Abstract
Histone deacetylase 7 (HDAC7) is highly expressed in CD4+/CD8+ thymocytes and functions as a signal-dependent repressor of gene transcription during T-cell development. In this study, we expressed HDAC7 mutant proteins in a T-cell line and use DNA microarrays to identify transcriptional targets of HDAC7 in T cells. The changes in gene expression levels were compared to differential gene expression profiles associated with positive and negative thymic selection. This analysis reveals that HDAC7 regulates an extensive set of genes that are differentially expressed during both positive and negative thymic selection. Many of these genes play important functional roles in thymic selection, primarily via modulating the coupling between antigen receptor engagement and downstream signaling events. Consistent with the model that HDAC7 may play an important role in both positive and negative thymic selection, the expression of distinct HDAC7 mutants or the abrogation of HDAC7 expression can either enhance or inhibit the signal-dependent differentiation of a CD4+/CD8+ cell line.
The thymic development of T cells is a highly regulated process involving multiple extracellular signals. A complex integration of these signals results in changes in gene expression that can lead to the adoption of different developmental fates. Thymocytes pass two main developmental checkpoints that involve antigen receptor signaling. The first, called β-selection, occurs after the first of the two chains of the T-cell receptor (TCR) has been productively rearranged and is required for further developmental progression of thymocytes (24, 55, 75). After β-selection, thymocytes proliferate extensively, rearrange their antigen receptor α-chains, and become CD4+, CD8+, or double-positive (DP) thymocytes. The subsequent developmental fate of DP thymocytes is determined by signaling interactions with antigen-presenting cells (APCs) in the thymic cortex and medulla, most critically between the TCRs of thymocytes and self peptides bound to the major histocompatibility complex (MHC) molecules of the APCs. Broadly, DP thymocytes can adopt three different developmental fates based on the character of this interaction. The large majority of the thymocytes have no functional interaction between their TCRs and the MHC-self peptide complexes on the APCs, and die of neglect within a few days. Thymocytes expressing TCRs that have an interaction of intermediate strength undergo positive selection, after which they downregulate either the CD4 or CD8 coreceptor and exit the thymus as mature CD4+ or CD8+ T cells. Potentially autoreactive thymocytes, with TCRs that interact strongly with MHC-self peptide complexes, are deleted by an apoptotic process termed negative selection. How this continuum of different antigen receptor signal strengths leads to a discrete set of developmental outcomes, in terms of both lineage determination and survival, is still poorly understood.
Numerous extracellular signals not involving the TCR have been implicated in the positive and negative selection of thymocytes. The receptors mediating these signals belong to multiple gene families and include CD28 and PD1 (7, 37, 60, 67), CD5 and CD6 (63, 80), CCR7 (39, 89), and CD43 (37). Among the intracellular factors thought to contribute to positive selection are the mitogen-activated protein (MAP) kinases (MAPKs) extracellular signal-regulated kinases 1 and 2 (84), BCL2 and BCLx (43, 49), NF-κB (21, 79), and EGR1 (5). Factors thought to be associated with negative selection include the Bcl-2 family member BIM (27), the Jun N-terminal protein kinase (JNK) and p38 MAP kinases (70, 74, 84), PTEN (86), and the orphan steroid receptors Nur77 and NOR1 (8, 13, 96).
Histone deacetylase 7 (HDAC7) is a class IIa HDAC that is highly expressed in DP thymocytes, suggesting a possible role in thymic T-cell development (22, 31). The class IIa HDACs play important roles in the developmentally regulated expression of genes involved in the differentiation and function of muscle, immune, and neural cells (reviewed in reference 92). Mice deficient in HDAC4, -5, -7, and -9 display abnormalities involving osteogenesis in the case of HDAC4 (91), cardiac stress response in the case of HDAC5 and -9 (9, 98), and angiogenesis in the case of HDAC7 (10). HDACs catalyze the removal of acetyl groups from lysine residues in the N-terminal regions of the core histone octamer subunits. The class IIa HDACs, comprising the subfamily members HDAC4, -5, -7, and -9, are defined by the presence of a large N-terminal domain that mediates both recruitment to specific promoters and signal-dependent shuttling between the nucleus and the cytoplasm. Nucleocytoplasmic shuttling is regulated by the phosphorylation of two or three conserved serine residues in the N-terminal domain (25, 32, 95) by protein kinase D (PKD) and other kinases. PKD phosphorylation of class IIa HDACs causes their removal from the nucleus in response to hypertrophic signals in cardiomyocytes (HDAC5) and in response to antigen receptor signals in T cells (HDAC7) (18, 64, 90). Mutations in the PKD target serine residues prevent class IIa HDACs from being exported to the cytoplasm in response to extracellular signals (32, 54, 95, 99, 101).
The recruitment of class IIa HDACs to various promoters can occur through interactions with different DNA-binding proteins. All four class IIa HDACs interact with myocyte enhancer-binding factor-2 (MEF2) family proteins through a highly conserved 17-amino-acid motif in their shared N-terminal domain (41, 83, 94). Through this signal-dependent interaction, the class IIa HDACs repress transcription at the MEF2-regulated promoters of genes, including myogenin, c-jun, and importantly, Nur77 (17, 20, 32). Other DNA-binding interaction partners of the class IIa HDACs that are of potential interest in the study of thymic selection are the Runx family of transcription factors (91) and the highly conserved calmodulin-binding transcriptional activator CAMTA2 (82).
Although HDAC7-deficient mice die too early to evaluate any phenotypic effects in the thymus, abundant in vitro data support the hypothesis that HDAC7 plays an important role in thymic T-cell development. Through MEF2D, HDAC7 regulates the expression of the Nur77 orphan steroid receptor, a protein that is thought to play an important role in negative selection (17). The release of HDAC7 from the Nur77 promoter is coupled to antigen receptor engagement by the activation of PKD (64), a protein that is also important in T-cell development (50). Thus, HDAC7 may be a key mediator of the antigen receptor-dependent changes in gene expression associated with T-cell development.
In this study, we analyzed the gene expression profiles of T cells in which the function of HDAC7 had been perturbed by the overexpression of two HDAC7 mutant proteins. The first HDAC7 mutant converts HDAC7 from a transcriptional repressor into an activator by substituting a potent transcriptional activation domain for its HDAC domain. The second mutant is not phosphorylated in response to TCR activation and thus retains its activity as a transcriptional repressor in activated T cells. We compared these data to the differential gene expression profiles of thymocytes undergoing either positive or negative selection. Unexpectedly, we discovered that significantly more of the putative targets of HDAC7 were associated with positive selection than with negative selection. We further performed experiments to position our putative targets within the transcriptional cascade downstream of HDAC7 and to determine which genes are direct targets. We also investigated the possible role of HDAC7 in T-cell differentiation in a tissue culture model. Our findings suggest a role for HDAC7 in multiple aspects of the antigen receptor-dependent thymic development of T cells, including both antigen-induced deletion and the differentiation of DP thymocytes into mature T cells.
MATERIALS AND METHODS
Plasmids.
Murine stem cell virus (MSCV)-internal ribosome entry site (IRES)-green fluorescent protein (GFP) (MSCVIG) and MSCV-IRES-Puro (MSCVIP), containing the packaging, insertion, and expression elements of MSCV and an expression cassette encoding GFP or a puromycin resistance gene, respectively, were a gift from William Sha (University of California, Berkeley, CA). MSCVG HDAC7-VP16 and MSCVG HDAC7 ΔP are described elsewhere (17). MSCVIG Nur77 was constructed by PCR amplification of the coding sequence of murine Nur77 from the plasmid pBluescript N10 (a gift from Astar Winoto, University of California, Berkeley, CA) with primers containing recognition sites for XhoI and NotI. The resultant fragment was subcloned into the corresponding sites of MSCVIG. MSCVIP MEF2D-VP16 was constructed by subcloning the sequence encoding a fusion of the N-terminal 92 amino acids of murine MEF2D to residues 412 to 490 of herpes simplex virus (HSV) VP16 from the plasmid pEF BOS MEF2D-VP16 (described in reference 33) into MSCVIP.
Mice and peptide injections.
