Abstract
Herpes simplex virus glycoprotein B (gB) is one of four glycoproteins essential for viral entry and cell fusion. Recently, an x-ray structure of the nearly full-length trimeric gB ectodomain was determined. Five structural domains and two linker regions were identified in what is probably a postfusion conformation. To identify functional domains of gB, we performed random linker-insertion mutagenesis. Analyses of 81 mutants revealed that only 27 could fold to permit processing and transport of gB to the cell surface. These 27 mutants fell into three categories. Insertions into two regions excluded from the solved structure (the N terminus and the C-terminal cytoplasmic tail) had no negative effect on cell fusion and viral entry activity, identifying regions that can tolerate altered structure without loss of function. Insertions into a disordered region in domain II and the adjacent linker region also permitted partial cell fusion and viral entry activity. Insertions at 16 other positions resulted in loss of cell fusion and viral entry activity, despite detectable levels of cell surface expression. Four of these insertion sites were not included in the solved structure. Two were between residues exposed to a cavity that is too small to accommodate the 5-amino acid insertions, consistent with the solved structure being different from the native prefusion structure. Ten were between residues exposed to the surface of the trimer, identifying regions that may be critical for interactions with other viral proteins or cellular components or for transitions from the prefusion to postfusion state.
Keywords: cell fusion, glycoprotein B, viral entry, mutation
Herpes simplex virus (HSV) is a neurotropic virus that can cause recurrent mucocutaneous lesions of the oral or genital epithlelium, lesions on the cornea and, rarely, encephalitis. Infection of host cells occurs through virus attachment to the cell surface and subsequent membrane fusion to deliver the nucleocapsid containing the viral genome into the host cell. Virus attachment is mediated by binding of glycoproteins C (gC) or B (gB) to cell surface glycosaminoglycans, primarily heparan sulfate (1). Subsequent fusion between the virion envelope and a host cell membrane requires gB, glycoprotein D (gD), and the heterodimer glycoprotein H (gH)–glycoprotein L (gL) and one of the cellular receptors for gD. These receptors include herpesvirus entry mediator (HVEM), a member of the TNF receptor family; nectin-1 and nectin-2, cell adhesion molecules of the Ig superfamily; and specific sites in heparan sulfate generated by 3-O-sulfotransferases (2). It has been proposed that binding of gD to one of its cellular receptors induces gD to undergo a conformational change, resulting in interactions with gB and/or the gH–gL complex to trigger membrane fusion (3, 4).
The exact roles for gB and gH–gL in the membrane fusion process have yet to be elucidated. Recently, it was shown (5) that gH–gL, along with gD and a gD receptor, were sufficient to induce hemifusion, mixing of the outer leaflets of two lipid bilayers. However, gB was required in addition for formation of a fusion pore to permit mixing of cytoplasmic contents in cell fusion or mixing of virion contents and cytoplasm in viral entry. It has been proposed that certain regions in gH might function as a fusion peptide and as heptad repeats that fold up into six-helix bundles (6–8).
For gB, mutations in the cytoplasmic tail have been shown to modulate (enhance or reduce) cell fusion activity (described and reviewed in ref. 9), as has also been observed with the class I viral fusion proteins, HIV gp41 (10) and paramyxovirus F protein (11). An x-ray structure of a portion of the HSV type 1 (HSV-1) gB ectodomain exhibits characteristics of both class I and class II viral membrane fusion proteins (12). Surprisingly, HSV-1 gB is structurally homologous to glycoprotein G of vesicular stomatitis virus (VSV), the glycoprotein solely responsible for entry of this virus. The solved structure of gB resembles the postfusion form of G rather than the prefusion form (13, 14). Similar to the postfusion forms of class I fusion proteins, gB has a central coiled coil. Like class II fusion proteins and by analogy with VSV G protein, the presumed fusion domain of gB is a very elongated 3-stranded beta sheet with the putative bipartite fusion loops at the tip. Overall, the crystal structure of gB reveals five distinct structural domains (I, II, III, IV, and V) and two linker regions in each monomer of the trimeric ectodomain. The apparent sequential roles of gH–gL and gB in HSV-induced membrane fusion and properties of each protein suggest a previously undescribed paradigm for fusion mediated by the herpesviruses.
Previous mutagenesis studies of HSV gB to identify functional domains have had limited success because of protein misfolding and aberrant processing of most mutants (15–17). The fraction of mutants that were expressed on the cell surface and incorporated into virions could be increased by targeting mutations to regions predicted to lack secondary structure but, as a result, most mutants retained significant function or were misfolded despite the targeting. The study presented here was designed to identify functional domains of HSV-1 gB by using a transposon-based random linker-insertion mutagenesis strategy to generate a large library of mutants spanning the entire length of HSV-1 gB, a 904-amino acid type I membrane protein.
