Abstract
The concept of a tight integration of transcription and splicing of mRNA precursors has been supported with increasing evidence in recent years. However, the mechanism and functional consequences of this integration remain largely unknown. We have examined how these processes impact upon one another when they occur together in HeLa nuclear extract. While both processes do in fact occur in parallel reactions in the extracts, we found no evidence that one process affects the other, under a variety of conditions tested. For example, neither the kinetics nor efficiency of splicing is significantly enhanced by de novo RNA polymerase II-mediated transcription, relative to that of presynthesized RNA added exogenously to the extract. Our results indicate that the act of transcription by RNA polymerase II in vitro is not sufficient to enhance splicing of the newly made RNA.
Keywords: transcription–splicing in vitro, coupled transcription-splicing system
INTRODUCTION
The gene expression pathway in eukaryotes involves extensive modification of primary transcripts prior to their export to the cytoplasm. In the case of mRNA, these include attachment of a 7mGppp group to the mRNA 5′-end (capping), removal of noncoding introns (splicing), and cleavage of the nascent transcript at the 3′ end followed by synthesis of a poly(A) tail (polyadenylation). All of these reactions can take place in isolation, in the absence of transcription and the other RNA processing events in vitro, and each was initially assumed to be functionally independent of the others. However, increasing evidence has emerged suggesting that these processes are tightly linked and are initiated and coordinated during transcription (for review, see Hirose and Manley 2000; Maniatis and Reed 2002; Proudfoot et al. 2002; Bentley 2005).
Early evidence consistent with close integration of transcription and splicing came from the studies of chromatin spreads of Drosophila melanogaster embryos (Osheim et al. 1985), which revealed assembly of protein particles on nascent transcripts and cotranscriptional shortening of these RNAs, presumably through splicing. Further studies generated a wealth of evidence of close interaction between the two processes. Microscopic observations revealed that splicing factors are recruited to sites of active transcription (Jiménez-García and Spector 1993), and that introns can be detected only in proximity of the original gene (Zhang et al. 1994), suggesting that intron removal takes place at, or in the immediate vicinity of, sites of transcription. In addition, functional studies in vivo demonstrated that promoter sequences can affect both constitutive splicing efficiency (Rosonina et al. 2003) and the alternative splicing pattern of the transcript (Cramer et al. 1997; Kadener et al. 2002; Nogues et al. 2002), as well as modulate the effect of protein factors involved in splicing regulation (Cramer et al. 1999). These and other results suggest that the molecular events that underlie integration of transcription and processing take place within, or at least involve, the RNA polymerase II (RNAP II) transcription elongation complex.
The premise that transcription and splicing are coupled in vivo is reinforced by the dramatic differences in properties of splicing in vitro and in vivo. Thus, in cells, multiple introns, some longer then 20,000 nucleotides (nt), are efficiently removed from all transcripts. In addition, the half-life of an intron in vivo has been estimated as 0.4–7.5 min (Audibert et al. 2002). In contrast, in the currently available in vitro splicing systems, few substrates have introns longer than 300 nt, and splicing reactions must be allowed to proceed for over an hour in order to process more than 50% of available substrate.
Splicing presents a particular challenge in the study of integration of transcription and RNA processing. Unlike capping or 3′ processing, splicing is typically repeated multiple times in every transcription event. Furthermore, the splicing reaction is considerably more complex than either of the other two processing reactions. Thus the mechanisms and functional consequences of coupling of splicing and transcription are not well understood. While certain problems, such as the order of cotranscriptional spliceosome assembly, have been addressed using chromatin immunoprecipitation assays in vivo (Görnemann et al. 2005; Tardiff and Rosbash 2006), further details, such as the identity of factors that mediate this process, cannot be elucidated without in vitro biochemical assays.
Coupling of transcription and splicing has the potential to alter several of the properties of the splicing reaction. First, transcription may target RNA to splicing, ensuring that transcribed introns will be recognized, committed to the splicing pathway, and removed, resulting in an improved overall efficiency of splicing. Such an effect has been observed in vivo, where splicing of a synthetic RNA injected into X. laevis oocytes was less efficient than cotranscriptional processing (Bird et al. 2004). Second, splicing kinetics may be improved, if the transcription elongation complex delivers some or all components of the spliceosome to splicing signals; this could reduce the time needed for splicing complex formation and speed up all the steps of the process that precede the catalytic steps. Third, transcription could stimulate splicing of “difficult” substrates, or substrates that contain long introns or multiple introns. Some models of transcription–splicing integration postulate that splice sites are committed to each other and even brought into close proximity within the transcription-processing complex, which will compensate for the initial distance between the splice sites on presynthesized splicing substrates that contain long introns (Goldstrohm et al. 2001). In addition, if the introns are committed to splicing cotranscriptionally, the transcription-processing complex can recognize several consecutive introns with equal efficiency. It is likely that any of these properties arising in a transcription-specific manner in a combined in vitro transcription–splicing reaction are the result of functional coupling between the two processes.
In the experiments described in this study, we characterized the transcription–splicing reaction in HeLa cells nuclear extracts and explored whether this reaction displays any properties expected of a functionally coupled reaction. Our experiments demonstrate that multiple properties of the splicing reaction in this transcription–splicing system are indistinguishable from those of a splicing reaction that uses a presynthesized substrate. Thus the linkage of RNAP II transcription and splicing is by itself not sufficient to enhance splicing in a detectable way.
RESULTS
Due to limitations of in vivo techniques, strategies for studying the integration of transcription and processing have shifted to development of combined in vitro transcription–processing systems that recapitulate the coupling mechanisms. In the past few years successful attempts were made to develop systems that coupled transcription and capping (Moteki and Price 2002), as well as transcription and polyadenylation (Adamson et al. 2005; Rigo et al. 2005). In vitro systems that combine transcription and splicing have also been described (Ghosh and Garcia-Blanco 2000; Ibrahim et al. 2005; Das et al. 2006). However, while both transcription and splicing were shown to take place in these studies, it is unclear whether there was a functional coupling between the two processes, that is, whether the properties of the splicing reaction were affected in a significant way by transcription.