BALB/c mice, C57BL/6 MHC null mice, and BALB/c DO11.10 TCR-transgenic mice were obtained from the Jackson Laboratory. The mice were maintained in a pathogen-free barrier facility. Thymocytes were harvested from 5- to 6-week-old mice. The chicken ovalbumin antigenic peptide for the DO11.10 TCR, KISQAVHAAHAEINEAG, was obtained by custom synthesis from Invitrogen. The peptide was injected intraperitoneally in a volume of 200 μl of phosphate-buffered saline, at a concentration of 50 μM.
Cell culture, transfections, and viral transductions.
DO11.10 T-cell hybridomas were grown in RPMI 1640 medium with 10% fetal bovine serum (FBS), 2 mM glutamine, and 50 U/ml streptomycin-penicillin. DPK thymomas were grown in Click's medium supplemented with 12% FBS, 4 mM glutamine, and 50 U/ml streptomycin-penicillin. DCEK-ICAM cells and the 293T-based BOSC23 and Phoenix Ampho cell lines (gifts from William Sha and Gerry Nolan, respectively) were grown in Dulbecco's modified Eagle's medium supplemented with 10% FBS, 2 mM glutamine, and 50 U/ml streptomycin-penicillin.
DPK cells were transfected with small interfering RNAs (siRNAs) using the Amaxa Nucleofection technology (Amaxa GmbH). Totals of 8 × 106 DPK cells were resuspended in 100 μl Amaxa Nucleofector solution V containing 150 pmol of siGlo Green fluorescently labeled RNA oligonucleotide (Dharmacon) together with 225 pmol of control or HDAC7-specific siRNA duplex. The recognition sequence of the HDAC7-specific siRNA corresponded to bases 579 to 597 of the sense strand of the murine HDAC7 mRNA (sequence, AGACAAGAGCAAGCGAAGU). The sequence of the control siRNA was identical to that of the HDAC7-specific siRNA, except for two mismatches at positions 9 and 10 of the recognition sequence (sequence, AGACAAGAUUAAGCGAAGU). The resuspended cells were transferred to a cuvette and pulsed using program B13 of the Amaxa Nucleofector device. Three hours after transfection, the cells were sorted for the levels of incorporation of the fluorescent oligonucleotide using a FACSDiva cell sorter (Becton Dickinson). This process was repeated three times at 48-h intervals, and the cells were plated on DCEK-ICAM cell layers as described below on the day following the final transfection, 5 days after the initial transfection.
Both DPK and DO11.10 cells were virally transduced by spin infection, as described in reference 65, using supernatants from BOSC23 or Phoenix Ampho cells 2 days after transfection with viral constructs. DO11.10 cells were transduced with ecotropic viral particles (BOSC23), and DPK cells with amphotropic viral particles (Phoenix Ampho). DO11.10 cells expressing HDAC7-VP16 were subjected to four cycles of spin infection at 12-h intervals, sorted for levels of GFP expression with a FACSDiva cell sorter (Becton Dickinson), cultured for 24 to 48 h, and harvested for microarray hybridization or Northern blotting. DO11.10 cells expressing HDAC7-ΔP were made as described previously (17). DO11.10 cells expressing MEF2D-VP16 were spin infected three times at 12-h intervals, cultured in the presence of 10 μg/ml puromycin (Sigma) for 5 days and then for 48 h in medium without puromycin, and harvested for microarray experiments. Cells expressing Nur77 were spin infected four times at 12-h intervals, sorted for GFP expression, and harvested the following day for microarray analysis or assays of levels of apoptosis, GFP expression, and Nur77 expression. The levels of apoptosis of Nur77-transduced DO11.10 cells were measured by staining with allophycocyanin-conjugated annexin V (Invitrogen) followed by flow cytometric analysis.
DPK cells were spin infected with empty MSCVIG or MSCVIG encoding HDAC7-VP16 or HDAC7-ΔP six times over 4 days, sorted for GFP expression, and cultured for 3 more days before use in the described experiments.
Microarray probe preparation, hybridization, and data analysis.
Total thymocytes for microarray analysis were harvested from 5- to 6-week-old untreated MHC−/− mice and DO11.10 TCR transgenic mice that were untreated or had been injected with antigenic peptide 3, 6, or 12 h previously. RNA was prepared from two independent experiments for each condition studied. RNA for microarray hybridization and blotting was prepared from DO11.10 cells that were untreated or treated for 2.5 h with 10 ng/ml phorbol myristate acetate (PMA) (Sigma) plus 0.5 mM ionomycin (Sigma). RNA was prepared from two independent cell populations for each experiment. Total RNA was prepared from thymocytes or DO11.10 cells by using guanidinium isothiocyanate, as described in reference 13a. For each probe, 30 μg of total RNA was reverse-transcribed using ImProm II reverse transcriptase (Promega), 500 μM dATP, dCTP, and dGTP, 350 μM aminoallyl-dUTP (Sigma), and 150 μM dTTP. The RNA was degraded by alkaline hydrolysis, and cDNA was recovered by ethanol precipitation. Labeled cDNA was coupled to N-hydroxysuccinimide-Cy3 or N-hydroxysuccinimide-Cy5 (Amersham-Pharmacia) purified with a QIAquick PCR purification kit (QIAGEN). The probes were resuspended in GeneTAC microarray hybridization buffer (Perkin-Elmer) with 30% formamide and applied to glass slides spotted with the Operon version 2.0 mouse oligonucleotide set (QIAGEN). Hybridization was performed with a GeneTAC Hybstation (Perkin-Elmer) for 3 h at 45°C, followed by 17 h at 42°C. Slides were washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.08% sodium dodecyl sulfate (SDS), twice with 0.1× SSC-0.04% SDS, and twice with 0.05× SSC, and scanned with a GenePix 4000B scanner (Axon Instruments). The fluorescence data for microarray features were acquired with the GenePix 6.0 software (Axon Instruments). The median fluorescence ratios were normalized by LOWESS and print-tip normalization, without background subtraction, by using the Acuity 4.0 software package (Axon Instruments). The ratio data were then analyzed for statistical significance using the Significance Analysis of Microarrays (SAM) analysis package (88) (www-stat.stanford.edu/∼tibs/SAM/).
Statistical significance was assessed with the two-class SAM t test. For DO11.10 cells, all arrays hybridized had cDNA from unmanipulated DO11.10 cells in one fluorescence channel and cDNA from virally transduced cells in the other. Thus, two-class comparisons were made between arrays hybridized with unmanipulated DO11.10 cDNA and cDNA from empty virus-transduced DO11.10 cells (unstimulated or PMA-ionomycin stimulated) as a negative control and arrays hybridized with unmanipulated DO11.10 cDNA and cDNA from HDAC7-VP16, HDAC7-ΔP, MEF2-VP16, or Nur77-transduced cells as the experimental condition. This allows for statistically valid comparisons to be made between any two arrays. For HDAC7-VP16 versus empty virus, 10 replicate arrays were analyzed. For HDAC7-ΔP versus stimulated empty virus, eight replicate arrays were analyzed. For MEF2-VP16 and Nur77 expression versus empty virus, four replicate arrays were analyzed. For thymocytes, SAM two-class comparisons were made by using ratio data derived from separate arrays hybridized with RNA from untreated DO11.10 thymocytes in different fluorescence channels as the control condition and ratios from arrays hybridized with DO11.10 thymocyte cDNA in one channel and MHC null thymocyte cDNA or treated DO11.10 thymocyte cDNA in the other. Six replicate arrays were analyzed for each of the experimental and control conditions. In all cases, an equal number of dye-swapped replicates were analyzed for each sample. Full ratio and statistical data for all experiments are provided in data supplements which may be downloaded as indicated below. A side-by-side comparison of all array results presented here is available at http://www.gladstone.ucsf.edu/gladstone/files/verdin/TableS10.xls.
Northern blotting.
