Abstract
Messenger RNA transcripts are coated from cap to tail with a dynamic combination of RNA binding proteins that process, package, and ultimately regulate the fate of mature transcripts. One class of RNA binding proteins essential for multiple aspects of mRNA metabolism consists of the poly(A) binding proteins. Previous studies have concentrated on the canonical RNA recognition motif-containing poly(A) binding proteins as the sole family of poly(A)-specific RNA binding proteins. In this study, we present evidence for a previously uncharacterized poly(A) recognition motif consisting of tandem CCCH zinc fingers. We have probed the nucleic acid binding properties of a yeast protein, Nab2, that contains this zinc finger motif. Results of this study reveal that the seven tandem CCCH zinc fingers of Nab2 specifically bind to polyadenosine RNA with high affinity. Furthermore, we demonstrate that a human protein, ZC3H14, which contains CCCH zinc fingers homologous to those found in Nab2, also specifically binds polyadenosine RNA. Thus, we propose that these proteins are members of an evolutionarily conserved family of poly(A) RNA binding proteins that recognize poly(A) RNA through a fundamentally different mechanism than previously characterized RNA recognition motif-containing poly(A) binding proteins.
Keywords: CCCH zinc finger, poly(A) binding protein, RNA binding
The fate of an mRNA transcript is largely determined by its associated RNA binding proteins. A wide variety of RNA binding proteins associate with the nascent transcript cotranscriptionally and act as processing factors involved in capping, splicing, cleavage, and polyadenylation of the transcript (1). Additional RNA binding proteins package the mRNA into complexes that regulate transcript stability (2), promote export from the nucleus (3), and modulate translation (4). Accordingly, the protein constituents of these mRNA ribonucleoprotein (mRNP) complexes have been accurately equated to posttranscriptional activators and repressors (5, 6) of gene expression.
One family of proteins that are key posttranscriptional regulators of gene expression is composed of the poly(A) binding proteins (Pabs). Functional studies in a wide variety of organisms ranging from yeast to humans have demonstrated that members of this evolutionarily conserved protein family (reviewed in refs. 7–9) directly contact the poly(A) tail of mRNA transcripts to regulate transcript polyadenylation (10, 11), translation (12–16), stability (17, 18), and possibly nuclear export (19, 20). All known Pab family members specifically bind to poly(A) RNA via at least one RNA recognition motif (RRM) (21, 22). For example, the primary cytoplasmic Saccharomyces cerevisiae Pab, poly(A) binding protein 1 (Pab1), contains four RRMs that can each bind RNA with varying specificity and affinity (21–23).
Although all conventional Pabs interact with RNA through RRM domains (24), there is evidence to suggest that at least one other type of RNA binding motif may confer specific binding to polyadenosine RNA (9, 25–27). The yeast protein, nuclear poly(A) binding protein 2 (Nab2), lacks RRM domains and instead contains two other potential RNA binding motifs, an arginine–glycine–glycine (RGG) repeat domain and seven tandem CCCH zinc fingers (25, 28). Nab2 was originally identified as an essential heterogeneous nuclear ribonucleoprotein (hnRNP) that copurified with polyadenylated RNA transcripts (25). Subsequent studies revealed that Nab2 shuttles between the nucleus and the cytoplasm and is required for both nuclear export and proper polyadenylation of mRNA transcripts (26, 28).
The original copurification of Nab2 with polyadenylated RNA transcripts merely indicated that Nab2 associates with RNA transcripts that contain poly(A) sequences and did not provide any information about the sequence specificity of this class of zinc finger proteins. Further characterization revealed that Nab2 bound to homopolymeric RNA and single-stranded DNA (25, 26). Domain analyses also suggested that the zinc finger domain could confer binding to poly(A) sepharose (25). A later study that purified several yeast hnRNPs and analyzed the copurified RNA for consensus binding motifs revealed a Nab2 consensus of (A)11G (27) but did not directly examine binding specificity by using purified Nab2 protein. Thus, although Nab2 association with poly(A) sequences has been observed, the specificity of this interaction has not been thoroughly examined. Given that Nab2 modulates poly(A) tail length in vivo, specific recognition of polyadenosine could be a key aspect of Nab2 function. Taken together, these results suggest that Nab2 may be a member of a new class of poly(A)-specific RNA binding proteins that recognizes poly(A) sequences in an RRM-independent manner.
To directly test whether a protein that lacked an RRM domain could bind specifically to polyadenosine RNA, we exploited a combination of conventional gel-shift assays and fluorescence correlation spectroscopy (FCS) (29–32) to measure the interaction between Nab2 and a variety of oligonucleotides in vitro. FCS measures the translational diffusion of fluorescently labeled oligonucleotides in solution and can distinguish between rapidly diffusing free oligonucleotides and the more slowly diffusing fluorescent oligonucleotides bound to protein (31, 32). The relative concentration of bound and free oligonucleotides can be recovered from these measurements, allowing for the direct determination of binding constants. Results of these studies indicate that Nab2 binds with nanomolar affinity to fluorescently labeled poly(A) RNA oligonucleotides. We have also investigated the specificity of this interaction through a series of competition experiments and find that Nab2 specifically binds to polyadenosine RNA as compared with other RNA or DNA sequences. Importantly, domain analyses reveal that the zinc finger domain of Nab2 mediates this sequence-specific RNA binding. To extend this study, we provide the first characterization of a human protein, ZC3H14 (zinc finger protein with CCCH motif #14), that contains CCCH zinc fingers similar to those found in Nab2. This zinc finger protein also specifically binds to polyadenosine RNA. Thus, our studies provide evidence to support the existence of evolutionarily conserved zinc finger polyadenosine RNA binding proteins.