A panel of 81 unique linker-insertion mutants was generated. Characterization of these mutants permits conclusions that (i) the N terminus and distal cytoplasmic tail of gB (neither included in the x-ray structure) tolerate 5-aa insertions without loss of cell surface expression or function, indicating flexibility in structural requirements in these regions; (ii) a disordered region and adjacent linker region within the ectodomain also tolerate insertions but with some loss of function; (iii) all other insertions that permitted cell surface expression were in regions exposed to the trimer surface or to an internal cavity of the trimer (or were outside the solved structure) and were nonfunctional. These results permit predictions about the prefusion form and functional domains of gB.
Results and Discussion
Cell Surface Expression, Conformation and Processing of the gB Mutants.
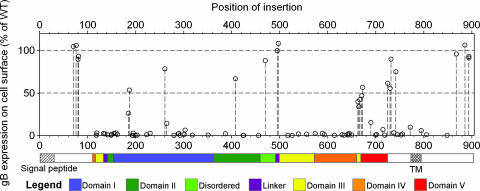
The 81 linker-insertion mutants are listed in supporting information (SI) Table 3 along with a summary of data for each. The position of each insertion is denoted by the name and number of the amino acid 5′ of the 5-aa insertion, counting from the first Met of the gB precursor. To assess expression levels on the cell surface, plasmids encoding each of the mutants were transfected into CHO cells, along with plasmids expressing gD, gH and gL, and the live cells were incubated with an anti-gB rabbit antiserum R74, for detection of gB by a cell-based ELISA (CELISA). Fig. 1 shows the levels of cell surface gB detected for each mutant, as a percentage of WT gB, in relation to the amino acid position of the insertion. A linear representation of gB is shown below the x axis and color-coded to identify regions comprising the five structural domains (I, II, III, IV, and V) and two linker regions identified in the x-ray structure (12). Mutants detectable on the cell surface were clustered at several distinct regions or positions: near the N terminus in a region not included in the x-ray structure, at two regions in domain I, at one position in domain II; at one position in the disordered central region, at two positions in the adjacent linker 2, at three positions in the small C-terminal segment of domain III, at two adjacent positions in domain V, in a region just downstream of domain V outside the solved x-ray structure, and at the membrane-distal region of the cytoplasmic tail. All other insertions abrogated cell surface expression of the mutant gBs or rendered them unrecognizable by the rabbit antiserum. Table 1 summarizes the positions of mutations that were expressed on the cell surface and indicates domains and secondary structures into which they were inserted.
Fig. 1.
Effects of insertional mutations on HSV-1 gB cell surface expression. CHO cells were transfected as for use as effector cells in cell fusion assays (with plasmids expressing the T7 RNA polymerase, gD, gH, and gL and plasmids expressing either WT or mutant gB) but not mixed with target cells. Cell surface expression of gB or gB mutant was quantified by CELISA. A linear representation of the gB polypeptide is shown below the graph. The colored bars represent the structural domains of a crystallized portion of the gB ectodomain (12); the signal peptide and transmembrane (TM) domain are indicated by hatched regions on uncolored portions of gB not included in the crystal structure. The values presented for cell surface expression of each mutant gB are means from three independent experiments expressed as percentage of WT gB values (after subtraction of background values obtained in the absence of gB expression) and as a function of position of the insertion. SDs are presented in Fig. 2 and SI Table 3.
Table 1.
Positions of HSV-1 gB linker-insertion mutations relative to structural domains and elements and listing of the mutants expressed on cell surfaces
| Region* | Expressed mutants/total† | Position of insertion for expressed mutants | Secondary structure* |
|---|---|---|---|
| N terminus | 4/4‡ | K70, K76, P80, P81 | — |
| Linker 1 | 0/2 | — | — |
| Domain I | 4/22 | I185, E187 | β5 |
| A261, Y265 | β11 | ||
| Domain II | 1/8 | D408 | — |
| Disordered | 1/1‡ | R470 | — |
| Linker 2 | 2/2‡ | I495, T497 | Linker 2 |
| Domain III | 3/9 | D663 | β36 (proximity) |
| T665, V667 | β36 | ||
| Domain IV | 0/10 | — | — |
| Domain V | 4/8 | I671, L673 | — |
| T690 | αE | ||
| A725 | — | ||
| Membrane-proximal | 4/8 | A730, F732, M742, S772 | — |
| TM | 0/1 | — | — |
| C-terminal tail | 4/6‡ | T868, N886, N894, N894 | — |
| Total | 27/81 |
Dashes indicate that no mutants expressed on the cell surface were mapped to the region indicated or to a secondary structured element.
*Domains and secondary structure from Heldwein et al. (12).
†Mutants expressed on cell surfaces at levels ≥10% of WT gB levels, as assessed by CELISA with rabbit antiserum R74.
‡Expressed mutants that retained cell fusion and viral entry activities.