Total RNA for Northern blotting was prepared as described above from DO11.10 cells transduced with empty or HDAC7 mutant-encoding viral constructs. Where indicated, cells were treated with PMA and ionomycin as in the microarray screen. RNA (25 μg/lane) was resolved by formaldehyde agarose gel electrophoresis and transferred to Hybond XL nylon membranes (Amersham). The membranes were hybridized using ULTRAhyb-Oligo hybridization buffer (Ambion). 32P-labeled oligonucleotide probes were prepared by end labeling using 70 μCi of [γ-32P]ATP (Amersham), 2 pmol 40-mer oligonucleotide (Elim Biopharmaceuticals, Inc.), and 20 U T4 polynucleotide kinase (New England Biolabs) in a volume of 30 μl. Probes were purified by size exclusion chromatography, briefly denatured at 70°C, and hybridized to membranes overnight. After being washed, the membranes were imaged with a FUJIX BAS1000 phosphor imaging system (Fuji, Tokyo, Japan). After being probed for the expression of putative HDAC7 targets, the membranes were stripped by incubation at 80°C in 0.05% SSC-1% SDS and reprobed as described above with a probe specific for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The probe sequences for genes yielding positive results were as follows: Actn1, 5′-TGGTCCCTCCGGGGCACTAGCTGCCGTACATGGTCCCATT-3′; Asb6, 5′-CCCGGCACAGGTGTTTGAGAGGTGGCGGGTAGCTCTCGAG-3′; Bpgm, 5′-ATAGGAGGTGGGGTCACGTTGTAGCTTCTCCTCCAGAGCC-3′; Cd28, 5′-CCATTGCTCCTCTCGTTGTCTAGGTAAGGCGGAGGGTACA-3′; Cd96, 5′-CAGAAAAAGAGCAAAGCCGCGACAAGCACAGGCCAGGACA-3′; Dgkz, 5′-CGCTGCCGGGGAGTGTCGCCCTGCAGATCTGTCTTCATGA-3′; Gpr146, 5′-TTCAGCGTGTCTCCTGGGCTGGAAGCTCCATGTGCCTGGG-3′; Gsn, 5′-CCAGCCCACGAAGGAAGGAGGCTCAAAGCCCTGCCTAACG-3′; Hdac5, 5′-AGCCGAGACACAGGCCTCAGAGGCATCACAGATGGCGGTC-3′; Hipk3, 5′-GGTCTCTTGCAAGGGGACGGGCTGGACTGCCCTGAGGAGC-3′; Itgb2, 5′-GTTGCATTGGCAGTGGCCACGGCCACTGCACTCTACCAGC-3′; Pglyrp2, 5′-GCGCGTTCCCGGGGCAGTGGGTGAGCACTAGCTGGCGGTG-3′; Siat8d, 5′-TGTTATGATTCCTTATGCCATCATGTGTGTTAAGTCCTAA-3′; Trim35, 5′-GGACCGTCACCTCGAGAGGCTCCCAGGTAGAAGTAGGGC-3′; Zap70, 5′-CACATGGTGACCCCATAGCTCCAGACGTCACTGCGGCTGG-3′; 4631408o11rik, 5′-GGCACAGATCTGGTGTGTGGCTCCTGGCTTCTACACAGCC-3′; BCL2, 5′-AGTTCACCCCATCCCTGAAGAGTTCCTCCACCACCGTGGC-3′; Ccr8, 5′-GGCTGTGACAGCAGCCAGCCACACTGTCAGACTCAGGGCT-3′; Cd5, 5′-GGTCACAGTGGCTCCTAGAGGATTGGGACTCCTGGATACT-3′; Cd6, 5′-GAGATAAGGCCTCAGGGTCACCCAGCATGACCCTTCACCA-3′; Dusp2, 5′-ACGCGCTGCTTTCATCCAGCACCACGGCACGGGCCAGCTC-3′; Dusp10, 5′-GTCATCCGTGTGTGCTTCATCAAGTAGGCGATGACGATGG-3′; Gpr65, 5′-TGAAGACAGACGTATACAGAGGAGAAGACTGATGCAGGCAG-3′; Lta, 5′-TCGGTGTGGGTGGACAGCTGGTCTCCCTTACTGAGCAGGA-3′; Ltb, 5′-ACAACAGGGTCACCAGGGAAGTAGCTCCTGCCACAGCCAG-3′; Nab2, 5′-CCAGTCTCAGAGTGAAGGTTCCACTGTAAGTCGCTAGACC-3′; Pdcd1, 5′-AGCTTTTGCCTGGTAAGTGCTCATGCTTCCCAGACCCCCG-3′; Pscdbp, 5′-TTGATCCTGAGATTTCTGGCCTTGGCATGAATGTTCAGGC-3′; Scotin, 5′-TTCAAGTGAGGAAGAAAACAGGTTGGTCCCGACCCACACT-3′; Sh2d2a, 5′-GCCCTGTTGGAGGAGCAGGCTGAGCTGGGTGTCTGGGTCC-3′; Zfp36l1, 5′-GCAACAACTCTCTGAGGGAGGTGAGTAACTATAAGGAAGA-3′; Cdkn1a, 5′-CTCTGGGAGGCCTGTCTCACCACCAAGGACCCACCCTAGA-3′; Cish, 5′-CAGGTGGCAAGGGCAGGCAAGTGAGTGGAGACCTAGGTCC-3′; Eno3, 5′-GCCTTTGGATTACGGAACTTTCTTCCAGCAAAGACAGCTT-3′; Gadd45b, 5′-AGGAGCTGCGCCAGCCTCTGCATGCCTGATACCCGGACGA-3′; Icsbp1, 5′-GCAGAGCACCTAGGCTTCTCTAGCTGCGTGGAGCATGTAA-3′; Irf4, 5′-TTCACTTACGGTGCAGGCTTCTTCAATGAGTGAAGAAGCA-3′; Lfng, 5′-TAGGGCATCAGAGGGTGGGGCTGAAGACTTCGCCTCCACC-3′; Socs1, 5′-TGAGAGGTGGGATGAGGTCTCCAGCCAGAAGTGGGAGGCA-3′; Tnfrsf9, 5′-CAGGGATGCATGGGTCTCCCCCTTCAGACAGTGGAGGTGT-3′; Trib1, 5′-AGTGCTTCCTTCCCATTCAAATGTTCAAGTTCACTACTAC-3′; and Gapdh, 5′-GTCATTGAGAGCAATGCCAGCCCCGGCATCGAAGGTGGAA-3′.
ChIP.
The results shown are representative of at least three independent experiments. Chromatin immunoprecipitations (ChIPs) were performed by using a modified version of the protocol specified in the Upstate cell signaling solutions acetyl-histone H3 ChIP assay kit (Upstate Biotechnology). Briefly, thymocytes (8 × 108) were harvested from mice as specified above and resuspended in RPMI medium with 10% FBS at 37°C, in a total volume of 350 ml, in two flasks. One flask was untreated, and the other one was treated with 10 ng/ml PMA and 0.25 μM ionomycin for 1 h. Thymocytes were fixed with 1% formaldehyde for 15 min at room temperature, quenched with 1.25 M glycine, washed, and resuspended in swelling buffer (25 mM HEPES, pH 7.8, 1.5 mM MgCl2, 10 mM KCl, 0.5% NP-40, 1 mM dithiothreitol, and 1% mammalian protease inhibitor cocktail [Sigma]), at a density of 1 × 107 cells/ml. After a 10-min incubation on ice, the cells were pelleted and resuspended in sonication buffer (0.4% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1, protease inhibitors) at a density of 4 × 107cells/ml. The cells were sonicated in 5-ml aliquots until the DNA was reduced to less than 3 kb in size. After this step, the protocol was as described in the Upstate Biotechnology ChIP assay kit, except that the cleared chromatin solutions were diluted 1:4 in dilution buffer, rather than 1:10. Aliquots of diluted chromatin (12 ml) were precleared as described above and immunoprecipitated overnight with antibodies to the hemagglutinin epitope tag (rabbit polyclonal, Y-11; Santa Cruz Biotechnology), MEF2D (goat polyclonal, P-17; Santa Cruz Biotechnology), or HDAC7 (rabbit polyclonal, H-273; Santa Cruz Biotechnology), at 2 μg/ml, washed, and eluted as described above. An amount of 100 μl of the final input or output chromatin represented 2 × 107 thymocytes. Output chromatin (5 μl) and threefold serial dilutions of input chromatin (1.35, 0.45, 0.15, or 0.05 μl) were amplified in a volume of 50 μl for 34 cycles, resolved by agarose gel electrophoresis, and stained with ethidium bromide. The following primers were used for amplification: BCL2 −30 kb 5′, 5′-CAGCCTCAGATGTATGAAACTCTGTCTTAAAA-3′; BCL2 −30 kb 3′, 5′-GGAATAAGGCCTAGAATCTATATGGATCAATAA-3′; Nur77 promoter 5′, 5′-GCGGGCACGGATTTACAACACC-3′; Nur77 promoter 3′, 5′-GGGTTCCATTGACGCAGGGAG-3′; HDAC5 promoter 5′, 5′-CGACGGCTCCTCCATCTTTGC-3′; and HDAC5 promoter 3′, 5′-CTATTGGTCCCGCCCATCTGAG-3′.