Results
To test the hypothesis that a protein lacking an RRM domain could specifically bind polyadenosine RNA, we performed in vitro FCS-based binding experiments with purified, recombinant Nab2 and a Cy3-labeled 25-nt poly(A) RNA oligonucleotide [Cy3-poly(rA)25]. As described in Materials and Methods, a sample of concentrated Nab2 and Cy3-poly(rA)25 was prepared. Nab2 was then serially diluted while the concentration of Cy3-poly(A)25 RNA remained constant. FCS measurements were taken for each concentration of Nab2, resulting in a series of autocorrelation curves (Fig. 1A). As Nab2 is serially diluted, a smaller fraction of the Cy3-poly(rA)25 is bound to the protein, resulting in shorter average diffusion times and the corresponding leftward shift of the correlation curves. FCS analysis of free oligonucleotide in solution and oligonucleotide bound to Nab2 yields diffusion coefficients of 1.25 × 10−6 and 4.5 × 10−7 cm2/s for free RNA and Nab2-bound RNA, respectively. Global fitting of correlation curves to a multicomponent diffusion model by using these recovered diffusion coefficients returns the bound and free concentrations of Cy3-poly(rA)25 at each protein concentration [supporting information (SI) Methods]. The recovered concentration dependence of the protein-bound fraction of Cy3-poly(rA)25 is shown in Fig. 1B. By using a least-squares fitting routine, these data were fit to Eq. 2 (SI Methods) to recover an average value for the dissociation constant, Kd, of 29 ± 10 nM. This value is consistent with a previous study that examined GST-Nab2-His6 binding to poly(A)25 RNA by using a filter binding assay and measured a Kd of ≈10 nM (26). As a control, titration experiments with both Pab1, a known yeast poly(A) binding protein (9), and ovalbumin, which is not expected to interact with nucleic acids, were performed (SI Fig. 5). The Pab1 data yields a slower diffusion time (D = 2.7 × 10−7 cm2/s) because of the interaction of Pab1 with the fluorescent oligonucleotides. In contrast, FCS experiments with ovalbumin are indistinguishable from pure oligonucleotide in buffer even at high protein concentrations (2.5 μM), indicating no interaction.
Fig. 1.
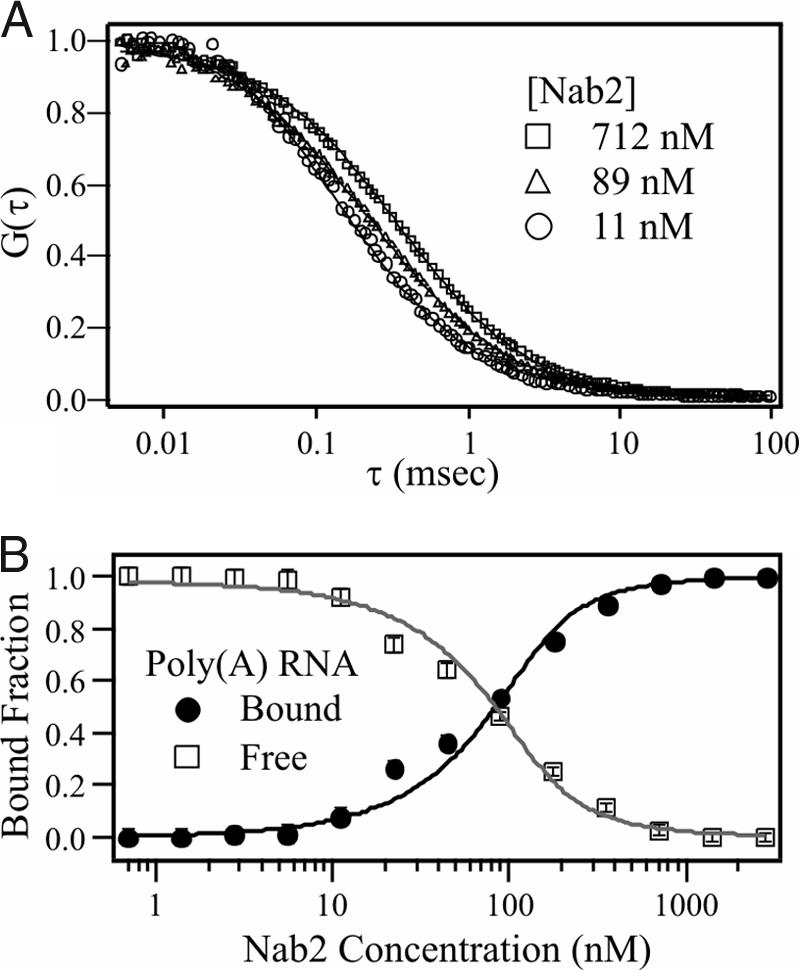
Nab2 binds polyadenosine RNA with high affinity. (A) Representative normalized FCS curves from a binding titration experiment where a concentrated sample of Nab2 (2.5 μM) was serially diluted while the Cy3-poly(A)25 RNA concentration (≈140 nM) remained constant. As Nab2 is diluted, the FCS decay curves shift to the left, indicating a decrease in the fraction of bound oligonucleotide. (B) FCS decay curves from A were used to determine the fraction of Nab2-bound Cy3-poly(A)25 RNA. Data were fit with Eq. 1 (SI Methods) by using global fit analysis to yield a Kd of 29 ± 10 nM.