Previously it was shown that certain 2-aa insertions into HSV-1 gB or the closely related HSV-2 gB (85% identity) also permitted cell surface expression (15, 16). These insertions map to the N-terminal region outside the solved structure (S48, A104: insertion sites in HSV-2 gB at positions equivalent to HSV-1 gB T53 and T109) and to the small N-terminal segment of domain III (P130), linker 1 (R136), domain I (R189, Y254, R304, T313, P358), domain II (G381, S403, G437), the disordered region between domain II and linker 2 (P483), domain V (D680), and the cytoplasmic tail (E816). (The numbering for HSV-1 gB is used for HSV-1 and HSV-2 mutants unless indicated otherwise.) In these other studies and the one described here, all insertions into the central portion of domain III and into domain IV abrogated cell surface expression. Presumably these domains and large portions, but not all, of domains I, II, and V cannot contribute to proper folding of gB if disrupted by insertions.
To assess whether mutant gBs expressed on the cell surface were grossly altered in conformation, the CELISA was repeated with a panel of conformation-dependent anti-gB mAbs (Table 2). For most mutants, the ratio of mAb binding to polyclonal antibody (R74) binding was essentially equivalent to that observed for WT gB, indicating retention of at least some aspects of WT conformation. Exceptions included T665, V667, and L673, which exhibited reduced or no binding of most or all of the mAbs, relative to binding of R74, and whose insertions are located in the smallest segment of domain III and in the adjacent region of domain V. Also, insertional mutations into the cytoplasmic tail of gB (T868 and N886) resulted in reduced binding of some or all of the mAbs, indicating that these mutations can alter conformation of the ectodomain.
Table 2.
Binding of monoclonal antibodies to selected gB mutants.
| Region* | Position of insertion† | Monoclonal antibodies‡ |
|||||
|---|---|---|---|---|---|---|---|
| II-105 | I-84-5 | II-125-4 | I-I-7 | I-144-2 | I-252-4 | ||
| N terminus | P80 | +++ | +++ | +++ | +++ | +++ | ++ |
| Domain I | I185 | +++ | +++ | +++ | +++ | ++ | +++ |
| E187 | +++ | +++ | +++ | +++ | ++ | ++ | |
| A261 | +++ | +++ | +++ | +++ | ++ | ++ | |
| Domain II | D408 | +++ | +++ | +++ | ++ | ++ | ++ |
| Disordered | R470 | +++ | +++ | +++ | +++ | +++ | +++ |
| Linker 2 | I495 | +++ | +++ | +++ | ++ | ++ | +++ |
| T497 | +++ | +++ | +++ | +++ | +++ | +++ | |
| Domain III | D663 | +++ | +++ | +++ | +++ | +++ | ++ |
| T665 | + | + | – | + | – | ++ | |
| V667 | + | – | – | – | – | – | |
| Domain V | I671 | +++ | +++ | ++ | +++ | + | ++ |
| L673 | +++ | + | + | – | – | + | |
| A725 | +++ | +++ | +++ | +++ | +++ | +++ | |
| Membrane-proximal | A730 | +++ | +++ | +++ | +++ | +++ | +++ |
| F732 | +++ | +++ | +++ | +++ | +++ | +++ | |
| M742 | +++ | +++ | +++ | +++ | +++ | +++ | |
| C-terminal tail | T868 | ++ | +++ | + | ++ | + | +++ |
| N886 | ++ | ++ | ++ | ++ | ++ | ++ | |
*From Heldwein et al. (12).
†List includes mutants for which rabbit antiserum R74 binds at >25% of WT gB levels by CELISA.
‡CELISA data were normalized to the reactivity of each mAb to WT gB. The normalized values for each mAb were then compared with values obtained using rabbit antiserum R74 for each gB mutant to determine relative changes in reactivity. The symbols represent >80% (+++), 50–80% (++), 20–50% (+), and <20% (–) reactivity of each mAb compared to R74 values for each gB mutant.
HSV-1 gB is synthesized and released into the ER as a 110-kDa high-mannose, precursor form (pgB), which associates into heat-labile oligomers (18). Transport of this trimeric precursor through the Golgi apparatus is associated with the addition of O-linked glycans and processing of N-linked glycans and with an increase in apparent molecular mass of the monomer to a 120-kDa mature form (mgB) (19). To test effects of the linker-insertion mutations on the processing of gB, Western blot analysis, using rabbit serum R74, was performed for all of the mutants. Lysates of transfected CHO cells were prepared, either heated to dissociate oligomeric forms or not heated, and then fractionated by SDS/PAGE. Levels of the two monomeric species (pgB and mgB) produced by each mutant, relative to WT gB, were assessed by using the heated samples (SI Fig. 5). Oligomeric forms could be detected in the unheated samples (SI Fig. 6) and, in some cases, in the heated samples. For all but three mutants detectable on cell surfaces by CELISA, both of the monomeric forms (pgB and mgB) as well as oligomeric forms could be detected, as expected, indicating relatively normal processing and transport to the cell surface (SI Table 3). Two of the exceptions (V667 and L673) were not detected at all by Western blot with the rabbit antiserum despite detection on the cell surface by CELISA using the same antiserum. These mutants exhibited reduced or no binding to all of the mAbs (Table 2), indicating disruption of multiple epitopes. Probably, epitopes retained by the unfixed and undenatured cell surface protein were able to bind to a subset of antibodies in the rabbit serum but these epitopes were not retained by the proteins after denaturation for Western blot analysis. The third exception (T665) was detected on Western blots as pgB and mgB, but not as oligomers, and was impaired for binding to several mAbs (Table 2).