DPK cell differentiation.
The results shown are representative of three separate experiments. DCEK-ICAM cells were grown to confluence in T-175 flasks and treated with 100 μg/ml mitomycin C (Sigma) in phosphate-buffered saline for 30 min at 37°C. Mitomycin B-treated cells were plated on 6-cm dishes at 3.5 × 106 cells/dish in 5 ml of Click's medium with 12% FBS and incubated overnight at 37°C. DPK cells, unmanipulated, transduced, or transfected as described above, were added to the dishes (final concentration, 0.35 × 106 cells/ml). Pigeon cytochrome c-derived antigenic peptide (KAERADLIAYLKQATAK; Research Genetics) was added to some of the dishes (at a concentration of 1 μM for the experiment whose results are shown in Fig. 9, panels A to D, or as indicated for the experiment whose results are shown in panel F). The cells were harvested from the dishes for analysis by gentle washing to remove adherent DPK cells and gated on GFP expression to exclude any DCEK-ICAM cells removed from the dishes. The levels of CD4/CD8 expression were assayed 48 or 72 h after plating, as indicated in the figure legend, by staining with phycoerythrin-conjugated antibodies to CD4 (RM2504; Caltag Laboratories) and allophycocyanin-conjugated antibodies to CD8 (MCD0805; Caltag Laboratories). The levels of CD5 expression were assayed at 0, 24, 48, and 72 h after plating by staining with biotin-conjugated antibody to CD5 (01032A; Becton Dickinson) and allophycocyanin-conjugated streptavidin (SA1005; Caltag Laboratories). Flow cytometric analysis was performed by using a FACSCalibur flow cytometer (Becton Dickinson) and FlowJo version 7.13 analysis software (Treestar, Inc.).
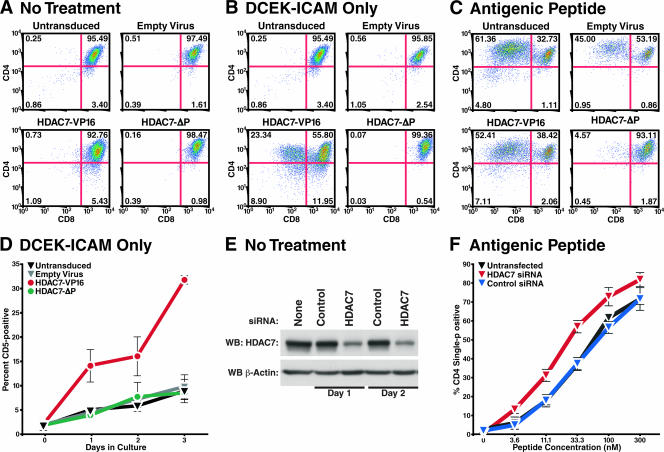
FIG. 9.
Effects of HDAC7 mutants or HDAC7 knockdown on antigen-induced differentiation of DPK cells. (A to D) DPK cells were transduced with either empty virus or virus encoding HDAC7-VP16 or HDAC7-ΔP, as indicated. The cells were then left untreated (A) or plated on DCEK-ICAM cell layers alone (B, D) or with antigenic peptide at a concentration of 1 μM (C). Panels A to C depict the fluorescence intensities of CD4 (vertical axis) and CD8 (horizontal axis) staining after 3 days of treatment. Panel D depicts the percentages of CD5-expressing cells over 3 days in culture. Error bars indicate the standard deviations of three replicate cultures. (E, F) DPK cells were transfected with mismatched control siRNA oligonucleotides or oligonucleotides specific for HDAC7 and plated on DCEK-ICAM cell layers in the presence of various concentrations of antigenic peptide. (E) Levels of HDAC7 expression, measured by Western blotting (WB), in DPK cells at 24 h (day 1) and 48 h (day 2) after plating on DCEK-ICAM cells. (F) Percentages of CD4 single-positive cells in DPK cultures after 2 days of culture in the presence of the indicated peptide concentration. Error bars indicate the standard deviations of four replicate cultures.
Microarray data accession number.
The results of all DNA microarray experiments associated with this work have been deposited in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) at the National Center for Biotechnology Information under the accession number GSE7648.
RESULTS
Regulation of gene expression in DO11.10 cells by HDAC7.
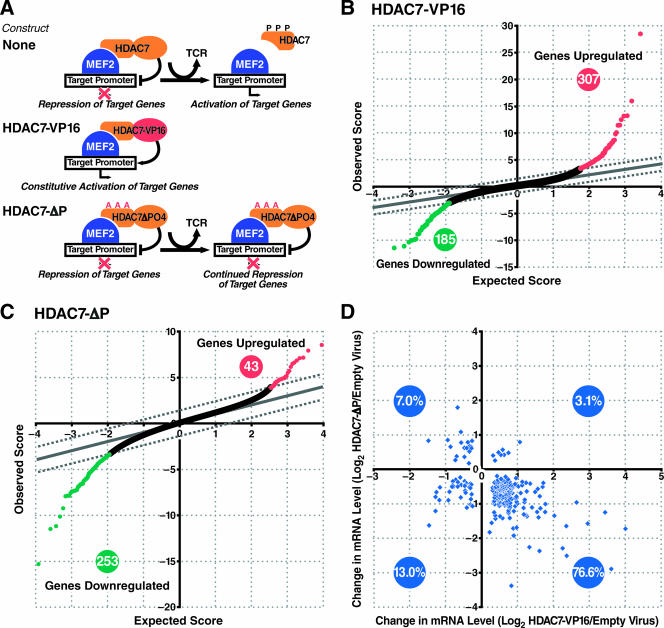
To identify genes regulated by HDAC7, we examined the global changes in gene expression in DO11.10 hybridoma cells caused by the expression of two mutant versions of HDAC7: HDAC7-VP16 and HDAC7-ΔP. HDAC7-VP16 is a fusion of the N-terminal regulatory domain of HDAC7 (residues 1 to 482) to the C-terminal activation domain of HSV VP16 (residues 412 to 490). This molecule, in which the repressive HDAC domain of HDAC7 has been replaced with the activation domain of VP16, activates rather than represses transcription (Fig. 1A). This is demonstrated by its activation of Nur77 expression in DO11.10 cells in the absence of antigen receptor signaling (17), as well as the ability of analogous constructs to induce differentiation in other systems independently of signals that are normally required (26, 47). HDAC7-ΔP is a mutant of HDAC7 in which three conserved serine residues at positions 155, 321, and 448 have been changed to alanine. This molecule is no longer subject to signal-dependent nuclear export upon phosphorylation by PKD (18, 64, 90) and continues to repress target genes in the presence of extracellular signals from target promoters that would normally induce its phosphorylation and removal (Fig. 1A). DO11.10 cells expressing HDAC7-ΔP are deficient in Nur77 expression in response to TCR signaling and are protected from activation-induced apoptosis (17). Similarly, a phosphorylation-deficient mutant of HDAC5 inhibits cardiac hypertrophy when expressed as a transgene in mice (98).
FIG. 1.