To analyze the sequence specificity of the Nab2 interaction with polyadenosine RNA, a series of RNA competition experiments were performed (SI Table 1). For these experiments, a sample containing Nab2 and Cy3-poly(A)25 RNA oligonucleotide was incubated with increasing amounts of a nonfluorescent competitor oligonucleotide. Oligonucleotides that compete with the Cy3-RNA for binding to Nab2 will displace the fluorescent RNA from the protein, resulting in faster average diffusion times and smaller bound fractions of Cy3-poly(rA)25 in FCS measurements. Oligonucleotides that do not compete for binding do not produce any change in measured diffusion rates or bound fraction.
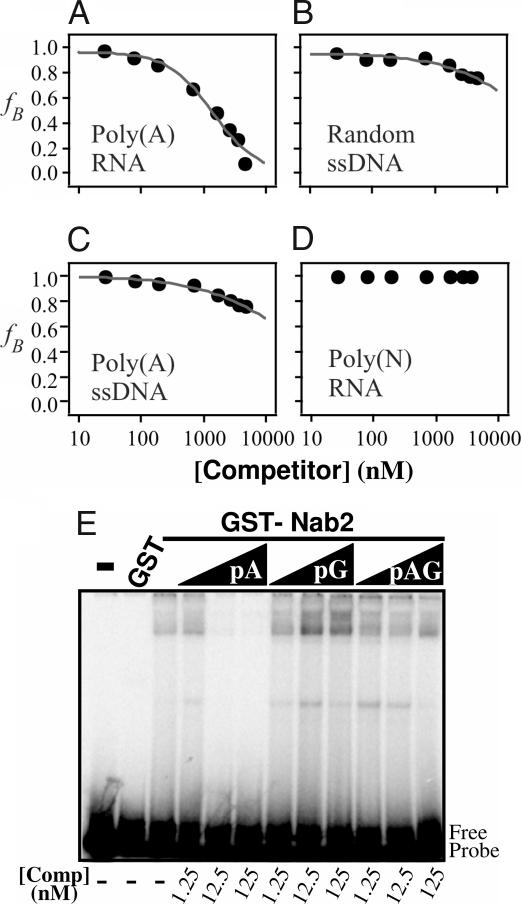
We first tested whether an unlabeled 25-nt poly(A) RNA oligonucleotide could efficiently compete with Cy3-poly(A)25 RNA for binding to Nab2. As shown in Fig. 2A, unlabeled poly(rA)25 competes efficiently for binding to Nab2. The amount of competitor needed to displace 50% of the bound fluorescent oligo (IC50) was determined by fitting the competitor concentration dependence of the bound fraction of Cy3-RNA to Eq. 3 (SI Methods). Once the IC50 has been determined, the Ki of the competitor oligonucleotide can then be computed by using Eq. 4 (SI Methods). In several independent experiments, the Ki calculated for the unlabeled poly(A) RNA oligonucleotide (Ki = 33 ± 12 nM) was virtually identical to the Kd calculated for the labeled poly(A)25 RNA oligonucleotide (Kd = 29 ± 10 nM). This analysis confirms that FCS-based competition assays can be used to assess binding to unlabeled oligonucleotides. Furthermore, these results demonstrate that the Cy3 label appears to have only minimal impact on the binding of the poly(A) oligonucleotide to Nab2.
Fig. 2.
Nab2 binds preferentially to polyadenosine RNA. Nucleic acid binding properties of Nab2 were investigated by competition experiments. Nab2 was incubated in binding buffer with Cy3-poly(A)25 RNA and increasing amounts (up to 5 μM) of an unlabeled 25-nt competitor oligonucleotide. (A) Poly(A) RNA. (B) Random sequence ssDNA (CTTCTCTAGTTCAATCTTAGCATCG). (C) Poly(A) DNA. (D) Poly(N) RNA (a pool of random 25-nt RNA oligonucleotides). Unlabeled poly(A)25 RNA competes for binding to Nab2. Both DNA oligonucleotides showed very limited competition. No competition was observed when using poly(N)25 RNA oligonucleotide. (E) GST-Nab2 (50 nM) was incubated with a radioactively labeled poly(A)25 RNA oligonucleotide probe (≈30 pM), and increasing amounts of unlabeled poly(A)25, poly(G)25, or poly(AG)12 RNA competitor were added as indicated. RNA–protein complexes were then resolved from free probe by electrophoresis on a 5% nondenaturing polyacrylamide gel. No significant competition for Nab2 binding was observed upon addition of either poly(G)25 or poly(AG)12 RNA competitor oligonucleotide.