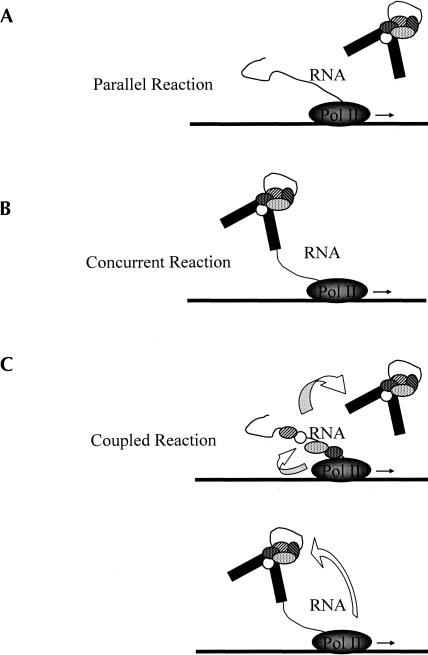
Indeed, there exists some confusion as to the definition of functional coupling (Ares and Proudfoot 2005). We therefore propose the following definitions for transcription–splicing reactions displaying different levels of integration. We term the reaction parallel if pre-mRNA is released from the transcription elongation complex before splicing and enters the splicing pathway in the same manner as a typical presynthesized splicing substrate used in in vitro splicing assays (Fig. 1A). In a concurrent reaction nascent RNA enters the splicing pathway in the same fashion as presynthesized RNA but prior to release from the transcription elongation complex (Fig. 1B). Finally, we term the reactions coupled if properties of the splicing reaction are specifically altered in a transcription-dependent manner (Fig. 1C).
FIGURE 1.
Possible levels of integration between transcription and splicing. In a parallel reaction, the splicing-related interaction occurs after pre-mRNA has been released from DNA template. In a concurrent reaction, the splicing-related interactions occur before release, while transcription is still in process, but independently of transcription elongation. In a coupled reaction, splicing is stimulated in a transcription-specific manner.
General properties of transcription–splicing in vitro
In the earliest in vitro studies of splicing the RNA substrate was generated in a transcription reaction by endogenous RNAP II from viral promoters (Kole and Weissman 1982). In subsequent studies the splicing substrate was presynthesized using bacterial or phage promoters and polymerases (Green et al. 1983; Padgett et al. 1983). Separation of the two processes made it possible to find conditions that are optimal specifically for splicing. We chose these conditions as the starting point in our attempt to analyze coupled transcription–splicing using HeLa nuclear extract (NE).
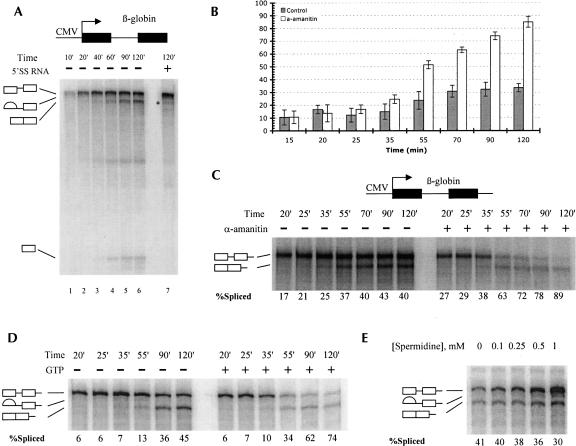
Transcription was readily obtained under conditions optimized for RNA splicing (see Materials and Methods). Several promoters and substrates in different combinations have been tested (data not shown), and the most efficient transcription and splicing were obtained with a construct containing the cytomegalovirus (CMV) promoter fused to the 5′ portion of the human β-globin gene. This template generates a transcript consisting of a 282 nt exon I followed by a 129 nt intron I and a 364 nt fragment of exon II (see Fig. 2A). When analyzed in a time course, bands corresponding to splicing products and intermediates were easily identifiable (Fig. 2A). These species disappeared when splicing was specifically inhibited by addition of a 5′ splice site RNA oligonucleotide (see below for details) or by mutation of the 5′ splice site (data not shown; Fig. 2A, lane7). Under the initial conditions tested, the NE maintained both transcription and splicing activity for at least 3 h at 30°C (data not shown). Over this time period the total yield of RNA reached 2–3 fmol, as determined by quantifying gel bands against known amounts of synthetic β-globin RNA. The overall splicing efficiency typically ranged from 25% to 45% after 2 h depending on the preparation of NE. Attempts to improve both transcription and splicing by further optimization of the reaction conditions were unsuccessful. While changing some of the conditions had little effect on the efficiency of either reaction (for example, polyethylene glycol could be substituted for polyvinyl alcohol), altering other conditions resulted in opposite effects on transcription or splicing. For example, increasing the concentration of MgCl2 to 5–6 mM dramatically increased the yield of transcription, consistent with earlier data, but it was detrimental to splicing. Conversely, increasing the ATP concentration markedly improved splicing while inhibiting transcription (data not shown).
FIGURE 2.
Properties of in vitro transcription-splicing reactions. (A) Time course of RNAP II transcription in HeLa nuclear extract. Transcription–splicing reactions have been incubated for indicated times. In the right lane, 5′SS RNA was added to a final concentration of 4 μM. (B) Summary of transcription–splicing pulse-chase assays. Here and in all other experiments the extent of splicing was determined by measuring the intensities of relevant phosphorimager bands and calculating the percentage of spliced mRNA relative to the total mRNA at the end of the reaction. The graph represents cumulative data from four independent experiments. (C) Sample transcription–splicing pulse-chase assay. Transcription from elongated β-globin templates (containing a downstream 5′ splice site) was allowed to proceed for 15 min, and the incubations were interrupted; α-amanitin was added to a final concentration of 1 ng/μL in order to inhibit RNAP II in the indicated reactions. The samples were incubated further to the times indicated. (D) Same as C, except that GTP and MgCl2 were added to final concentrations of 1 mM and 4 mM, respectively, in order to compensate for chelating of Mg2+ by GTP. (E) Spermidine concentration screen. Standard transcription–splicing reactions were incubated for a total of 2 h in the presence of different concentrations of spermidine as indicated on the top of the panel.
We have also tested the properties of transcription–splicing reactions under conditions that were very similar to those used in other efforts to develop coupled transcription–splicing systems (Ghosh and Garcia-Blanco 2000; Hicks et al. 2006). We found that relative efficiencies of transcription and splicing were not significantly different from those observed under our initial conditions (data not shown). After exhaustive testing, we concluded that the conditions described above gave rise to the best relative efficiencies of transcription and splicing.
Splicing in the presence of downstream signals
According to the exon definition model (Berget 1995), mutation of the 5′ splice site of an internal exon will result in skipping of that exon. A more dynamic version of this model (Goldstrohm et al. 2001) posits that the upstream 5′ splice site may be tethered to the RNAP II transcription complex, and the downstream 5′ splice site may act as signal for initiation of splicing of the preceding intron. In such a case, the model β-globin construct used above would provide a poor substrate for transcription-coupled splicing, since it lacks a downstream 5′ splice site. Accordingly, an elongated version of the CMV-β-globin template containing an additional 148 nt at its 3′ end, including a downstream 5′ splice site and portion of β-globin intron II, was tested in a transcription–splicing reaction. This elongated transcription product was spliced to the same extent and with similar kinetics as the β-globin product lacking the downstream 5′ splice site. In fact, the properties of the two RNAs were so similar in both splicing and transcription–splicing reaction that they could be used interchangeably (for example, the experiment in Fig. 2B used this template). We conclude that the presence of a downstream 5′ splice site does not confer any advantage in splicing efficiency in a transcription–splicing reaction in NE.