Identification of putative HDAC7 targets in T cells. DO11.10 cells were transduced with viruses encoding HDAC7-VP16 or HDAC7-ΔP and examined for changes in gene expression in the absence of stimulation for HDAC7-VP16 or after PMA-ionomycin stimulation for HDAC7-ΔP. (A) Diagram depicting how HDAC7 mutants are expected to perturb gene expression in DO11.10 cells. (B, C) SAM two-class t test significance results for the differential gene expression induced by HDAC7-VP16 (B) or HDAC7-ΔP (C). Dashed lines represent the SAM score cutoff corresponding to an 0.85% overall false-discovery rate. (D) Scatter plot depicting the log2-fold differential expression for HDAC7-ΔP (vertical axis) and HDAC7-VP16 (horizontal axis) for genes that showed significantly differential expression for both HDAC7 screen conditions.
DO11.10 T-cell hybridomas were infected with viral vectors encoding HDAC7-VP16 or HDAC7-ΔP. Gene expression profiles were generated comparing the basal levels of gene expression in cells infected with empty virus with the levels in cells infected with HDAC7-VP16 and in PMA-ionomycin-stimulated cells infected with empty virus with the levels in cells infected with HDAC7-ΔP. The arrays were spotted at the Gladstone genomics core facility, using the Operon version 2.0 oligonucleotide set which comprises 16,463 mouse transcripts. Each array was hybridized with RNA from DO11.10 cells infected with viral expression vectors in one fluorescence channel and RNA from unmanipulated DO11.10 cells in the other, providing a universal standard of comparison between arrays. The statistical significance of differences between the arrays was measured using the Stanford University SAM data analysis package (88). Details are provided under “Microarray probe preparation, hybridization, and data analysis” in Materials and Methods.
Using 10 replicate arrays, we observed that 307 genes were upregulated by HDAC7-V16, while 185 genes were downregulated, at an overall false-discovery rate of 0.77%, according to the SAM algorithm (Fig. 1B) (additional data may be downloaded at http://www.gladstone.ucsf.edu/gladstone/files/verdin/TableS1.xls). For HDAC7-ΔP, using 8 replicate arrays, 43 genes were upregulated and 253 genes downregulated at a false-discovery rate of 0.85% (Fig. 1C) (additional data may be downloaded at http://www.gladstone.ucsf.edu/gladstone/files/verdin/TableS2.xls). Of the 704 total genes that meet these statistical significance criteria in either screen, 257 genes were differentially expressed in both HDAC7 target screens, using the requirement that both genes have a false-discovery rate of less than 20%. Of these 257 genes, 197 (77%) were found to be both upregulated by HDAC7-VP16 and downregulated by HDAC7-ΔP, as would be expected for direct targets of HDAC7, based on the design of the screen (Fig. 1D). Conversely, 18 of these genes (7%) were upregulated by HDAC7-ΔP and downregulated by HDAC7-VP16, 3% of the genes were upregulated by both constructs, and 13% were downregulated by both, as might be expected for some indirect targets of HDAC7. The large number of genes downregulated by HDAC7-VP16 in comparison to those upregulated by HDAC7-ΔP is probably due to nonspecific squelching of gene expression by the VP16 activation domain. This idea is supported by a comparison of the differential gene expression profiles for cells transduced with HDAC7-VP16 and HDAC7 (Δ75-91)-VP16, a construct in which the sequence mediating the interactions with MEF2, Runx, and CAMTA has been disrupted. The expression of HDAC7 (Δ75-91)-VP16 in DO11.10 cells downregulates many of the same genes as HDAC7-VP16, but upregulates only one gene that is also upregulated in response to HDAC7-VP16 (data not shown). For this reason, we did not further consider genes that are downregulated by HDAC7-VP16 unless they are also differentially expressed in response to HDAC7-ΔP.
Ontological analysis of putative HDAC7 targets.
To obtain a broad view of the potential physiologic significance of these differentially expressed genes, we used MAPPFinder, a module of GenMAPP (Gene Map Annotator and Pathway Profiler, http://www.genmapp.org). MAPPFinder is a tool for the ontological analysis of gene expression data, which allows users to identify statistically overrepresented ontological categories within a set of genes of interest. These categories are then organized as a hierarchical tree.
As input for MAPPFinder, we used the 257 genes identified as differentially expressed in both screens (Fig. 1D), as well as another 44 genes (∼10% of the total) that were identified by only one screen but were differentially expressed by more than twofold and at a false detection rate of less than 0.1%. Among these putative HDAC7 targets, MAPPFinder identified genes in 104 of 2,996 ontological categories relating to physiologic processes as statistically overrepresented (at a P value of <0.05) (data may be downloaded at http://www.gladstone.ucsf.edu/gladstone/files/verdin/tables3.xls). Fifty-five of these categories can be assigned to four branches of the MAPPFinder ontology term hierarchy. These categories (Table 1) are development, defense/immune response, signal transduction, and cell death/apoptosis. Interestingly, the category of development includes proteins that play key roles in known physiologic functions of the class IIa HDACs, including muscle and bone development (reviewed in reference 52) (91), as well as hemopoiesis, T-cell development, and thymic selection. The category of the immune response contains genes involved in T-cell activation, T-cell costimulation, and the TH1 response, supporting the idea that HDAC7 might play a role in TCR signaling. Putative HDAC7 targets in the category of cell death, in which HDAC7 has a known function in T cells (17), include a variety of known regulators of apoptosis in thymocytes: Nur77, BCL2, CD28, and CD5.
TABLE 1.
Summary of MAPPFinder gene ontology analysis of putative HDAC7 targetsa
| Gene ontology term | No. of genesb | Z scorec | P valued |
|---|---|---|---|
| Development | 64 | 2.68 | 0.004 |
| Lymph node development | 3 | 4.18 | 0.008 |
| Hemopoiesis | 10 | 2.84 | 0.005 |
| T-cell differentiation | 3 | 2.37 | 0.042 |
| Positive regulation of αβ T-cell differentiation | 1 | 4.31 | 0.047 |
| Negative thymic T-cell selection | 2 | 4.43 | 0.007 |
| B-cell differentiation | 3 | 2.44 | 0.036 |
| Myeloid-cell differentiation | 5 | 2.43 | 0.039 |
| Regulation of myeloid-cell differentiation | 3 | 2.44 | 0.042 |
| Muscle development | 8 | 2.49 | 0.013 |
| Regulation of striated muscle development | 3 | 5.01 | 0.001 |
| Negative regulation of striated muscle development | 2 | 4.43 | 0.015 |
| Regulation of ossification | 5 | 7.67 | 0 |
| Odontogenesis (sensu Vertebrata) | 3 | 3.47 | 0.016 |
| Negative regulation of ossification | 3 | 6.85 | 0.002 |
| Negative regulation of bone mineralization | 2 | 7.13 | 0.003 |
| Positive regulation of bone mineralization | 1 | 4.31 | 0.048 |
| Eye photoreceptor cell differentiation | 2 | 3.55 | 0.023 |
| Compartment specification | 2 | 4.85 | 0.009 |
| Anterior compartment specification | 1 | 4.31 | 0.048 |
| Posterior compartment specification | 1 | 4.31 | 0.048 |
| Defense response | 34 | 4.54 | 0 |
| Immune response | 33 | 4.77 | 0 |
| Inflammatory response | 11 | 3.08 | 0.010 |
| Regulation of inflammatory response | 3 | 3.15 | 0.015 |
| Positive regulation of inflammatory response | 2 | 3.15 | 0.025 |
| Cellular defense response | 6 | 2.37 | 0.025 |
| T-cell activation | 8 | 4.07 | 0.003 |
| Regulation of T-cell activation | 7 | 4.88 | 0 |
| T-cell costimulation | 2 | 5.38 | 0.003 |
| T-helper-1-type immune response | 4 | 2.99 | 0.014 |
| Positive regulation of interleukin 2 biosynthesis | 3 | 5.01 | 0.003 |
| Positive regulation of interleukin 13 biosynthesis | 1 | 4.31 | 0.038 |
| Positive regulation of interleukin 10 biosynthesis | 1 | 4.31 | 0.041 |
| Humoral immune response | 7 | 2.34 | 0.022 |
| B-cell activation | 7 | 4.63 | 0 |
| Complement activation and alternative pathway | 2 | 3.80 | 0.025 |
| Signal transduction | 77 | 3.93 | 0.001 |
| Intracellular signaling cascade | 43 | 4.38 | 0 |
| Protein kinase cascade | 22 | 6.06 | 0 |
| Negative regulation of protein kinase activity | 4 | 2.87 | 0.032 |
| MAPKK kinase cascade | 7 | 2.54 | 0.015 |
| Protein kinase C activation | 3 | 4.01 | 0.011 |
| I-κB kinase/NF-κB cascade | 6 | 2.78 | 0.02 |
| Insulin receptor signaling pathway | 3 | 3.25 | 0.017 |
| Negative regulation of insulin receptor pathway | 2 | 6.10 | 0.003 |
| Cell death | 30 | 4.78 | 0 |
| Apoptosis | 28 | 4.47 | 0 |
| Regulation of apoptosis | 21 | 4.15 | 0.001 |
| Negative regulation of apoptosis | 10 | 3.48 | 0.006 |
| Antiapoptosis | 8 | 3.20 | 0.008 |
| Induction of apoptosis | 11 | 3.73 | 0.001 |
| Apoptotic program | 4 | 2.28 | 0.034 |
| Release of cytochrome c | 2 | 5.38 | 0.005 |
The four overrepresented branches of the MAPPFinder ontology hierarchy with the largest number of overrepresented child nodes are listed in bold type. The entries in plain type are all children of the nodes listed in bold type above them. The indentation of entries indicates the nesting of subcategories.