To assess the binding specificity of the zinc finger-containing protein, Nab2, we tested the ability of a 25-nt single-stranded DNA oligonucleotide (Random ssDNA) to compete with Cy3-labeled poly(A)25 RNA oligonucleotide for binding to Nab2. As illustrated by Fig. 2B, the DNA oligonucleotide showed only minimal competition at very high concentrations of unlabeled competitor. This result indicates that Nab2 does not bind indiscriminately to nucleic acids. To determine whether Nab2 preferentially binds to RNA or single-stranded DNA, we analyzed binding of Nab2 to Cy3-labeled poly(A)25 RNA when unlabeled poly(A)25 DNA was added as competitor (Fig. 2C). Again, only minimal competition was observed, suggesting that Nab2 preferentially binds RNA rather than DNA. Because some weak non-sequence-specific binding of DNA oligonucleotides to Nab2 was observed (Fig. 2 B and C), we used FCS to directly examine Nab2 binding to single-stranded DNA using a 25-nt Cy3-labeled poly(A) DNA oligonucleotide (SI Fig. 6). The results of this analysis reveal that the interaction between Nab2 and DNA highly depends on salt concentration. In agreement with the competition experiments, Nab2 binding to Cy3-poly(A)25 DNA was observed in buffer containing 50 mM NaCl (Kd = 400 ± 170 nM). However, in buffer containing 100 mM NaCl, only very weak binding to DNA could be detected and the Kd was too weak to be determined. In contrast, binding to Cy3-poly(A)25 RNA was virtually identical in buffer containing 50 mM (Kd = 29 ± 10 nM) or 100 mM (Kd = 39 ± 16 nM) NaCl. Thus, Nab2 does display some weak binding to single-stranded DNA, but it displays a much stronger affinity for poly(A) RNA.
To determine whether Nab2 binds in a sequence-nonspecific manner to RNA, we investigated the ability of an unlabeled poly(N)25 RNA competitor oligonucleotide to compete for Nab2 binding. The poly(N) RNA sample consists of a pool of randomized 25-nt RNA oligonucleotides. Upon addition of increasing amounts of poly(N)25 RNA, no competition was detected (Fig. 2D). To determine whether Nab2 binds polyadenosine RNA specifically or merely stretches of polypurine, we used an RNA gel-shift assay to determine whether a 25-nt unlabeled poly(G) or poly(AG) competitor oligonucleotide could compete with poly(A)25 RNA for binding to Nab2. As indicated by the shift from free probe to bound complex, Nab2 binds to a radioactively labeled 25-nt poly(A) RNA oligonucleotide (Fig. 2E). This binding is specific for poly(A) because unlabeled poly(A)25 RNA oligonucleotide, but not poly(G) or poly(AG) RNA oligonucleotides, compete for binding to Nab2. These results further strengthen the argument that Nab2 is a specific polyadenosine RNA binding protein.
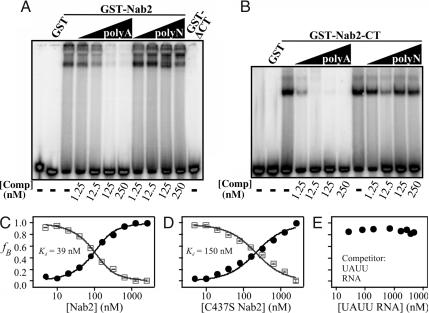
Nab2, unlike other poly(A) binding proteins, lacks an RRM RNA binding domain but instead contains two other domains, an RGG domain (33) and seven tandem CCCH zinc fingers (34), previously implicated in RNA binding. Hence, specific binding of either domain to poly(A) RNA constitutes a fundamentally different mechanism for polyadenosine RNA recognition than has been previously characterized. To determine which domain of Nab2 confers specific poly(A) RNA binding, we used an RNA gel-shift assay. Both full-length Nab2 (Fig. 3A) and the zinc finger domain alone (Fig. 3B) bind to a radioactively labeled 25-nt poly(A) RNA oligonucleotide specifically because unlabeled poly(A)25 RNA competitor, but not unlabeled poly(N)25 RNA competitor, can compete for binding. In contrast, a C-terminal truncation of Nab2 lacking the zinc fingers but still containing the RGG domain shows no binding to poly(A)25 RNA (Fig. 3A, GST-ΔCT).
Fig. 3.
The zinc finger domain of Nab2 mediates polyadenosine RNA binding. Gel-shift assays were used to determine which domain of Nab2 confers RNA binding. Fifty nanomolar full-length GST-Nab2 (A), GST-Nab2-ΔCT (amino acids 1–261) (A), or GST-Nab2-CT (amino acids 262–473) (B) was incubated with a radioactively labeled poly(A)25 RNA oligonucleotide probe (≈30 pM). RNA–protein complexes were then resolved from free probe by electrophoresis on a 5% nondenaturing polyacrylamide gel. To investigate binding specificity, either unlabeled poly(A)25 RNA or poly(N)25 RNA competitor oligonucleotides were added as indicated. No binding was observed to GST alone or Nab2 lacking the C-terminal zinc finger domain (GST-ΔCT). (C and D) Binding curves for wild-type (C) and C437S (D) Nab2 binding to Cy3-poly(A)25 RNA generated from FCS analysis. (E) An oligonucleotide containing repeats of the TTP/TIS11 target sequence, UAUU, cannot compete with Cy3-labeled poly(A)25 RNA for binding to Nab2.