RNAP II-generated and presynthesized RNAs undergo splicing with equal efficiency
In the time course experiments described, the primary transcription products accumulated earlier and at a greater rate than splicing products. The constant production of potential splicing substrate could have resulted in obscuring the true efficiency of splicing in this reaction. In some experiments, the increase of RNA production sufficiently outpaced splicing to result in an apparent stall of splicing (see, for example, Fig. 2B, time points 70′,90′,120′). In order to obtain a more accurate measure of the overall efficiency of splicing, a pulse-chase assay was used. In these experiments production of labeled RNA was stopped at an early time point by inhibiting RNAP II with α-amanitin and the efficiency of splicing was monitored. A significant increase in splicing efficiency was observed, averaging 80%–85% after 2 h (Fig. 2B). Moreover, in some assays nearly all RNA (>90%) was spliced after 2 h (Fig. 2C, time point 120′ in the presence of α-amanitin). To ensure that this result was not due to the presence of α-amanitin or the inhibition of ongoing transcription per se, the experiment was repeated chasing the labeled RNA with a 100-fold molar excess of unlabeled GTP. The increase in splicing efficiency was still detected, although its extent was slightly less dramatic (Fig. 2D). These results show that efficient splicing was indeed masked by even more efficient transcription in our initial experiments.
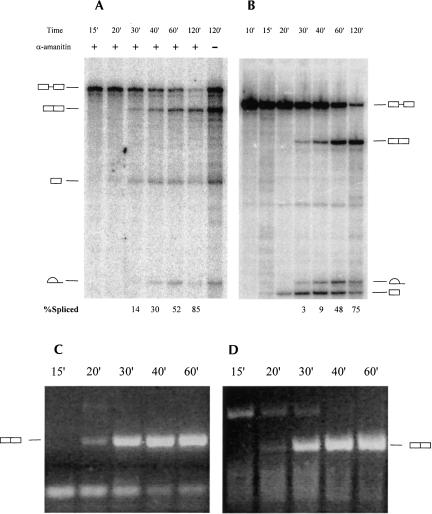
One possible explanation for the increase in splicing efficiency detected above was that RNA is specifically targeted for splicing in the coupled reaction. If this were the case, then the highly efficient splicing, such as observed in the above experiments, would be detected only in the presence of RNAP II transcription. However, presynthesized β-globin pre-mRNA displayed a splicing efficiency as great as that observed in the pulse-chase experiments described above (Fig. 3B, time point 120′). It is important to note that the initial amount of splicing substrate in these reactions (∼600 fmol) was at least two orders of magnitude greater than the total amount of RNA produced in 2 h of an uninterrupted transcription–splicing reaction (Fig. 3A,B), indicating that overall RNAP II transcription yield is insufficient to saturate the splicing machinery. Decreasing the amount of presynthesized substrate in splicing assays had only a negligible effect on splicing efficiency (results not shown). Thus while RNAP II synthesized pre-mRNA can be spliced with high efficiency during a chase, this efficiency is no greater than observed with the identical presynthesized substrate.
FIGURE 3.
Comparative time course of transcription–splicing and splicing reactions. (A) Transcription–splicing time course with standard CMV-β globin template. Transcription was allowed to proceed for 15 min and the incubations were interrupted; α-amanitin was added to a final concentration of 1 ng/μL in order to inhibit RNAP II in the indicated reactions. The samples were incubated further for the times indicated. Precursors, products, and intermediates of the transcription–splicing reactions are indicated to the left of the panel. (B) Splicing of T7-synthesized standard β-globin substrate. Precursors, products, and intermediates of the splicing reactions are indicated to the right of the panel. (C) RT-PCR analysis of reactions shown in panel B. (D) RT-PCR analysis of reactions shown in panel A.
It is important to note that a significant fraction of the RNA is degraded in both splicing reactions with presynthesized pre-mRNA and during a chase in transcription–splicing reactions. Degradation, however, appears to be nonspecific, as it affects RNA in both reactions equivalently (for example, see Fig. 3). We conclude that in our experiments RNAP II transcription does not confer any additional stability to the RNA (see Discussion).
In the course of our attempts to improve the general properties of the transcription–splicing reaction, we discovered that the efficiency of transcription markedly increased in the presence of spermidine. The maximal stimulation (approximately fivefold) was achieved at a final concentration of 1 mM (Fig. 2E). Importantly, spermidine had no effect on splicing of the T7- generated pre-mRNA at that concentration (data not shown). Thus it was surprising that the spermidine-mediated increase of RNA production was accompanied by a small but significant decrease in splicing efficiency in transcription–splicing reactions (Fig. 2E). In addition, when the α-amanitin-mediated pulse-chase assay was repeated in the presence of 1 mM spermidine, the extent of splicing fell to ∼50% from 80% to 90% (data not shown). This indicates that either increasing the amount of RNAP II-transcribed RNA saturates some component of the splicing machinery or, alternatively, that this RNA enters the splicing pathway less efficiently than presynthesized RNA. The former possibility appears to be less likely, as the total amount of RNAP II-transcribed RNA (15–20 fmol) is still much lower than the initial amount of presynthesized splicing substrate used in splicing reactions. We therefore conclude that under these conditions RNAP II-transcribed and presynthesized pre-mRNAs are directed to splicing through different pathways, of which the latter is somewhat more efficient. These results strengthen our conclusion that the production of splicing substrate via RNAP II transcription does not enhance the overall extent of splicing relative to that of presynthesized substrates.
Splicing rates are unaffected by the origin of the substrate
Next we examined whether RNAP II transcription improves the kinetic characteristics of splicing. When the progress of splicing in a transcription-splicing reaction was monitored in time course assays, the cleaved 5′ exon was first observed after 20 min and splicing products were first observed after 30 min (Fig. 3A). In splicing of presynthesized RNA, the intermediates were first observed as early as 15 min, although products were detected, like in the transcription–splicing reaction, after 30 min (Fig. 3B). However, the total amount of labeled RNA is very small at the earliest stages of the transcription–splicing reaction, making detection of possible intermediates and/or products difficult. Therefore, samples from the time course experiments were analyzed by RT-PCR specifically targeting the spliced RNA products. In contrast to the autoradiographic data, the PCR products derived from spliced β-globin RNA were first observed at the 20-min time point in both splicing of the presynthesized substrate (Fig. 3C) and in the transcription–splicing reaction (Fig. 3D). The apparently similar kinetics of splicing in the presence or absence of transcription under such conditions leads us to conclude that RNAP II transcription neither enhances the rate of the splicing reaction nor affects a rate-limiting step of splicing under the conditions tested.