The number of putative HDAC7 targets that are found in the corresponding categories and their children.
The distance in standard deviation units of the observed representation of the category from its expected representation.
The permuted P value for the observed level of enrichment.
In order to confirm that the overrepresentation of these gene ontology categories is specific to perturbation of HDAC7 function and not simply the result of querying a gene expression profile in a T-cell line, we did similar MAPPFinder analyses with two sets of 301 randomly selected genes that were expressed in DO11.10 cells, according to our microarray data. These randomly selected gene sets identified 28 and 35 ontological categories as overrepresented, respectively, but none of the categories related to T cells or immune function, and only 5 of the 104 categories identified by the HDAC7 screen were identified by either of the random gene sets (data may be downloaded at http://www.gladstone.ucsf.edu/gladstone/files/verdin/tables3.xls).
Comparison of putative HDAC7 targets to genes differentially expressed during T-cell development.
HDAC7 is expressed at high levels in CD4+/CD8+ thymocytes, a lymphocyte developmental stage characterized by positive and negative selection. To elucidate the potential role of HDAC7 in these developmental events, we compared our list of putative HDAC7 targets to genes that are differentially expressed during either positive or negative selection of thymocytes. We generated differential gene expression profiles associated with positive selection by comparing the levels of gene expression for thymocytes from MHC-deficient mice, which have no ligand for the TCR, and thymocytes from DO11.10 TCR-transgenic mice, which receive positively selecting TCR signals (57) (data may be downloaded at http://www.gladstone.ucsf.edu/gladstone/files/verdin/TableS4.xls). Gene expression profiles associated with antigen receptor signals leading to deletion (negative selection) were generated by comparing the levels of gene expression in positively selected thymocytes from untreated DO11.10 mice to the levels of gene expression in thymocytes from DO11.10 mice injected with a deletion-inducing antigenic peptide (3, 6, and 12 h postinjection) (data may be downloaded at http://www.gladstone.ucsf.edu/gladstone/files/verdin/TableS5.xls, http://www.gladstone.ucsf.edu/gladstone/files/verdin/TableS6.xls, and http://www.gladstone.ucsf.edu/gladstone/files/verdin/TableS7.xls). With respect to differential levels of expression during thymic selection, our criteria for inclusion in the comparison were that the genes be differentially expressed by at least 1.75-fold and at a false-discovery rate of <0.1% for positive selection and <0.5% for negative selection. With respect to putative HDAC7 targets, we used the same criteria as those employed for our MAPPFinder analysis described above, with the added constraint that genes identified by both screens must also be differentially expressed by at least 1.75-fold in at least one of them.
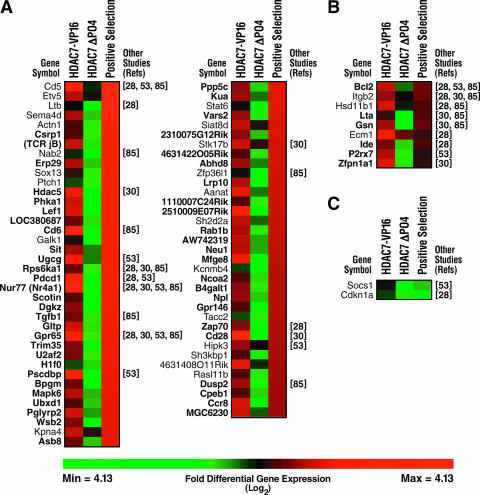
We identified 142 putative HDAC7 targets that met the above criteria and were also upregulated by HDAC7-VP16 and/or downregulated by HDAC7-ΔP. Of these, 73 genes were upregulated during positive selection of DO11.10 TCR transgenic thymocytes (Fig. 2A). Eighteen of these genes were also found to be upregulated during positive thymic selection according to at least one of four other studies that recently examined differential levels of gene expression during positive selection (Fig. 2A) (28, 30, 53, 85). An additional nine of our putative HDAC7 targets were identified as being upregulated during positive selection according to one of these published studies, but not according to ours (Fig. 2B). We found that only two putative HDAC7 targets were downregulated during positive selection according to our study and/or those of others (Fig. 2C). We identified 29 HDAC7 targets that met the statistical and fold-differential expression criteria for the comparison but did not respond to introduction of the HDAC7 mutants in the manner expected for direct targets of HDAC7. Of these, 8 genes were upregulated by HDAC7-ΔP and downregulated by HDAC7-VP16, 8 were upregulated by HDAC7-ΔP alone, 12 were downregulated by both constructs, and 1 was upregulated by both constructs. Only four of these genes were upregulated during positive thymic selection, according to our screen and/or those of other groups, and two were downregulated (data not shown). We observed a remarkable preponderance of signaling molecules with known functions in T cells among the putative targets of HDAC7 that are upregulated in positive selection. These include Nur77 (Nr4a1), TDAG8 (Gpr65), RSK1 (Rps6ka1), PAC-1 (Dusp2), CD5, CD6, PD-1 (Pdcd1), CD28, BCL2, and LFA-1 (Itgb2), suggesting that HDAC7 might play an important role in coupling antigen receptor signaling to downstream cell differentiation events during positive selection.
FIG. 2.
Comparison of HDAC7 putative target list to genes differentially expressed during positive selection. The colors represent the log2-fold differential regulation of genes, as indicated on the scale bar. The references listed at the ends of the rows indicate other studies that found the corresponding genes to be upregulated during positive selection and thymocyte differentiation. Genes listed in bold type were differentially expressed in both HDAC7 screen conditions, and genes listed in plain type were differentially expressed in only one HDAC7 screen condition. (A) Putative HDAC7 targets that are upregulated during positive thymic selection, according to our study. (B) Putative HDAC7 targets upregulated during positive thymic selection, according to the studies of other groups. (C) Putative HDAC7 targets downregulated during positive thymic selection.
We observed a strikingly different pattern of gene regulation in negative selection than in positive selection among putative HDAC7 targets. Among the 142 genes that met our criteria and were upregulated by HDAC7-VP16 and/or downregulated by HDAC7-ΔP, we found that 20 were also upregulated during the antigen-induced deletion of DO11.10 TCR-transgenic thymocytes (Fig. 3A). Thirteen of these genes were upregulated during negative selection, according to at least one of three published studies of gene regulation during thymic selection (Fig. 3A) (19, 77, 102). An additional 10 genes were upregulated during negative selection according to one of these studies, but not our own (Fig. 3B). Among the 29 genes that did not have the expected profile of regulation by our HDAC7 constructs (upregulated by HDAC7-VP16 and/or downregulated by HDAC7-ΔP), we found 4 genes that were upregulated during negative selection, according to our study (data not shown).
FIG. 3.