To further probe the role of zinc fingers in mediating specific binding to polyadenosine RNA, we exploited a Nab2 variant, C437S, which contains a single conservative cysteine-to-serine amino acid change in the first cysteine of the sixth zinc finger. Because the last three zinc fingers of Nab2 have been implicated in Nab2 cross-linking to polyadenylated RNA transcripts in vivo (28), we predict that this substitution should disrupt the sixth zinc finger and alter the RNA binding properties of Nab2. To test this prediction, we used FCS to compare the binding affinity of wild-type Nab2 and C437S Nab2 for Cy3-poly(rA)25 (Fig. 3 C and D). This analysis yielded a binding affinity of C437S Nab2 for Cy3-poly(A)25 RNA (Kd = 150 ± 40 nM) that is almost 4-fold weaker than the affinity of wild-type Nab2 for Cy3-poly(A)25 RNA (Kd = 39 ± 3 nM). Together, these gel-shift and FCS experiments establish that a functional Nab2 zinc finger domain is both necessary and sufficient to confer preferential binding of Nab2 to polyadenosine RNA compared with random RNA.
Zinc fingers are common RNA binding motifs found in many proteins (34). One family of proteins that contain two CCCH zinc fingers, similar to those found in Nab2, consists of the human tristetraproline (TTP)/Tis11 proteins, which specifically bind to the sequence UAUU to regulate the stability of specific mRNA transcripts (35, 36). To test whether the CCCH zinc finger motifs in Nab2 might also display affinity for the sequence recognized by this family of proteins, we used FCS-based competition assays to examine binding of Nab2 to a 25-nt RNA oligonucleotide (UAUU RNA) containing tandem repeats of the TTP/Tis11 target sequence. As shown in Fig. 3E, no competition was observed, demonstrating that the zinc fingers of Nab2 show preferential binding to poly(A) RNA as compared with the target sequence of other proteins containing CCCH zinc fingers.
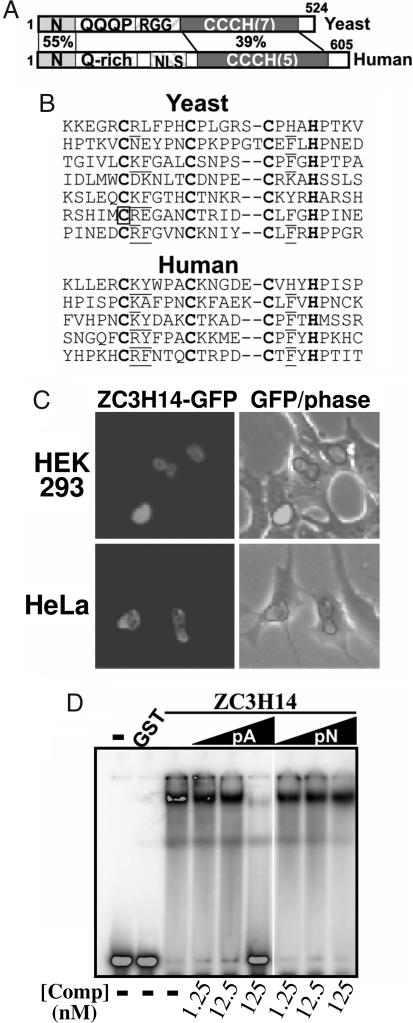
To begin to assess whether Nab2 is part of an evolutionarily conserved family of CCCH zinc finger proteins that preferentially binds polyadenosine RNA, we performed a BLAST search to identify other eukaryotic orthologues containing CCCH zinc fingers with similar spacing to those found in Nab2 (CX5CX4–6CX3H). A survey of the database reveals a single human protein with zinc finger motifs that are closely related to Nab2, ZC3H14 [also known as NY-REN-37 (37) or UKp68]. Whereas the yeast protein contains seven zinc finger domains, the human protein contains only five (Fig. 4 A and B). However, only a subset of the yeast zinc fingers has been directly implicated in binding to RNA in vivo (28), suggesting that all seven zinc fingers in the yeast protein may not be critical for RNA binding. Nab2 and ZC3H14 also share homology within their N-terminal regions (Fig. 4A), a domain required for Nab2 export from the nucleus (28) and association with other mRNA export factors (38). Whereas Nab2 contains an arginine–glycine–glycine (RGG) repeat domain that mediates nuclear import (39), ZC3H14 lacks an RGG domain and instead contains a predicted classical bipartite NLS. To test whether ZC3H14 is localized to the nucleus, we created a plasmid encoding ZC3H14-GFP and transiently transfected both HeLa and HEK cells. Similar to Nab2, the steady-state localization of ZC3H14 is nuclear (Fig. 4C). Finally, to test whether the human CCCH zinc finger protein, ZC3H14, displays nucleic acid binding properties similar to Nab2, we examined the interaction of GST-ZC3H14 with a radioactively labeled poly(A) RNA using a gel-shift assay. As shown in Fig. 4D, GST-ZC3H14, but not GST alone, binds a radioactively labeled 25-nt poly(A) RNA oligonucleotide. Furthermore, unlabeled 25-nt poly(A) RNA competitor competes for binding, whereas unlabeled 25-nt poly(N) RNA competitor does not, indicating that, like the yeast Nab2, ZC3H14 preferentially binds poly(A) RNA.