Splicing in the transcription-splicing reaction is susceptible to competitive inhibition
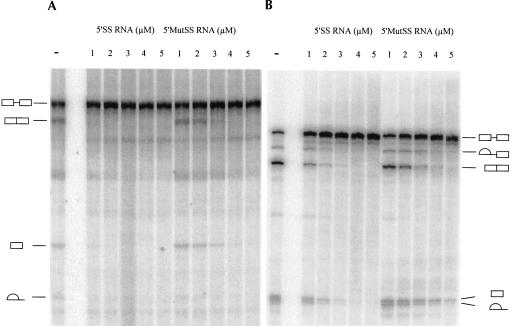
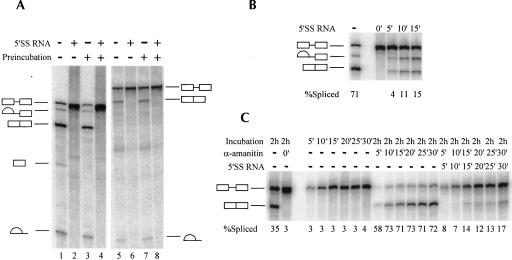
As mentioned above, one of the possible features of a functionally coupled transcription–splicing reaction might be enhanced resistance to competitive inhibition of splicing. Among the best characterized competitive inhibitors of splicing is a short RNA oligonucleotide containing the consensus sequence of the mammalian 5′ splice site (Hall and Konarska 1992). A 5′ splice site oligonucleotide (5′SS RNA) specifically inhibits splicing at the earliest steps (E complex formation) by assembling U2/U4/U5/U6 snRNPs into a stable complex and thus sequestering these snRNPs from the splicing substrate. If the RNAP II elongation complex is involved in delivery of splicing factors to the nascent pre-mRNA and actively facilitates recognition and binding of splicing signals, one would expect that splicing factors associated with the elongation complex would bind nascent RNA and recruit splicing factors from solution with greater specificity than 5′SS RNA. However, when 5′SS RNA was titrated into a transcription–splicing reaction, splicing was completely abolished at concentrations greater than 1 μM (Fig. 4A). In contrast, some of the presynthesized RNA substrate still underwent splicing when the concentration of 5′SS RNA was as high as 4 μM (Fig. 4B).
FIGURE 4.
5′SS-Mediated inhibition of splicing. Transcription–splicing (A) or splicing (B) reactions were carried out for 2 h in the presence of increasing concentrations of 5′SS RNA oligonucleotide (UCACAGGUAAGUACUUAUUUUCCCAGGCC) or mutated 5′MutSS RNA oligonucleotide (UCACAGCCAAGUACUUAUUUUCCCAGGCC). Oligonucleotide concentrations are indicated on the top of the panels. Precursors, products, and intermediates of the transcription–splicing reactions are indicated to the left of the panel (A), and those of the splicing reactions are indicated to the right of the panel (B).
The above results indicate unexpectedly that an RNAP II-synthesized pre-mRNA was more susceptible to competitive inhibition of splicing than a presynthesized pre-mRNA. The apparent increase in sensitivity to inhibition can be explained by the lower amount of pre-mRNA in transcription–splicing reactions, by less efficient commitment of RNAP II-synthesized pre-mRNA to the splicing pathway, or by the existence of a different mechanism of 5′SS-mediated inhibition of splicing of RNAP II-synthesized pre-mRNA. We tested the last possibility by repeating the above experiments with an RNA oligonucleotide in which the sequence was altered to substitute the GU residues critical to the 5′ splice site consensus. In line with previous findings (Hall and Konarska 1992), inhibition was significantly weakened in both assays (Fig. 4A,B). In addition, splicing did not recover in the presence or absence of RNAP II transcription after the 5′SS–U2/U4/U5/U6 complex was allowed to form by preincubating the 5′SS RNA in NE without RNA substrate or transcription template (Fig. 5A, lanes 2,4,6,8). These results suggest that 5′SS RNA inhibits splicing via the same mechanism in both reactions. We therefore conclude that RNAP II transcription confers no resistance to 5′SS-mediated splicing inhibition.
FIGURE 5.
5′SS-Mediated splicing inhibition and commitment to splicing pathway. (A) Transcription does not facilitate recruitment of splicing factors from complexes that form on 5′SS RNA oligonucleotides. Splicing (left panel) or transcription–splicing reactions (right panel) were either carried out normally (lanes 1,2,5,6) or preincubated for 15 min in the presence (lanes 4,8) or absence of 5′SS RNA oligonucleotide (lanes 3,7). Following preincubation either β-globin RNA or CMV-β globin DNA template was added and the samples were incubated further for a total of 2 h. Precursors, products, and intermediates of the splicing reactions are indicated to the left of the panel, and those of the transcription–splicing reactions are indicated to the right of the panel. (B) Commitment of presynthesized RNA to the splicing pathway. Splicing reactions were allowed to proceed for 5, 10, or 15 min; the incubations were interrupted and 5′SS RNA oligonucleotide was added to 5 μM final concentration, and the samples were incubated further for a total of 2 h. 5′SS RNA was added to one of the reactions at the start of the incubation (0′). Precursors, products, and intermediates of the splicing reactions are indicated to the left of the panel. (C). Same assay as in B with transcription–splicing reactions. Reactions were carried out for the indicated periods of time and were either stopped or the incubation was interrupted and α-amanitin was added to a final concentration of 1 ng/μL or α-amanitin and 5′SS RNA oligonucleotide were added together to 1 ng/μL and 5 μM final concentrations, respectively. Samples were incubated further for a total of 2 h. 5′SS RNA was added to one of the reactions at the start of the incubation (0′). Precursors, products, and intermediates of the transcription splicing reactions are indicated to the left of the panel.