Comparison of HDAC7 putative target list to genes differentially expressed during negative selection. The colors represent the log2-fold differential regulation of genes, as indicated on the scale bar. The references listed at the ends of the rows indicate other studies that found the corresponding genes to be upregulated during antigen-induced deletion. Genes listed in bold type were differentially expressed in both HDAC7 screen conditions, and genes listed in plain type were differentially expressed in only one HDAC7 screen condition. (A) Putative HDAC7 targets that are upregulated during peptide-induced deletion, according to our study. (B) Putative HDAC7 targets upregulated during peptide-induced deletion, according to the studies of other groups. (C) Putative HDAC7 targets downregulated during peptide-induced deletion.
In contrast to what we observed in the case of positive selection, 29 putative HDAC7 targets were downregulated during negative selection, according to our study and/or those of others (Fig. 3C). Thus, while candidate HDAC7 targets that are differentially expressed during positive selection are 98% upregulated and only 2% downregulated in that process, HDAC7 targets that are differentially expressed in negative selection are almost evenly divided between those that are upregulated and those that are downregulated (51% versus 49%). Regulation by HDAC7 is, therefore, much better correlated with upregulation during positive selection than with upregulation during negative selection.
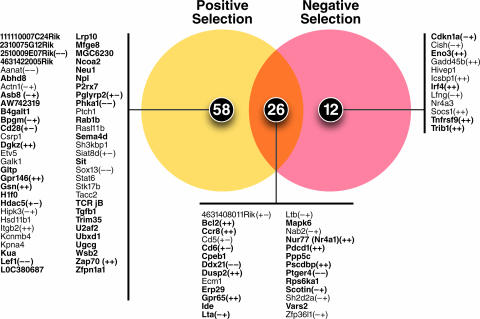
Thus, based on this clustering of the 142 genes showing the expected profiles of differential expression in response to the HDAC7 constructs, we identified 82 genes that were upregulated during positive selection (Fig. 2A and B) and 30 genes that were upregulated during negative selection (Fig. 3A and B). In order to draw conservative boundaries around genes that were upregulated during only positive or negative selection, we relaxed the criteria for differential expression within this group slightly (to 1.5-fold), while maintaining the same criteria for statistical significance. With these criteria, we found that 26 genes were upregulated during both positive and negative selection (Fig. 4, intersection), while 58 were upregulated only during positive selection (Fig. 4, left) and 12 were upregulated only during negative selection (Fig. 4, right). This distribution of putative HDAC7 targets, substantially favoring positive over negative selection, suggests that TCR signals that induce positive selection are sufficient to release HDAC7 from its target promoters, leading to their derepression.
FIG. 4.
Summary of putative HDAC7 targets associated with positive and negative selection. The Venn diagram indicates the distribution of genes upregulated by HDAC7-VP16 and/or downregulated by HDAC7-ΔP between studies identifying genes differentially expressed during positive and negative selection. Symbols in parentheses indicate a positive or negative result in the Northern blot analysis confirming differential expression of the gene. The first symbol indicates the result with HDAC7-VP16, and the second symbol the result with HDAC7-ΔP. Genes listed in bold type were differentially expressed in both HDAC7 screen conditions, and genes listed in plain type were differentially expressed in only one HDAC7 screen condition.
Verification of regulation by HDAC7 in DO11.10 cells.
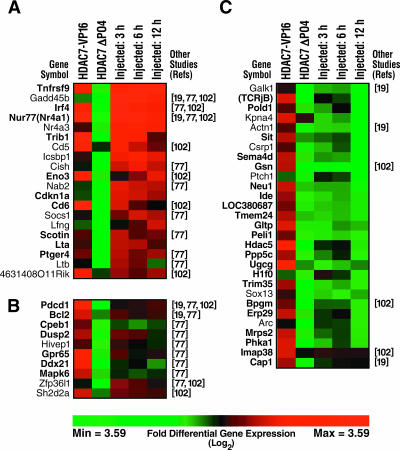
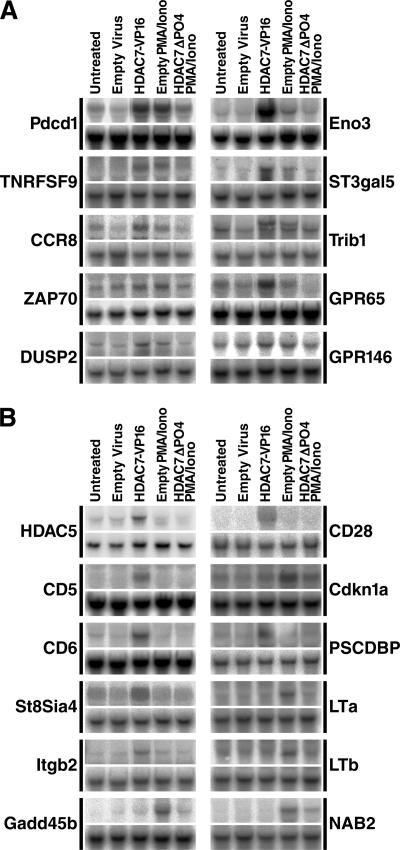
To verify by independent means that the putative HDAC7 targets are differentially expressed in DO11.10 cells in response to perturbation of HDAC7 function, we selected 46 of the 96 genes upregulated during positive or negative selection (Fig. 4) and tested them by Northern blotting. RNA was prepared from all the cell populations used in the original screen, blotted, and probed with oligonucleotides corresponding to sequences near, but not overlapping, those present on the microarray (Fig. 5A and B, upper gels). GAPDH RNA levels were used as an internal control (Fig. 5A and B, lower gels). We obtained concordant results with our microarray data for 39 genes and negative results for 7 genes, resulting in an overall false-positive rate of 15% for the genes tested (summarized in Fig. 4 by symbols in parentheses after gene names). Northern blotting results are shown for 22 of these genes, 10 genes confirmed to be differentially expressed in response to the expression of both HDAC7 mutants (Fig. 5A) and 12 genes confirmed to be differentially expressed in response to the expression of only one HDAC7 mutant (Fig. 5B). Twelve confirmed HDAC7 targets were upregulated solely during positive selection, 16 during both positive and negative selection, and 10 during negative selection only, again indicating that antigen receptor signals associated with both positive and negative selection can release HDAC7 from its target promoters.
FIG. 5.
Northern blot analysis confirming differential expression of putative HDAC7 targets in DO11.10 cells. DO11.10 cells were transduced with empty virus or virus encoding HDAC7 mutants and left untreated or treated with PMA and ionomycin, as indicated. Total RNA from these cells was resolved by agarose gel electrophoresis, blotted, and hybridized with 32P-labeled oligonucleotide probes specific for the indicated genes. (A) Genes differentially expressed in response to both HDAC7 mutant constructs. (B) Genes differentially expressed in response to only one HDAC7 mutant construct.
Regulation of HDAC7 targets in T cells by MEF2 and Nur77.
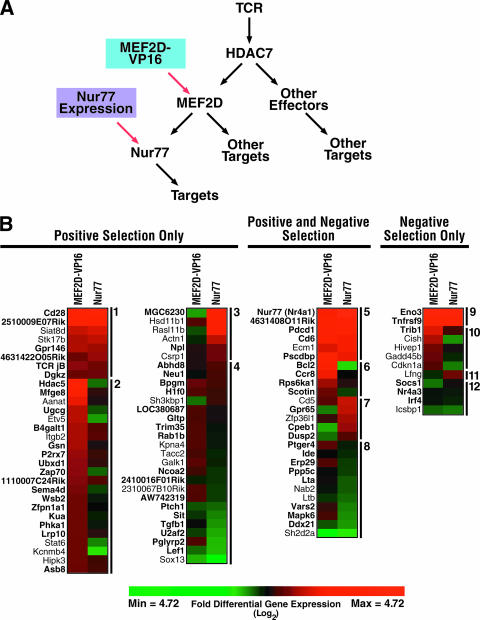
HDAC7 and other class IIa HDACs are recruited to specific promoter elements via their association with DNA-bound transcription factors such as MEF2. After TCR engagement, the phosphorylation and nuclear export of HDAC7 allows the derepression of its target genes. HDAC7 target genes can be transcriptional activators themselves, such as Nur77, which activates its own genomic targets (Fig. 6A). The genes that we have identified in our screen described above could, therefore, represent either direct, primary HDAC7 targets, bound by HDAC7 under basal conditions, or secondary targets, activated by a primary target of HDAC7, such as Nur77. To distinguish between these two sets of genes, we analyzed gene expression changes induced in DO11.10 cells after the overexpression of either a MEF2-VP16 fusion protein or Nur77.