Fig. 4.
A human CCCH zinc finger protein, ZC3H14, binds specifically to polyadenosine RNA. (A) Domain alignment of Nab2 and a putative human orthologue, ZC3H14. The percentage of similar amino acid residues between the N- and C-terminal zinc finger domains is indicated. (B) The cysteine and histidine residues in the zinc fingers of Nab2 (Upper, S. cerevisiae) and ZC3H14 (Lower, human) have a similar spacing pattern (CX5CX4–6CX3H). Cysteine and histidine residues are shown in bold. The first cysteine of the sixth zinc finger, which is changed to serine in C437S Nab2, is boxed, and conserved residues are underlined. (C) Localization of ZC3H14-GFP is shown in both HEK (Upper) and HeLa (Lower) cells. (D) RNA binding properties of ZC3H14 analyzed by gel-shift assay. GST-ZC3H14, but not GST, binds poly(A)25 RNA in a gel-shift assay. GST-ZC3H14 (1.2 μM) was incubated with a radioactively labeled poly(A)25 RNA probe (≈30 pM), and increasing amounts of unlabeled poly(A)25 or poly(N)25 RNA competitor oligonucleotides were added as indicated. RNA–protein complexes were then resolved from free probe by electrophoresis on a 5% nondenaturing polyacrylamide gel. Unlabeled poly(A)25 RNA competitor efficiently competes for binding to ZC3H14, whereas unlabeled poly(N)25 competitor does not.
Discussion
In this study, we report three important findings. First, the yeast zinc finger protein, Nab2, binds with high affinity (≈30 nM) to polyadenosine RNA oligonucleotides in vitro. This interaction is specific for polyadenosine RNA, because poly(N), poly(G), poly(AG) RNA oligonucleotides, or RNA oligonucleotides containing the TTP/TIS11 binding sequence, UAUU, could not compete for binding to Nab2. Second, we show that the zinc fingers of Nab2 are necessary and sufficient to mediate this specific interaction. The sixth zinc finger at least partially contributes to this interaction because a conservative amino acid change in the first cysteine of this zinc finger causes a 4-fold decrease in the affinity of Nab2 for poly(A)25 RNA in vitro. Finally, we also present evidence to support the existence of a Nab2 orthologue in higher eukaryotes that contains highly homologous CCCH zinc finger motifs to those found in Nab2 and also preferentially binds polyadenosine RNA. Together, these results provide evidence for a previously uncharacterized family of CCCH zinc finger-containing poly(A)-specific RNA binding proteins. To the best of our knowledge, these are the only poly(A) RNA binding proteins that lack RRM domains and bind specifically to poly(A) RNA, suggesting that they may be founding members of a new class of zinc finger-containing polyadenosine RNA binding proteins.
Numerous studies have demonstrated that the poly(A) tail and its associated proteins greatly influence the fate of an mRNA transcript and hence gene expression (5, 7–9). Transcripts lacking poly(A) tails are not properly translated and are quickly degraded (40). Additionally, S. cerevisiae transcripts that are terminated by a self-cleaving ribozyme element and thereby lack poly(A) tails are at least partially retained in the nucleus (41), suggesting that the poly(A) tail may also play a role in mRNA export from the nucleus. By regulating the stability, translatability, and even subcellular localization of mRNA transcripts, the poly(A) tail and its associated proteins can act as potent posttranscriptional regulators of gene expression. We speculate that Nab2 and possibly other zinc finger-containing poly(A) binding proteins, such as ZC3H14, could function in some of the roles previously attributed to the canonical family of RRM-containing Pabs.
Poly(A)-specific RNA binding proteins were once considered histone-like proteins that merely packaged the mRNA and prevented it from being prematurely degraded. More recent work has provided a better understanding of posttranscriptional control of gene expression and has convincingly established a direct link between the poly(A) tail, poly(A) binding proteins, and the stability and translatability of numerous mRNA transcripts. Interestingly, all recent studies investigating the role of the family of poly(A) binding proteins in translation and transcript stability have solely focused on RRM-containing poly(A) binding proteins. With the identification of a zinc finger protein that displays poly(A) binding, the repertoire of this family of proteins has expanded. Given that zinc finger domains are one of the most abundant domains found in the human genome (42), this finding raises the possibility that there are additional proteins that could interact with polyadenylated mRNA transcripts and modulate gene expression.
Materials and Methods
Chemicals and Plasmids.
Chemicals were obtained from Fisher Scientific (Pittsburgh, PA), Sigma–Aldrich (St. Louis, MO), or USBiological (Swampscott, MA) unless otherwise noted. DNA manipulations were performed according to standard methods (43) and all media were prepared by using standard procedures (44). Plasmids for protein expression are described in SI Methods.