It is perhaps not surprising that these insertions had such a large effect on conformation because they are targeted within or near residues that form many of the essential trimer contacts. Residues R661 to T669 in one protomer donate one strand to a four-strand mixed β sheet comprised mostly of strands from another protomer (12). Although oligomers of mutants T665, V667, and L673 could not be detected on Western blots, perhaps because of instability to detergent or loss of epitopes, presumably they must form to enable transport of gB to the cell surface. All mutants that failed to be detected on the cell surface by CELISA produced pgB but not mgB and, in some cases, also failed to produce oligomers, indicating significant disruption of gB folding and/or processing.
Functional Activities of the gB Mutants.
To assess cell fusion activity by a quantitative luciferase-based assay, all 81 gB mutants were coexpressed with HSV-1 gD, gH, and gL in CHO cells (effectors), which were mixed with nectin-1-expressing or HVEM-expressing CHO cells (targets). CELISAs were performed in parallel to quantitate cell surface expression of gB in the effector cell populations. The results obtained with HVEM as the fusion receptor did not differ from those obtained with nectin-1, and therefore only the latter results are presented here. Failure of a mutant to be detected on the cell surface by rabbit serum R74 correlated with absence of activity in the cell fusion assay (SI Table 3), as expected. These mutants were not studied further.
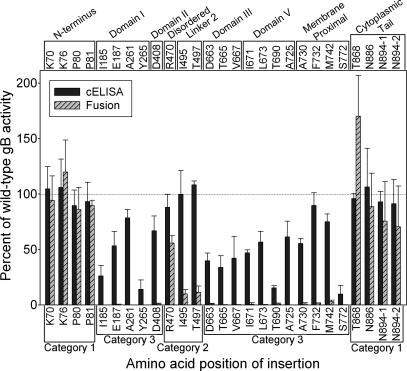
Fig. 2 presents the cell fusion results for all mutants that were detected on the cell surface, in comparison with the CELISA results obtained with the rabbit antiserum. Category 1 mutants (insertions in the N terminus or C terminus of gB) were nearly indistinguishable from WT gB in both cell fusion activity and cell surface expression, with the exception of T868 which had enhanced cell fusion activity. Category 2 mutants were located centrally in the gB ectodomain and had slightly or severely reduced cell fusion activity but nearly WT levels of cell surface expression. Category 3 mutants had little or no detectable cell fusion activity and variable levels of cell surface expression.
Fig. 2.
Cell fusion activities of the gB insertion mutants in relation to cell surface expression. CHO cells were transfected with plasmids expressing WT gB or each of the gB mutants capable of cell surface expression, other HSV-1 glycoproteins (gD, gH, and gL) and T7 polymerase (effector cells). CHO-nectin-1 cells were transfected with a plasmid carrying the firefly luciferase gene under control of the T7 promoter (target cells). One set of effector cells was used for CELISA (results shown also in Fig. 1), and the other set was mixed with the target cells for assessment of cell fusion activity by quantification of luciferase activity. Positions of the mutations with respect to structural domains are indicated across the top. Categorization of the mutants with respect to phenotype is indicated across the bottom: Category 1, indistinguishable from WT gB or with enhanced fusion activity; Category 2, normal levels of cell surface expression but reduced fusion activity; Category 3, normal or reduced levels of cell surface expression but no fusion activity. The results are expressed as percentage of WT gB activity, after subtraction of background values obtained in the absence of gB expression, and are means and SDs of three independent experiments.
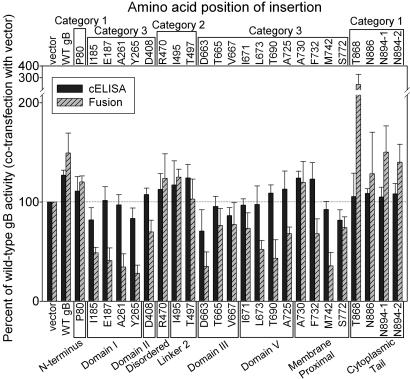
Most of the mutants shown in Fig. 2 were also coexpressed with WT gB (twice the amount of gB-expressing-plasmid total) in the fusion assay to determine whether they had dominant-negative effects on cell surface expression or cell fusion. Fig. 3 shows that the functional gB mutants (categories 1 and 2) exhibited slightly enhanced levels of cell surface expression and cell fusion activity as did samples with twice the amount of WT gB. Most of the nonfunctional category 3 mutants caused reduced cell fusion activity, to various levels, and also caused reduced cell surface expression in a subset of cases. A notable exception was mutant A730, in which the insertion is downstream of the last ordered residue in the x-ray structure (A725). It appears that WT gB may have rescued the reduced cell surface expression and undetectable activity of this mutant whereas most of the nonfunctional mutants had dominant-negative effects on WT gB cell surface expression and function.