The presynthesized and RNAP II-transcribed RNAs are committed to the splicing pathway with similar efficiencies
The potency and highly specific nature of 5′SS-mediated splicing inhibition enabled us to measure the rate of the steps of spliceosome formation that take place prior to catalysis and thus to gauge the efficiency of commitment of RNA to the splicing pathway. Standard splicing reactions were allowed to proceed for 5, 10, or 15 min, and then all further splicing was inhibited by addition of 5′SS RNA and the reaction was allowed to proceed further to 2 h. In these assays, after 5 min, ∼4% of input RNA has progressed through the splicing pathway past the stage where the reaction was susceptible to 5′SS RNA-mediated inhibition. This fraction increased to ∼15% after 15 min (Fig. 5B) and increased further at longer incubation times (data not shown). In the corresponding transcription–splicing experiments, transcription was inhibited, along with splicing, by addition of α-amanitin. If RNAP II transcription efficiently targets RNA to splicing, then one would expect that most of the newly transcribed RNA would be committed to the splicing pathway regardless of the duration of transcription. In our experiments, however, the data did not agree with this prediction: When both processes were inhibited at early time points, most of the RNA remained unspliced upon further incubation (Fig. 5C). The extent of commitment to splicing during the pulse in the transcription–splicing reaction (∼12%–14%) showed no substantial variation with the length of the pulse and was close to the value observed with presynthesized pre-mRNA (∼15%). The apparent lack of commitment of RNA to splicing can be explained by significantly faster rates of transcription compared to commitment to splicing, similar to the relatively low level of splicing observed in the transcription–splicing time course experiments (Fig. 2C). We conclude that RNAP II transcription is not sufficient to stimulate the early steps of splicing or the commitment of the pre-mRNA to the splicing pathway.
Splicing of long introns is equivalent in synthetic and RNAP II-generated substrates
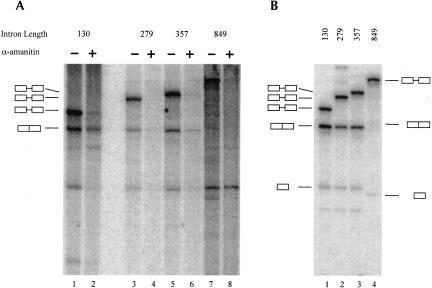
Another way that transcription could positively affect splicing would be to facilitate the excision of longer introns. It has been previously shown that the efficiency of splicing in NE drops sharply when intron length exceeds 200 nt (Fox-Walsh et al. 2005). Consequently, we investigated whether splicing of long introns is enhanced by RNAP II transcription. Rather than use natural constructs with longer introns, we chose to extend the first intron of the model β-globin substrate while minimally altering the surrounding sequences. The intron was extended by inserting a fragment of the second intron of human β-globin into a unique restriction site in the first intron. The novel constructs contained introns that were extended to 279 and 357 nt from the original 129 nt. Since this set of constructs carries exactly the same splice sites and other splicing signals as the natural β-globin construct used in previous experiments, any change in splicing patterns is expected to be primarily due to the intron length. In addition, the insert was subcloned in both orientations to ensure that any effects observed were not due to the presence of cis-acting splicing signals in the insert.
The results indicate that doubling and even tripling the length of the intron had only a modest (about 1.5-fold) negative effect on splicing (Fig. 6B, lanes 1–3). The inserted fragments do not appear to contain any functional cis-acting sequences, since inserting them in reverse orientation did not affect splicing efficiency (data not shown). Notably, the efficiency of splicing was not improved by transcription; in fact, the splicing of the 279- and 357-nt introns in transcription–splicing assays (Fig. 6A, lanes 3,5) was noticeably poorer than in splicing assays using presynthesized RNA (Fig. 6A, lanes 3,5). The final extent of splicing was higher in pulse-chase assays (Fig. 6A, lanes 4,6), but these results did not represent a significant improvement over those of control experiments using presynthesized substrate (Fig. 6B, lanes 1–4).
FIGURE 6.
Processing of expanded introns. (A) Transcription–splicing reactions were carried out for 2 h on the β-globin templates encoding RNAs with varying length of the intron (indicated on top of the panel). Where indicated, transcription was allowed to proceed for 15 min, at which point the incubations were interrupted and α-amanitin was added to a final concentration of 1 ng/μL. The samples were incubated further for a total of 2 h. The precursors and products of the splicing reactions are indicated to the left of the panel. (B) The corresponding splicing assay using substrates with varying length of the intron (indicated on top of the panel). Samples were incubated for 2 h. The precursors and products of the splicing reactions are indicated on the left of the panel, except in the case of ßglobin-ΔInt construct (849-nt intron), where precursors and intermediates are indicated on the right.
The splicing efficiency of a longer natural intron was also tested by examining splicing of the 849-nt intron II in its native context. To avoid any complications that may arise from splicing of multiple introns, intron I and the surrounding sequences were removed, generating a construct consisting of 270-nt chimeric 5′ exon, followed by β-globin intron II and exon III. In this context, splicing of intron II was detectable but inefficient (∼5%) in reactions using presynthesized substrate (Fig. 6B, lane 4). However, no splicing products or intermediates were detected in transcription–splicing assays (Fig. 6A, lanes 7,8). We conclude that the splicing of longer introns, at least in the size range tested, is not improved by RNAP II transcription.
DISCUSSION
In the experiments described here we tested whether a transcription–splicing reaction in HeLa NE displays any properties expected of a functionally coupled reaction. We find that, at least with our substrates and conditions, transcription does not affect the properties of splicing in a detectable way. Thus, both the overall extent of splicing and its kinetics were similar for RNAP II-transcribed and for presynthesized pre-mRNAs. In addition, RNAP II transcription neither confers inhibitor resistance to the splicing reaction nor improves removal of long introns that are poorly spliced in control reactions that use presynthesized RNA. Finally, we find that additional splicing signals downstream of the 3′ exon, proposed to be instrumental for integration of transcription and splicing, do not improve splicing of the upstream intron in our reactions.
These findings lead us to conclude that there is no significant functional coupling between transcription and splicing in HeLa NE under the variety of conditions tested. Although both processes take place concurrently, we could not detect any transcription-dependent effects on the splicing process and its properties. Thus, it seems that under these conditions the act of RNAP II transcription may not be involved with splicing beyond producing the substrate, and is certainly insufficient to stimulate splicing efficiency. We therefore conclude that RNAP II-synthesized pre-mRNA enters and progresses through the splicing pathway in nearly the same manner as presynthesized substrate. It follows that in this reaction RNAP II and other components of the transcription initiation and elongation complex either do not play a role in splice site recognition and spliceosome assembly or are involved in splicing in a manner that does not require a functioning RNAP II transcription elongation complex. In accordance with the terminology we proposed above, this is most likely either a parallel or concurrent reaction, but it is does not display the properties of a coupled reaction. The data presented here are not sufficient to classify this reaction as definitively either parallel or concurrent. However, we speculate that it is mostly the former, since most RNA was released into solution at early time points using DNA templates immobilized on streptavidin beads (unpublished data).
Several previous investigations of HeLa NE-based transcription–splicing have suggested the possible existence of functional coupling. However, in none of these experiments were the fundamental properties of the splicing reaction altered in an RNAP II transcription-specific manner. Although it is possible that some functional coupling does occur in one or more of these systems, by the definitions we use here, this has not been conclusively established.
