006;26:2758–2771. doi: 10.1128/MCB.26.7.2758-2771.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q, Yoshimatsu Y, Hildebrand J, Frisch SM, Goodman RH. Cell. 2003;115:177–186. doi: 10.1016/s0092-8674(03)00802-x. [DOI] [PubMed] [Google Scholar]
- 30.Kanei-Ishii C, Ninomiya-Tsuji J, Tanikawa J, Nomura T, Ishitani T, Kishida S, Kokura K, Kurahashi T, Ichikawa-Iwata E, Kim Y, et al. Genes Dev. 2004;18:816–829. doi: 10.1101/gad.1170604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierantoni GM, Fedele M, Pentimalli F, Benvenuto G, Pero R, Viglietto G, Santoro M, Chiariotti L, Fusco A. Oncogene. 2001;20:6132–6141. doi: 10.1038/sj.onc.1204635. [DOI] [PubMed] [Google Scholar]
- 32.Taylor G, Lehrer MS, Jensen PJ, Sun TT, Lavker RM. Cell. 2000;102:451–461. doi: 10.1016/s0092-8674(00)00050-7. [DOI] [PubMed] [Google Scholar]
- 33.Massague J. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 34.Balmain A, Ramsden M, Bowden GT, Smith J. Nature. 1984;307:658–660. doi: 10.1038/307658a0. [DOI] [PubMed] [Google Scholar]
- 35.Hennings H, Shores R, Mitchell P, Spangler EF, Yuspa SH. Carcinogenesis. 1985;6:1607–1610. doi: 10.1093/carcin/6.11.1607. [DOI] [PubMed] [Google Scholar]
- 36.Hall M, Peters G. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Q, Nottke A, Goodman RH. Proc Natl Acad Sci USA. 2005;102:2802–2807. doi: 10.1073/pnas.0409373102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, Cotsarelis G. Nat Med. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- 40.Fuchs E, Tumbar T, Guasch G. Cell. 2004;116:769–778. doi: 10.1016/s0092-8674(04)00255-7. [DOI] [PubMed] [Google Scholar]
- 41.van Genderen C, Okamura RM, Farinas I, Quo RG, Parslow TG, Bruhn L, Grosschedl R. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 42.Lowry WE, Blanpain C, Nowak JA, Guasch G, Lewis L, Fuchs E. Genes Dev. 2005;19:1596–1611. doi: 10.1101/gad.1324905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kemp CJ, Donehower LA, Bradley A, Balmain A. Cell. 1993;74:813–822. doi: 10.1016/0092-8674(93)90461-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.