Oligonucleotides.
RNA oligonucleotides were obtained from Dharmacon (Lafayette, CO). Fluorescent RNA oligonucleotides were labeled with Cy3 on the 5′ end and PAGE-purified. All RNA oligonucleotides were deprotected by using the manufacturer-supplied buffer and protocol. Deprotected oligonucleotides were then evaporated to dryness by using a speed-vac centrifuge and frozen at −20°C. Fluorescent and nonfluorescent competitor DNA oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). All oligonucleotides used were 25 nt long. Oligonucleotides were resuspended in binding buffer (20 mM Tris·HCl, pH 8.5/50 mM NaCl/2 mM MgOAC/2 μM ZnCl2/2% glycerol) before each experiment.
Protein Expression and Purification.
To express recombinant Nab2 in E. coli, the expression plasmid, pAC2133, was transformed into BL21(DE3)pLYS cells (Novagen). Single colonies were inoculated into 2 ml of media and grown to saturation overnight. This culture was then used to inoculate 1 liter of LB media. Cells were grown at 37°C until they reached an OD600 of 0.4–0.6 nm. Cultures were then shifted to 30°C and induced with 200 μM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 5 h. Untagged Nab2 was purified by anion-exchange and gel-filtration chromatography (see SI Methods). GST-tagged proteins were purified by incubation with glutathione Sepharose 4B (GE Healthcare Life Sciences) according to the manufacturer's directions. Recombinant His-Pab1 was expressed and purified essentially as described in ref. 20.
FCS Measurements.
Two-photon FCS data were acquired to measure the interaction between fluorescent oligonucleotides and Nab2 by using a previously described home-built instrument (45, 46). Multicomponent diffusion FCS analysis was applied to determine the fraction of fluorescent oligonucleotides bound to Nab2 for different experimental conditions (32, 46, 47). All FCS curves were fit by using a standard 3D Gaussian multicomponent diffusion model (29, 48) with the volume calibrated by using rhodamine 6G. Each series of FCS experiments used global analysis to simultaneously fit all FCS curves acquired at different protein or competitor concentrations for a given fluorescent oligonucleotide. The global fits return the concentrations of bound and free fluorescent oligonucleotides for each Nab2 protein or competitor oligonucleotide concentration (32, 49, 50). Binding constants were determined by measuring the fraction of Cy3-labeled oligonucleotide bound to Nab2 at different protein concentrations as described in SI Methods.
Gel-Shift RNA Binding Assays.
Synthetic 25-nt poly(A) RNA oligonucleotides (Dharmacon) were 5′-end-labeled with [γ-32P]ATP (GE Healthcare Life Sciences) by using T4 polynucleotide kinase (Promega). RNA electrophoretic mobility-shift assays were performed by incubating 50 nM recombinant GST, GST-Nab2, GST-Nab2-CT, or GST-Nab2ΔCT with ≈30 pM radioactively labeled poly(A) RNA oligonucleotide and an increasing amount of unlabeled 25-nt competitor RNA oligonucleotide in binding buffer for 30 min at 20°C. For binding reactions containing GST-ZC3H14, a solution of 1.2 μM GST or GST-ZC3H14 was incubated with ≈30 pM radioactively labeled poly(A) RNA oligonucleotide and an increasing amount of unlabeled competitor RNA oligonucleotide. Binding reactions were loaded onto a 5% native polyacrylamide gel and electrophoresed at 30 mA in 0.3× TBE for 30 min to separate free oligonucleotide from protein–RNA complexes. Gels were dried and exposed overnight by using a PhosphorImager (Amersham).
Supplementary Material
Acknowledgments
We thank members of the A.H.C. and K.M.B. laboratories for their contributions. This work was supported by grants from the National Institutes of Health (to A.H.C. and K.M.B.).
Abbreviations
- FCS
fluorescence correlation spectroscopy
- RRM
RNA recognition motif.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701244104/DC1.