The earliest study suggesting functional coupling based this conclusion on association of spliced RNA with immobilized DNA templates (Ghosh and Garcia-Blanco 2000). However, when we utilized such templates, we observed that RNA, spliced or unspliced, nonspecifically bound streptavidin beads (unpublished data), complicating any conclusions of such studies. In any event, such an association would at best only provide evidence for a concurrent reaction. In subsequent work, the authors reported that transcription could increase the extent of splicing, another characteristic of functional coupling (Natalizio and Garcia-Blanco 2005). However, the overall efficiency of splicing observed was very low, even with presynthesized RNA, making interpretation of this data difficult.
Another recent study proposed that coupling splicing to transcription enhances splicing via stabilization of the pre-mRNA (Hicks et al. 2006). Initial experiments suggested that splicing of an RNAP II-synthesized transcript displayed improved kinetic parameters compared to splicing of the identical transcript produced by T7 RNAP in NE. Subsequent analyses, however, revealed that the apparent enhancement of splicing, while dependent on splice sites and splicing factors, was due to increased stability of the RNAP II-synthesized transcript and not to any changes in the kinetics of the splicing reaction. While stabilization is likely an indication of an interaction between transcription and splicing, it does not confer the very high rates of splicing typically found in vivo. Hence stabilization can only partially explain the differences in properties of splicing in vivo and in vitro. Additionally, enhancement of splicing resulting from stabilization could be viewed as a passive or indirect effect of transcription, not the active effect envisioned by our definition of functional coupling. Although for unknown reasons we did not detect significant stabilization of RNAP II-synthesized transcripts, the results of Hicks et al. (2006) as they relate to functional coupling are consistent with those described here.
Functional coupling has also been inferred in experiments of Das et al. (2006). These authors reported an apparent improvement in the kinetic parameters of splicing, of RNAP II-synthesized transcripts, and rapid formation of a splicing- and RNAP II-specific high MW complex. The results of kinetic assays were very similar to those of Hicks et al. (2006), but the relative amounts of spliced and unspliced RNA were calculated in a way that did not account for possible alterations in RNA stability. Thus the possibility that RNAP II-specific stabilization may be the underlying cause of the apparent improvement in splicing has not been ruled out. Consequently, in the absence of conclusive evidence for an RNAP II transcription-specific enhancement of splicing, the observed RNAP II-specific high-MW complex may at best be viewed as evidence for a concurrent reaction. Furthermore, this complex appeared at early time points of the reaction, before splicing was detectable, and dissipated while nearly all RNA was still unspliced. Therefore, while the complex may represent RNAP II elongation complex components interacting with spliceosomes, or two complexes forming on the same RNA molecule, its functional significance is currently unclear.
Our data suggest that under a variety of conditions the NE based transcription–splicing reaction is insufficient to functionally couple the two processes to the extent that such coupling can be measured. One possible explanation for this insufficiency is that the high rate and efficiency of splicing in vivo are achieved in ways that are not directly related to transcription. Although the currently available data suggest that the two processes are integrated, direct proof exists only for concurrent transcription–splicing, as observed, for example, by Osheim et al. (1985) in D. melanogaster embryos. At the moment, there is no explicit functional evidence that RNAP II, or an RNAP II elongation complex, is directly involved in splice site recognition and selection or in spliceosome assembly. It is therefore possible that the factors responsible for efficient recognition of splicing signals and rapid spliceosome assembly do not associate with the transcription complex in a way that enhances their activity. By this scenario, it could be that whatever factor(s) delimit the efficiency of splicing in vitro are unrelated to transcription.
It is also possible that the absence of functional coupling between transcription and splicing represents an innate limitation of HeLa NE. The extraction procedure isolates the soluble fraction of the nucleoplasm, removing a large fraction of nuclear proteins that are either insoluble or are tightly associated with the insoluble fraction, which is discarded in the form of the nuclear pellet (NP). The NP in fact contains a large fraction of the cell's RNAP II, and may also contain a significant portion of RNA processing factors, possibly including the hypothetical factor(s) that couple transcription and processing. We have made attempts to recover proteins from the NP and add them to transcription–splicing reactions. While some fractions increased transcription, none improved splicing (unpublished data), suggesting, if coupling factors are indeed present in the NP, that either further fractionation or an entirely different recovery approach may be necessary.
The dramatic difference of splicing properties in vivo and in vitro may also be explained by the fundamental differences between the two environments. In NE, the template is linear naked DNA, while in cells DNA is packaged into chromatin. In fact, factors involved in chromatin remodeling have been suggested to be involved in regulation of splicing both in yeast (Xiao et al. 2005) and in humans (Batsché et al. 2005). Furthermore, splice sites have been found to preferentially localize in regions protected by nucleosomes in a genome-wide nucleosome mapping study (Denisov et al. 1997). So far, nucleosomes and other structural components of chromatin have not been shown to associate with splicing complexes. This is hardly surprising, since proteomic analyses of splicing complexes (Neubauer et al. 1998; Jurica et al. 2002; Makarov et al. 2002; Zhou et al. 2002) have been conducted on purified spliceosomes formed in HeLa NE, which lacks chromatin. One way to address the possibility that chromatin associated factors are responsible for enhanced splicing efficiency would be to use DNA templates on which chromatin has been reconstructed in vitro.
Potentially even more important for efficient coordination of transcription and splicing is the overall context of nuclear skeleton (matrix), which is entirely disrupted in NEs. There is a great deal of data pointing to a high level of organization in the spatial distribution of transcription and splicing factors relative to the sites of active transcription. Thus U snRNPs, SR proteins, and nascent RNAs concentrate in regions positioned in the immediate vicinity of the sites of active transcription, termed nuclear speckles (for review, see Misteli and Spector 1998). The defined structure of these domains suggests some sort of underlying scaffold, which is most likely to be the nuclear skeleton. Since splicing, unlike capping and polyadenylation, has to occur multiple times for each transcription event, its coordination with transcription may be more dependent on the local three-dimensional structure than the other processing events. Destroying this structure during extract preparation could disrupt the mechanism for delivery of necessary factors and spliceosomal components in response to splicing signals on the nascent transcripts. Presently there is no technology to replace or restore this structure once it is disrupted.
In summary, our experiments have demonstrated that HeLa NEs capable of simultaneous transcription and splicing do not provide the conditions that allow splicing to attain the parameters observed in vivo. Importantly, the act of promoter-dependent RNAP II transcription is not sufficient to bring about functional coupling of transcription and splicing. Development of an in vitro assay that recapitulates in vivo conditions will be essential for establishing the specifics of functional coupling of transcription and splicing.