Fig. 3.
Effects of gB insertion mutants on cell surface expression and cell fusion activity of WT gB. The CELISA and cell fusion assays were performed as described in Fig. 2 except that the transfection mixtures for each sample of effecter cells contained an additional 30 ng of plasmid expressing WT gB. Positions of the mutations with respect to structural domains are indicated across the bottom. Categorization of the mutants with respect to phenotype is indicated across the top. The results are expressed as percentage of WT gB activity (1× dose of gB plasmid mixed with added empty vector).
Selected mutants were tested for ability to complement the entry deficiency of a gB-negative viral mutant (SI Table 3). The only mutants that exhibited any viral entry activity were those that were expressed on the cell surface and had detectable levels of cell fusion activity. The two category 2 mutants with cell fusion activity reduced to 10% of WT gB levels (I495 and T497) were more active in viral entry (54% and 48% activity, respectively).
Locations of Insertions on gB Structure in Relation to Effects on Function.
The only insertions without negative consequences on cell surface expression of gB and function were outside the solved structure in the N terminus and distal portion of the C-terminal cytoplasmic domain. The N-terminal region from A31 to D110 appears to be quite flexible. To obtain well ordered crystals for the high resolution structure, the N terminus up to D103 was removed by trypsin from the expressed form of gB, A31 to A730. In a lower resolution structure of uncleaved gB (A31 to A730), the N-terminal residues from A31 to N108 remained disordered (12). The results presented here and elsewhere show that this region is also tolerant of insertions and deletions. Insertions after K70, K76, P80, and P81 had no effect on expression or function of gB, as assessed here. Similarly, insertions into HSV-2 gB at positions equivalent to HSV-1 gB T53 and T109 (S48 and A104 in HSV-2) had no effect on expression and only the insertion at A104 partially reduced function (16). Insertions K70 and K76 targeted a Lys-rich region from K68 to K76 shown to be critical for the binding of gB to heparin (20). Deletion of these amino acids impaired the binding of gB to heparin and also to heparan sulfate-expressing cells but had relatively minor effects on viral infectivity (20).
It is not surprising that insertions into the distal part of the cytoplasmic tail of gB had no negative effect of gB expression and function. Deletions of up to ≈40 aa from the C terminus of HSV-1 or HSV-2 gB either had no effect on or enhanced function in cell fusion, whereas larger deletions inhibited cell fusion (21, 22). The insertion that enhanced cell fusion activity (T868) also altered epitopes in the ectodomain, suggesting both that conformation of the cytoplasmic tail can influence conformation of the ectodomain and that this altered conformation in the ectodomain may have a role in the enhanced cell fusion activity.
Membrane-proximal insertions into the cytoplasmic tail (M806 and Y849) abrogated cell surface expression. This was found also for certain amino acid substitutions into conserved positions in the membrane-proximal region of the cytoplasmic tail of HSV-2 gB, although adjacent substitutions permitted cell surface expression and enhanced cell fusion activity (9). Amino acid substitutions at various positions in the cytoplasmic tails of HSV-1 and HSV-2 gB have been shown to enhance cell fusion (reviewed in ref. 9). The mechanistic basis for the positive and negative effects of the various mutations is not understood.
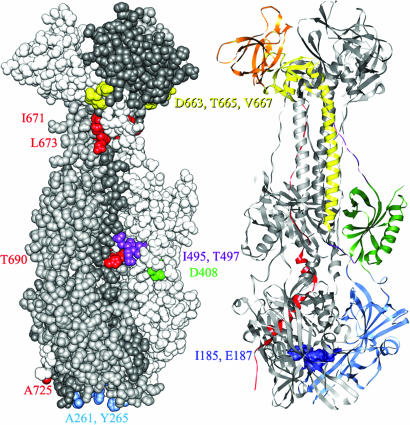
Only two of the insertions that map to the crystallized portion of the gB ectodomain (Fig. 4) permitted expression on the cell surface at WT levels and retention of cell fusion activity. Category 2 mutants I495 and T497 in the second linker region had significantly reduced cell fusion activity (10%) but higher complementing activity (≈50%). A third category 2 mutant, insertion R470, retained the highest level of fusion activity (50%) and maps to a disordered loop between domain II and linker 2 (Fig. 1). This loop was cleaved by trypsin in preparing the recombinant protein for crystallization and is a poorly conserved region where posttranslational cleavage occurs for members of the gB family encoded by other herpesviruses. Nine 2-amino acid insertions into HSV-1 or HSV-2 gB have been described to permit cell surface expression and at least partial function in cell fusion or viral entry (15, 16). With the exception of one in the cytoplasmic tail and one in the disordered central loop, all map to the surface of the crystallized portion of gB, in domain I (Y254, R304, T313), domain II (G381, G437), the N-terminal-most segment of domain III (P130) and linker 1 (R136).
Fig. 4.