References
- 1.Maniatis T, Reed R. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 2.Beelman CA, Parker R. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 3.Dimaano C, Ullman KS. Mol Cell Biol. 2004;24:3069–3076. doi: 10.1128/MCB.24.8.3069-3076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richter JD, Sonenberg N. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 5.Moore MJ. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 6.Keene JD, Tenenbaum SA. Mol Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- 7.Gorgoni B, Gray NK. Briefings Funct Genomics Proteomics. 2004;3:125–141. doi: 10.1093/bfgp/3.2.125. [DOI] [PubMed] [Google Scholar]
- 8.Mangus DA, Evans MC, Jacobson A. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhn U, Wahle E. Biochim Biophys Acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Amrani N, Minet M, Le Gouar M, Lacroute F, Wyers F. Mol Cell Biol. 1997;17:3694–3701. doi: 10.1128/mcb.17.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerwitz Y, Kuhn U, Lilie H, Knoth A, Scheuermann T, Friedrich H, Schwarz E, Wahle E. EMBO J. 2003;22:3705–3714. doi: 10.1093/emboj/cdg347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sachs AB, Davis RW. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 13.Tarun SZ, Jr, Sachs AB. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 14.Le H, Tanguay RL, Balasta ML, Wei CC, Browning KS, Metz AM, Goss DJ, Gallie DR. J Biol Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- 15.Gallie DR. Gene. 1998;216:1–11. doi: 10.1016/s0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- 16.Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caponigro G, Parker R. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 18.Caponigro G, Parker R. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunn EF, Hammell CM, Hodge CA, Cole CN. Genes Dev. 2005;19:90–103. doi: 10.1101/gad.1267005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brune C, Munchel SE, Fischer N, Podtelejnikov AV, Weis K. RNA. 2005;11:517–531. doi: 10.1261/rna.7291205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adam SA, Nakagawa T, Swanson MS, Woodruff TK, Dreyfuss G. Mol Cell Biol. 1986;6:2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sachs AB, Davis RW, Kornberg RD. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deardorff JA, Sachs AB. J Mol Biol. 1997;269:67–81. doi: 10.1006/jmbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- 24.Maris C, Dominguez C, Allain FH. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 25.Anderson JT, Wilson SM, Datar KV, Swanson MS. Mol Cell Biol. 1993;13:2730–2741. doi: 10.1128/mcb.13.5.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hector RE, Nykamp KR, Dheur S, Anderson JT, Non PJ, Urbinati CR, Wilson SM, Minvielle-Sebastia L, Swanson MS. EMBO J. 2002;21:1800–1810. doi: 10.1093/emboj/21.7.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guisbert KK, Duncan K, Li H, Guthrie C. RNA. 2005;11:383–393. doi: 10.1261/rna.7234205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marfatia KA, Crafton EB, Green DM, Corbett AH. J Biol Chem. 2003;278:6731–6740. doi: 10.1074/jbc.M207571200. [DOI] [PubMed] [Google Scholar]
- 29.Hess ST, Huang SH, Heikal AA, Webb WW. Biochemistry. 2002;41:697–705. doi: 10.1021/bi0118512. [DOI] [PubMed] [Google Scholar]
- 30.Maiti S, Haupts U, Webb WW. Proc Natl Acad Sci USA. 1997;94:11753–11757. doi: 10.1073/pnas.94.22.11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eigen M, Rigler R. Proc Natl Acad Sci USA. 1994;91:5740–5747. doi: 10.1073/pnas.91.13.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wohland T, Friedrich K, Hovius R, Vogel H. Biochemistry. 1999;38:8671–8681. doi: 10.1021/bi990366s. [DOI] [PubMed] [Google Scholar]
- 33.Burd CG, Dreyfuss G. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 34.Brown RS. Curr Opin Struct Biol. 2005;15:94–98. doi: 10.1016/j.sbi.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Hall TM. Curr Opin Struct Biol. 2005;15:367–373. doi: 10.1016/j.sbi.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Carrick DM, Lai WS, Blackshear PJ. Arthritis Res Ther. 2004;6:248–264. doi: 10.1186/ar1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scanlan MJ, Gordan JD, Williamson B, Stockert E, Bander NH, Jongeneel V, Gure AO, Jager D, Jager E, Knuth A, et al. Int J Cancer. 1999;83:456–464. doi: 10.1002/(sici)1097-0215(19991112)83:4<456::aid-ijc4>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 38.Suntharalingam M, Alcazar-Roman AR, Wente SR. J Biol Chem. 2004;279:35384–35391. doi: 10.1074/jbc.M402044200. [DOI] [PubMed] [Google Scholar]
- 39.Lee DC, Aitchison JD. J Biol Chem. 1999;274:29031–29037. doi: 10.1074/jbc.274.41.29031. [DOI] [PubMed] [Google Scholar]
- 40.Edmonds M. Prog Nucleic Acid Res Mol Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 41.Dower K, Kuperwasser N, Merrikh H, Rosbash M. RNA. 2004;10:1888–1899. doi: 10.1261/rna.7166704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andreini C, Banci L, Bertini I, Rosato A. J Proteome Res. 2006;5:196–201. doi: 10.1021/pr050361j. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 44.Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab Press; 1997. [Google Scholar]
- 45.Berland KM, So PTC, Gratton E. Biophys J. 1995;68:694–701. doi: 10.1016/S0006-3495(95)80230-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berland KM. J Biotechnol. 2004;108:127–136. doi: 10.1016/j.jbiotec.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 47.Van Craenenbroeck E, Engelborghs Y. J Mol Recognit. 2000;13:93–100. doi: 10.1002/(SICI)1099-1352(200003/04)13:2<93::AID-JMR492>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 48.Magde D, Elson EL, Webb WW. Biopolymers. 1974;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 49.Rigler R, Pramanik A, Jonasson P, Kratz G, Jansson OT, Nygren PA, Stahl S, Ekberg K, Johansson BL, Uhlen S, et al. Proc Natl Acad Sci USA. 1999;96:13318–13323. doi: 10.1073/pnas.96.23.13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berland KM. Methods in Molecular Biology. In: Fu H, editor. Protein–Protein Interactions: Methods and Applications. Vol 261. Totowa, NJ: Humana; 2004. pp. 383–397. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.