MATERIALS AND METHODS
Plasmids and transcription templates
A fragment of β-globin open reading frame that included exon I, intron I, exon II, and a portion of intron II was PCR-amplified using the forward primer CGGGGGCTCGAGGCTTCTGACACAACTGTG and the reverse primer CGGGGGCTCGAGAACGATCCTGAGACTTCC. The resulting fragment was subcloned into the unique XhoI restriction site of pcDNA3 expression vector (Invitrogen) to generate a pCMV-ßglobin vector. The resulting construct contained a T7 promoter positioned between the CMV promoter and the β-globin sequences. DNA templates for transcription experiments were generated by PCR using the forward primer CAATTGCATGAAGAATCTGCTTAGGGTTAGG and the reverse primer GGATCCACGTGCAGCTTGTCACAGTGCAGC to obtain the standard template or the reverse primer used in the initial amplification of β-globin sequences (see above) to obtain the elongated template that includes the downstream 5′ splice site. CMV transcription from these templates generated RNAs consisting of a 282 nt exon I, a 130 nt intron I, and either a 209 nt exon II (standard template) or a complete 222 nt exon II followed by 148 nt of β-globin intron II in the elongated template. The T7 promoter located between the CMV promoter and the β-globin sequences was used to generate splicing substrates for control assays; the T7 transcripts are ∼30 nt shorter at the 5′ end.
To extend intron 1 of the β-globin, a unique AflII site was inserted into the pcDNA3-ßglobin construct by site directed mutagenesis using the forward primer CAAGGTTACAAGACAGGCTTAAGGAGACCAATAGAAACTGGGC and reverse primer GCCCAGTTTCTATTGGTCTCCTTAAGCCTGTCTTGTAACCTTG. The inserts were amplified from the pCMVß-FL vector (pcDNA3 plasmid derivative containing the complete human β-globin gene subcloned downstream of the CMV promoter, generously provided by A. Tang, Columbia University). The 150-nt insert (final intron length 270 nt) was generated using the forward GGGGGCTTAAGGCTTATTTGCATATTCATAATCTCCC primer and the reverse GGGGGCTTAAGCAAATGCAAAATTACCCTGATTTGG primer. The same reverse primer was used with GGGGGCTTAAGTCTCTGAGATACATTAAGTAAC forward primer to generate the 228 nt insert (final intron length 357 nt). Both inserts were subcloned into the pCMV-ßglobin vector containing the AflII restriction site.
The construct containing only the second intron of β-globin was generated by digesting the pCMVß-FL plasmid with Bsu36I restriction enzyme, filling in the overhangs with Klenow fragment of DNA polymerase I (Invitrogen) and recircularizing the vector to generate the pCMVß-ΔInt plasmid. The DNA template was generated with forward CMV promoter primer and the reverse GCAATGAAAATAAATGTTTTTTATTAGGCAGAATCCAGATGC primer. The RNAP II transcript of this template consists of the chimeric 270 nt exon I–II followed by the 898 nt intron II and the 261 nt intron III. As before, T7 promoter was used to generate control splicing substrate, which is ∼30 nt shorter on the 5′ end.
Splicing substrates and transcription–splicing assays
Splicing substrates were generated by in vitro transcription with bacterial T7 polymerase (Promega) in the presence of [α-32P] GTP (Amersham). The transcripts were gel purified, and the amounts were estimated using specific activity calculations.
In vitro splicing assays were carried out at 30°C in 15 μL reactions containing 3 mM MgCl2, 20 mM Creatine Phosphate, 1 U of RNAsin (Promega), 3% PVA, 1 mM ATP, ∼600 fmol RNA substrate, and 7 μL HeLa cells Nuclear Extract prepared exactly as described by Dignam (1983). In transcription–splicing assays RNA substrate was replaced with 50μM rNTPs, 10 μCi [α32P]-GTP (3000 Ci/mmol), and 50 ng template DNA. Both splicing and transcription–splicing reactions were stopped by addition of 200 μL proteinase K Mix (50 mM Tris-HCl at pH 7.5, 10 mM EDTA, 100 mM NaCl, 1% SDS, 25 μg/mL proteinase K, 20 μg/mL tRNA) immediately after incubation at 30°C, and incubating for 30 min at 37°C. The RNA was extracted and resolved on 6% 7 M urea polyacrylamide gel. The bands were visualized using a PhosphorImager (Molecular Dynamics). The amounts of RNA generated by RNAP II transcription were estimated by loading known amounts of T7-generated RNA onto assay gels, and comparing the intensity of the relevant bands, after adjusting for different specific activity.
RT-PCR assays
The splicing and transcription–splicing reactions were carried out as described above, and the RNA products were resuspended in water. The samples were treated with RQ DNAse (Promega) for 30 min, at 37°C, and subsequently DNAse was deactivated by 10 min incubation at 65°C in the presence of 10 mM EGTA. The first DNA strand was reverse-transcribed using the GGTGCCCTTGAGGTTGTCCAGGTGAGCCAGGCC primer and the SuperScript First-Strand Synthesis kit (Invitrogen), according to the manufacturer's instructions. The DNA fragments were amplified using Taq DNA polymerase (Invitrogen) with the GGTGGTGAGGCCCTGGGCAGGC forward primer (the splice junction is underlined), and the reverse primer used in first strand synthesis.
ACKNOWLEDGMENTS
We would like to thank N. Rao for excellent technical assistance and S. Millhouse, F. Kleiman, and C. Wachtel for helpful discussions. This work was supported by NIH grant GM 48259 to J.L.M.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.595907.