Mapping of insertion mutations on the structure of gB. (Left) Space-filling model with each protomer colored a different shade of gray. The amino acids bounding each insertion site are colored according to the domain or region to which each maps, using the color code shown in Fig. 1. Insertion sites in all three protomers are colored so that all surface insertion sites can be seen in one view. (Right) Ribbon model with the domains and regions of one protomer colored according to the code in Fig. 1. The insertion sites into the cavity of the trimer are indicated by dark blue coloring of the residues displayed in space-filling mode.
Sixteen other insertions that permitted cell surface expression of gB (>10% of WT levels) were nonfunctional (category 3 mutants). Four map outside the solved structure just downstream of domain V (A730, F732, M742, and S772). The remainder were insertions between residues on the surface of the trimer or of an internal cavity (Fig. 4). Of these, only three mutants (T665 and V667 in the C-terminal-most segment of domain III and L673 in domain V) exhibited global alterations in conformation, as discussed above. The remainder could not be distinguished from WT gB in conformation and yet are nonfunctional. Four 2-aa insertions into HSV-1 or HSV-2 gB have been described to permit cell surface expression but with loss of function (15, 16). These map to domain I in the internal cavity (R189) or to the trimer surface in domains I (P358), II (S403), and V (D680).
Insertions A261 and Y265 are located in or near one of the two loops at the lower tip of domain I (Fig. 4), loops analogous to the bipartite fusion loops of VSV G (13). Loss of function caused by these insertions is consistent with recent findings that certain amino acid substitutions at W174, Y179, V259 and A261 also abrogated function in cell fusion; only V259R prevented cell surface expression of gB (23). It remains to be determined whether the two loops at the ends of domain I actually serve as internal fusion peptides. If so, it appears to be important that all protomers of the trimer have intact fusion loops because mutants A261 and Y265 were among those exerting the strongest dominant-negative effects when coexpressed with WT gB. The only other expressed insertions in domain I (I185 and E187) point toward a small cavity inside the trimer (Fig. 4). Similar cavities are also found in the postfusion forms of VSV G (13) and flavivirus protein E (24, 25). In gB this cavity is not large enough to accommodate three 5-aa insertions, supporting the idea that the solved structure of gB is different from the prefusion structure.
Insertion D408 is located within domain II which, as is also the case for a part of domain I and for an analogous domain in VSV G, resembles a canonical pleckstrin-homology (PH) domain (12, 13). PH domains in cytoplasmic molecules can serve as scaffolds for phosphoinositide and peptide binding, but it is unclear what roles the PH domains in the ectodomains of gB and VSV G may play.
HSV gB and VSV G.
Although gB cannot act alone to mediate membrane fusion, as VSV G does, it is clearly essential for HSV-induced membrane fusion. Its structural homology with VSV G invites comparisons in relating the effects of insertions in gB on function in cell fusion. Three domains of G (equivalent to domains I, II, and IV of gB) retain their folded structure in both the prefusion and postfusion forms, despite large rearrangements in their relative orientations (13, 14). Of interest, the G domain equivalent to domain II of gB is at the top of the prefusion structure but is relocated to the side of the trimer in the postfusion form, also the location for domain II in the solved structure of gB. Assuming this structure of gB is equivalent to the postfusion form of G, insertion D408 in domain II is likely to be at a site exposed also on the prefusion form, a site available for interactions with cell receptors or other viral proteins required for fusion. Insertions A261 and Y265, in one of the putative fusion loops of domain I, would presumably interfere with insertion of these loops into the target membrane but would point toward the virion envelope in both the prefusion and postfusion forms of gB, by analogy with VSV G. The VSV G monomers in the region of domain IV (elongated three-stranded β sheet with internal fusion loops equivalent to a similar region of gB domain I) are close together in the postfusion form (as in the solved gB structure) but separated in the prefusion form. Such separation would permit accommodation of insertions I185 and E187 in a prefusion structure. Insertions into gB that would likely affect hinge regions involved in reorientation of domains I, II, and IV include those in linker 2 (I495, T497) and those near the junction of domains III and V (D663, T665, V667, I671, L673). By analogy with VSV G, domain III in the postfusion form of gB is likely to differ in conformation from that in the prefusion form.
In summary, the results presented here provide support for the notion that the solved structure of gB is a postfusion form; identify regions of gB that are critical for function in membrane fusion and require further study using assays that can assess relevant protein–protein or protein–lipid interactions; and identify regions of gB that tolerate insertions without loss of function and may tolerate insertions of probes for fusion-associated alterations in conformation.
Materials and Methods
Cells and Viruses.
Cell lines used included CHO cells, CHO cells stably expressing human HVEM (26) or nectin-1 (27), Vero cells, Vero-B24 cells carrying the HSV-1 gB gene (28) and used for the propagation and titration of the gB-negative mutant HSV-1(KOS)K082 (29). The CHO cell line and derivatives were grown in Ham's F12 medium supplemented with 10% FBS, and the Vero and Vero-B24 cells were grown in DMEM supplemented with 10% FBS.
Random Linker-Insertion Mutagenesis of HSV-1 gB.
The GPS-LS linker scanning system (New England Biolabs, Ipswich, MA) was used, as recommended by the manufacturer, on the insert excised from pPEP98 (30) to generate random linker-insertion mutations of gB. After religation of the library of inserts into pCAGGS and transformation of bacteria, 81 unique gB linker-insertion mutants were isolated and sequenced by using PrimerN (30-mer) and PrimerS (30-mer) provided by the manufacturer. After removal of the transposon, each mutant plasmid was resequenced to verify the position of each insertion.
Western Blot Analysis.
CHO cells seeded in 24-well plates were transfected with 400 ng of empty vector (pCAGGS) or a plasmid expressing WT gB (pPEP98) or a gB mutant and 1.2 μl of Lipofectamine 2000 (Invitrogen, Carlsbad, CA) both diluted in Opti-MEM (Gibco, Carlsbad, CA). After 24 h of incubation, the cells were washed with PBS and lysed with 300 μl of lysis buffer (50 mM Tris, pH 8.0/150 mM NaCl/1% Nonidet P-40) containing protease inhibitor mixture (Roche Diagnostics, Indianapolis, IN). Nuclei and debris were removed by centrifugation and the supernatants mixed with sample buffer (without reducing agent) for SDS/PAGE and boiled for 5 min to detect monomeric forms of gB or not heated to detect high-molecular-weight gB oligomers. Proteins in the cell lysates were separated by electrophoresis on 4–15% gels under nonreducing conditions, and Western blot analyses were performed by using the rabbit anti-gB antiserum R74 (28).
CELISA.
CHO cells seeded in 96-well plates were transfected with 30 ng of empty vector or a plasmid expressing WT gB or a gB mutant and 0.15 μl of Lipofectamine 2000 both diluted in Opti-MEM. The cells were washed once with PBS 24 h after transfection, and CELISA was performed as described in ref. 31, using anti-gB serum R74 at 1:10,000 dilution or one of several anti-gB mAbs (32). In addition, CHO cells were transfected with all of the plasmids used to prepare effector cells for the cell fusion assays as described below, including plasmids expressing mutant or WT forms of gB, so that levels of gB expression could be assessed in replicates of the cell populations used in the fusion assays. The cells were washed once with PBS 16 h after transfection, and CELISA was performed with the anti-gB serum R74 as described above.
Cell Fusion Assay.
In parallel with CELISA, cell fusion activity of gB was measured by using a modification of a quantitative luciferase-based cell fusion assay, along with the plasmids for that assay described in ref. 30. CHO cells were seeded in 96-well plates and CHO cells stably expressing human HVEM or nectin-1 were seeded in six-well plates 1 day before transfection. CHO (effector) cells were transfected with 20 ng each of plasmids expressing the T7 RNA polymerase, gD, gH, and gL; 30 ng of empty vector or plasmid expressing either WT (pPEP98) or mutant gB; and 0.4 μl of Lipofectamine 2000. For interference assays, 30 ng of pPEP98 expressing WT gB was added to each well in addition. CHO-nectin-1 or CHO-HVEM (target) cells were transfected with 400 ng of a plasmid carrying the firefly luciferase gene under control of the T7 promoter (33), 1.8 μg of empty vector (pCAGGS) and 7 μl of of Lipofectamine 2000. Two hours after transfection, effector cells were washed once with Opti-MEM while each target cell population was washed, detached with versene (0.2 g/L EDTA in PBS), and suspended in Opti-MEM. The target cell population was overlayed (3 × 104 cells per well) on the effector population. After 10 h, the cells were washed once with PBS and lysed with 50 μl per well of 1× passive lysis buffer (Promega, Madison, WI). Expression of luciferase was quantified by adding 50 μl per well of luciferase substrate (Promega) and measuring light output with a Wallac 1420 plate reader (PerkinElmer Instruments, Norwalk, CT).
Complementation Assay.
This assay was done as described for complementation of a gD-negative virus by WT and mutant forms of gD (34), except that Vero cells in six-well plates were transfected with 1.0 μg of plasmid expressing WT or mutant forms of gB and 3.5 μl of Lipofectamine 2000 and later infected with the gB-negative mutant, HSV-1(KOS)K082. Virus stocks were prepared and titrations performed on Vero-B24 cells.
Supplementary Material
Acknowledgments
We thank N. Susmarski for excellent technical assistance. This work was supported by the U.S. Public Health Service, National Institutes of Health Grants CA 21776 and AI 36293 (to P.G.S.), National Research Service Award F30 NS049726 (to E.L.), and Medical Scientist Training Program 5T32 GM008152 (to E.L.).
Abbreviations
- CELISA
cell-based enzyme-linked immunosorbent assay
- gB
glycoprotein B
- gD
glycoproteins D
- gH
glycoprotein H
- gL
glycoprotein L
- HSV
herpes simplex virus
- HSV-n
herpes simplex virus type n
- HVEM
herpesvirus entry mediator
- mg
mature form glycoprotein
- pg
precursor form glycoprotein
- VSV
vesicular stomatitis virus.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705926104/DC1.