REFERENCES
- Adamson, T.E., Shutt, D.C., Price, D.H. Functional coupling of cleavage and polyadenylation with transcription of mRNA. J. Biol. Chem. 2005;280:32262–32271. doi: 10.1074/jbc.M505532200. [DOI] [PubMed] [Google Scholar]
- Ares, M., Proudfoot, N.J. The Spanish connection: Transcription and mRNA processing get even closer. Cell. 2005;120:163–166. doi: 10.1016/j.cell.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Audibert, A., Weil, D., Dautry, F. In vivo kinetics of mRNA splicing and transport in mammalian cells. Mol. Cell. Biol. 2002;22:6706–6718. doi: 10.1128/MCB.22.19.6706-6718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batsché, E., Yaniv, M., Muchardt, C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2005;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- Bentley, D.L. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Berget, S.M. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Bird, G., Zorio, D.A., Bentley, D.L. RNA polymerase II carboxyterminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol. Cell. Biol. 2004;24:8963–8969. doi: 10.1128/MCB.24.20.8963-8969.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer, P., Pesce, C.G., Baralle, F.E., Kornblihtt, A.R. Functional association between promoter structure and transcript alternative splicing. Proc. Natl. Acad. Sci. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer, P., Caceres, J.F., Cazalla, D., Kadener, S., Muro, A.F., Baralle, F.E., Kornblihtt, A.R. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell. 1999;4:251–258. doi: 10.1016/s1097-2765(00)80372-x. [DOI] [PubMed] [Google Scholar]
- Das, R., Dufu, K., Romney, B., Feldt, M., Elenko, M., Reed, R. Functional coupling of RNAP II transcription to spliceosome assembly. Genes & Dev. 2006;20:1100–1109. doi: 10.1101/gad.1397406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov, D.A., Shpigelman, E.S., Trifonov, E.N. Protective nucleosome centering at splice sites as suggested by sequence-directed mapping of the nucleosomes. Gene. 1997;205:145–149. doi: 10.1016/s0378-1119(97)00406-x. [DOI] [PubMed] [Google Scholar]
- Dignam, J.P., Lebovitz, R.M., Roeder, R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucl. Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox-Walsh, K.L., Dou, Y., Lam, B.J., Hung, S., Baldi, P.F., Hertel, K.J. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc. Natl. Acad. Sci. 2005;102:16176–16181. doi: 10.1073/pnas.0508489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, S., Garcia-Blanco, M.A. Coupled in vitro synthesis and splicing of RNA polymerase II transcripts. RNA. 2000;6:1325–1334. doi: 10.1017/s1355838200992537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstrohm, A.C., Greenleaf, A.L., Garcia-Blanco, M.A. Cotranscriptional splicing of pre-messenger RNAs: Considerations for the mechanism of alternative splicing. Gene. 2001;277:31–47. doi: 10.1016/s0378-1119(01)00695-3. [DOI] [PubMed] [Google Scholar]
- Görnemann, J., Kotovic, K.M., Hujer, K., Neugebauer, K.M. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol. Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Green, M.R., Maniatis, T., Melton, D.A. Human β-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell. 1983;32:681–694. doi: 10.1016/0092-8674(83)90054-5. [DOI] [PubMed] [Google Scholar]
- Hall, K.B., Konarska, M.M. The 5′ splice site consensus RNA oligonucleotide induces assembly of U2/U4/U5/U6 small nuclear ribonucleoprotein complexes. Proc. Natl. Acad. Sci. 1992;89:10969–10973. doi: 10.1073/pnas.89.22.10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, M.J., Yang, C.R., Kotlajich, M.V., Hertel, K.J. Linking splicing to Pol II transcription stabilizes pre-mRNAs and influences splicing patterns. PLoS Biol. 2006;4:e147. doi: 10.1371/journal.pbio.0040147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, Y., Manley, J.L. RNA polymerase II and the integration of nuclear events. Genes & Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Ibrahim, E.C., Schaal, T.D., Hertel, K.J., Reed, R., Maniatis, T. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc. Natl. Acad. Sci. 2005;102:5002–5007. doi: 10.1073/pnas.0500543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-García, L.F., Spector, D.L. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell. 1993;73:47–59. doi: 10.1016/0092-8674(93)90159-n. [DOI] [PubMed] [Google Scholar]
- Jurica, M.S., Licklider, L.J., Gygi, S.R., Grigorieff, N., Moore, M.J. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener, S., Fededa, J.P., Rosbash, M., Kornblihtt, A.R. Regulation of alternative splicing by a transcriptional enhancer through RNA pol II elongation. Proc. Natl. Acad. Sci. 2002;99:8185–8190. doi: 10.1073/pnas.122246099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole, R., Weissman, S.M. Accurate in vitro splicing of human β-globin RNA. Nucleic Acids Res. 1982;10:5429–5445. doi: 10.1093/nar/10.18.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarov, E.M., Makarova, O.V., Urlaub, H., Gentzel, M., Will, C.L., Wilm, M., Lührmann, R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., Reed, R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Misteli, T., Spector, D.L. The cellular organization of gene expression. Curr. Opin. Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- Moteki, S., Price, D. Functional coupling of capping and transcription of mRNA. Mol. Cell. 2002;10:599–609. doi: 10.1016/s1097-2765(02)00660-3. [DOI] [PubMed] [Google Scholar]
- Natalizio, B.J., Garcia-Blanco, M.A. In vitro coupled transcription splicing. Methods. 2005;37:314–322. doi: 10.1016/j.ymeth.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Neubauer, G., King, A., Rappsilber, J., Calvio, C., Watson, M., Ajuh, P., Sleeman, J., Lamond, A., Mann, M. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 1998;20:46–50. doi: 10.1038/1700. [DOI] [PubMed] [Google Scholar]
- Nogues, G., Kadener, S., Cramer, P., Bentley, D., Kornblihtt, A.R. Transcriptional activators differ in their abilities to control alternative splicing. J. Biol. Chem. 2002;277:43110–43114. doi: 10.1074/jbc.M208418200. [DOI] [PubMed] [Google Scholar]
- Osheim, Y.N., Miller O.L., Jr, Beyer, A.L. RNP particles at splice junction sequences on Drosophila chorion transcripts. Cell. 1985;43:143–151. doi: 10.1016/0092-8674(85)90019-4. [DOI] [PubMed] [Google Scholar]
- Padgett, R.A., Hardy, S.F., Sharp, P.A. Splicing of adenovirus RNA in a cell-free transcription system. Proc. Natl. Acad. Sci. 1983;80:5230–5234. doi: 10.1073/pnas.80.17.5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., Dye, M.J. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Rigo, F., Kazerouninia, A., Nag, A., Martinson, H.G. The RNA tether from the poly(A) signal to the polymerase mediates coupling of transcription to cleavage and polyadenylation. Mol. Cell. 2005;20:733–745. doi: 10.1016/j.molcel.2005.09.026. [DOI] [PubMed] [Google Scholar]
- Rosonina, E., Bakowski, M.A., McCracken, S., Blencowe, B.J. Transcriptional activators control splicing and 3′-end cleavage levels. J. Biol. Chem. 2003;278:43034–43040. doi: 10.1074/jbc.M307289200. [DOI] [PubMed] [Google Scholar]
- Tardiff, D.F., Rosbash, M. Arrested yeast splicing complexes indicate stepwise snRNP recruitment during in vivo spliceosome assembly. RNA. 2006;12:968–979. doi: 10.1261/rna.50506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y., Yang, Y.H., Burckin, T.A., Shiue, L., Hartzog, G.A., Segal, M.R. Analysis of a splice array experiment elucidates roles of chromatin elongation factor Spt4-5 in splicing. PLoS Comput. Biol. 2005;1:e39. doi: 10.1371/journal.pcbi.0010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G., Taneja, K.L., Singer, R.H., Green, M.R. Localization of pre-mRNA splicing in mammalian nuclei. Nature. 1994;372:809–812. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., Licklider, L.J., Gygi, S.P., Reed, R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]