References
- 1.Shukla D, Spear PG. J Clin Invest. 2001;108:503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spear PG, Eisenberg RJ, Cohen GH. Virology. 2000;275:1–8. doi: 10.1006/viro.2000.0529. [DOI] [PubMed] [Google Scholar]
- 3.Krummenacher C, Supekar VM, Whitbeck JC, Lazear E, Connolly SA, Eisenberg RJ, Cohen GH, Wiley DC, Carfi A. EMBO J. 2005;24:4144–4153. doi: 10.1038/sj.emboj.7600875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rey FA. EMBO Rep. 2006;7:1000–1005. doi: 10.1038/sj.embor.7400807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramanian RP, Geraghty RJ. Proc Natl Acad Sci USA. 2007;104:2903–2908. doi: 10.1073/pnas.0608374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gianni T, Martelli PL, Casadio R, Campadelli-Fiume G. J Virol. 2005;79:2931–2940. doi: 10.1128/JVI.79.5.2931-2940.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gianni T, Menotti L, Campadelli-Fiume G. J Virol. 2005;79:7042–7049. doi: 10.1128/JVI.79.11.7042-7049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gianni T, Piccoli A, Bertucci C, Campadelli-Fiume G. J Virol. 2006;80:2216–2224. doi: 10.1128/JVI.80.5.2216-2224.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruel N, Zago A, Spear PG. Virology. 2006;346:229–237. doi: 10.1016/j.virol.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Abrahamyan LG, Mkrtchyan SR, Binley J, Lu M, Melikyan GB, Cohen FS. J Virol. 2005;79:106–115. doi: 10.1128/JVI.79.1.106-115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong S, Li M, Vincent A, Compans RW, Fritsch E, Beier R, Klenk C, Ohuchi M, Klenk H-D. Virology. 2002;301:322–333. doi: 10.1006/viro.2002.1594. [DOI] [PubMed] [Google Scholar]
- 12.Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. Science. 2006;313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 13.Roche S, Bressanelli S, Rey FA, Gaudin Y. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 14.Roche S, Rey FA, Gaudin Y, Bressanelli S. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- 15.Cai W, Gu B, Person S. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Minova-Foster TJ, Norton DD, Muggeridge MI. J Virol. 2006;80:3792–3800. doi: 10.1128/JVI.80.8.3792-3800.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norton DD, Dwyer DS, Muggeridge MI. Virus Res. 1998;55:37–48. doi: 10.1016/s0168-1702(98)00030-6. [DOI] [PubMed] [Google Scholar]
- 18.Claesson-Welsh L, Spear PG. J Virol. 1986;60:803–806. doi: 10.1128/jvi.60.2.803-806.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson DC, Spear PG. Cell. 1983;32:987–997. doi: 10.1016/0092-8674(83)90083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laquerre S, Argnani R, Anderson DB, Zucchini S, Manservigi R, Glorioso JC. J Virol. 1998;72:6119–6130. doi: 10.1128/jvi.72.7.6119-6130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan Z, Grantham ML, Smith MS, Anderson ES, Cardelli JA, Muggeridge MI. J Virol. 2002;76:9271–9283. doi: 10.1128/JVI.76.18.9271-9283.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foster TP, Melancon JM, Kousoulas KG. Virology. 2001;287:18–29. doi: 10.1006/viro.2001.1004. [DOI] [PubMed] [Google Scholar]
- 23.Hannah BP, Heldwein EE, Bender FC, Cohen GH, Eisenberg RJ. J Virol. 2007;81:4858–4865. doi: 10.1128/JVI.02755-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, Heinz FX, Rey FA. EMBO J. 2004;23:728–738. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Modis Y, Ogata S, Clements D, Harrison SC. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 26.Montgomery RI, Warner MS, Lum BJ, Spear PG. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 27.Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ, Spear PG. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 28.Herold BC, WuDunn D, Soltys N, Spear PG. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai W, Person S, Warner SC, Zhou J, DeLuca NA. J Virol. 1987;61:714–721. doi: 10.1128/jvi.61.3.714-721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pertel P, Fridberg A, Parish ML, Spear PG. Virology. 2001;279:313–324. doi: 10.1006/viro.2000.0713. [DOI] [PubMed] [Google Scholar]
- 31.Geraghty RJ, Jogger CR, Spear PG. Virology. 2000;268:147–158. doi: 10.1006/viro.1999.0157. [DOI] [PubMed] [Google Scholar]
- 32.Para MF, Parish ML, Noble AG, Spear PG. J Virol. 1985;55:483–488. doi: 10.1128/jvi.55.2.483-488.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okuma K, Nakamura M, Nakano S, Niho Y, Matsuura Y. Virology. 1999;254:235–244. doi: 10.1006/viro.1998.9530. [DOI] [PubMed] [Google Scholar]
- 34.Manoj S, Jogger CR, Myscofski D, Yoon M, Spear PG. Proc Natl Acad Sci USA. 2004;101:12414–12421. doi: 10.1073/pnas.0404211101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.