FIG. 6.
Microarray screen for targets of known HDAC7 downstream effectors. (A) Diagram indicating the steps at which the expression of MEF2D-VP16 and Nur77 exerts their effects in the transcriptional cascade downstream of HDAC7. (B) Differential expression values due to the expression of MEF2D-VP16 or Nur77 for genes differentially expressed in our HDAC7 target screen and positive or negative selection. See Results for an explanation of the group numbers. Genes listed in bold type were differentially expressed in both HDAC7 screen conditions, and genes listed in plain type were differentially expressed in only one HDAC7 screen condition.
In T cells, MEF2D does not strongly activate transcription by itself, but depends on signal-dependent coactivators and corepressors to cause changes in gene expression levels (33). However, substituting the HSV-VP16 transcriptional activation domain for the C-terminal MEF2D transcriptional activation domain bypasses these mechanisms and causes constitutive activation of transcription. We therefore employed a MEF2D-VP16 fusion protein (residues 1 to 92 of MEF2D fused to residues 412 to 490 of HSV VP16) as a probe for MEF2-regulated genes in DO11.10 cells. Nur77 strongly activates transcription by itself, and its introduction into thymocytes as a transgene induces massive apoptosis (8, 13). We therefore expressed wild-type Nur77 to identify its transcriptional targets in T cells.
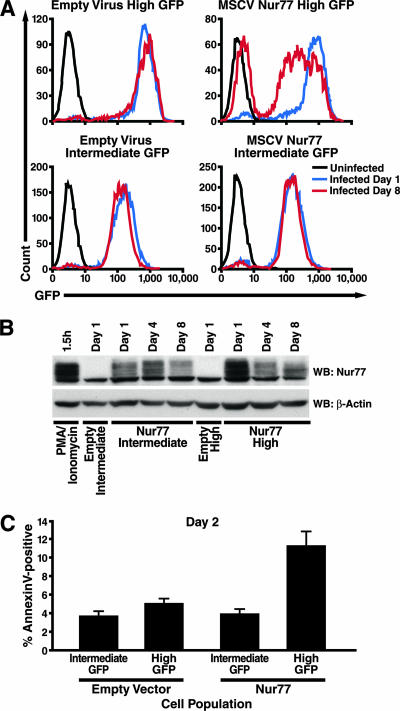
DO11.10 cells were infected with viral vectors encoding either MEF2D-VP16 or wild-type Nur77, and their gene expression profiles were compared to the expression profiles obtained after infection with empty virus. MEF2D-VP16 upregulated the expression of 575 genes (at a false detection rate of 0.5%), 169 of them by more than twofold (data may be downloaded at http://www.gladstone.ucsf.edu/gladstone/files/verdin/TableS8.xls). Similarly, the expression of Nur77 caused the upregulation of 266 genes (at a false detection rate of 0.5%), 160 of them by twofold or more (data may be downloaded at http://www.gladstone.ucsf.edu/gladstone/files/verdin/TableS9.xls). Among putative HDAC7 targets that are differentially regulated in positive and/or negative selection (Fig. 4), 59 genes (61%) are regulated via MEF2D or Nur77 (Fig. 6B, groups 1 to 3, 5 to 7, and 9 to 11), while 37 genes are not (Fig. 6B, groups 4, 8, and 12). Since the Nur77 promoter is itself regulated by MEF2D, our finding that 14 genes are regulated by Nur77 but not MEF2 (Fig. 6B, groups 3, 7, and 11) does not fit our model of how MEF2D and Nur77 expression levels are related. However, these observations can be explained by the fact that the expression of MEF2D-VP16 did not result in levels of Nur77 expression as high as those caused by its direct expression (data not shown). If some transcriptional targets of Nur77 require higher expression levels for their activation, these might not have been sufficiently activated by MEF2D-VP16 to be detected in our screen. Interestingly, we did not observe any marked bias towards positive or negative selection among HDAC7 targets regulated via MEF2 and Nur77. Overall, 61% of HDAC7 targets upregulated during thymic selection are upregulated only during positive selection, while 39% are upregulated during negative selection (Fig. 4). Similarly, 64% of MEF2-regulated HDAC7 targets are associated only with positive selection, while 36% are associated with negative selection. Nur77 regulation is only slightly more biased towards negative selection, with 50% of its target genes associated only with positive selection, and 50% associated with negative selection. While this finding may tend to contradict the prevailing notion that Nur77 expression is both a marker and a mediator of negative selection (8, 96), it is consistent with the observation that it is upregulated during positive selection in all published studies (see Fig. 2) (28, 30, 53, 85) Since Nur77 is also further upregulated during negative selection, according to all available microarray data (see Fig. 3), its level of expression may determine which targets it activates and, therefore, how it functions in thymic selection. This idea is also consistent with our observation that DO11.10 hybridoma cells can tolerate a moderate level of Nur77 expression indefinitely, but become progressively more likely to undergo spontaneous apoptosis as the level of Nur77 expression is increased. To demonstrate this, we infected DO11.10 cells with a viral construct encoding Nur77 and GFP in the same transcript, separated by an internal ribosome entry site (MSCV Nur77-IRES-GFP), or with the same vector lacking Nur77 (MSCVIG). The cells were then sorted by a fluorescence-activated cell sorter into populations with high levels of GFP expression (Fig. 7A, top) or intermediate levels of GFP expression (Fig. 7A, bottom) and maintained in culture for 8 days. In the cells infected with MSCV Nur77-IRES-GFP, a significant loss of GFP expression was observed after 8 days in culture (Fig. 7A, upper right), but no decrease in GFP expression was observed in the cells infected with MSCVIG (Fig. 7A, upper left). In the cells expressing GFP at an intermediate level, no change in GFP expression was seen for either construct (Fig. 7A, lower panels). This pattern was also observed when Nur77 expression was measured directly in these cell populations via Western blotting (Fig. 7B). Additionally, on the day following fluorescence-activated cell sorting, an increase in spontaneous apoptosis, as measured by annexin V staining, was observed in the cells expressing Nur77 at high levels compared to the levels in other cell populations (Fig. 7C).
FIG. 7.
Differential induction of apoptosis in DO11.10 cells by Nur77 expression. DO11.10 cells were transduced with either empty MSCVIG virus or MSCVIG encoding Nur77 upstream of the internal ribosome entry site and then sorted for intermediate or high GFP expression levels. (A) Changes in levels of GFP expression of sorted cells in culture. Black histogram represents untransduced cells; blue histogram represents cells on the day of sorting; and red histogram represents cells after 8 days in culture. (B) Changes in levels of Nur77 expression of sorted cells in culture, measured by Western blotting (WB) with antibodies to Nur77 and β-actin. (C) Percentages of early apoptotic cells, measured by annexin V staining, in cell populations on the day after sorting.
Direct association of HDAC7 with target promoters in thymocytes.
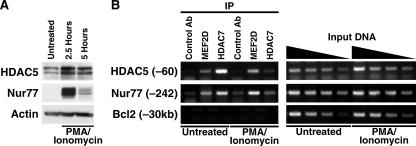
Our microarray data indicate that HDAC7 may regulate gene expression during both positive and negative thymic selection. If this is the case, HDAC7 should have an activation-dependent association with at least some promoters of genes that are differentially expressed in both of these processes. The set of HDAC7 targets that are regulated by MEF2D but not Nur77 (Fig. 6B, groups 2, 6, and 10) represents genes that may be direct transcriptional targets of HDAC7 via MEF2 sites in their promoters. We therefore analyzed the regions from 6 kb upstream to 4 kb downstream of the transcriptional start sites of 11 of these genes by using the Transcriptional Element Search System (TESS, http://www.cbil.upenn.edu/cgi-bin/tess/tess) and identified potential MEF2 sites in the flanking regions of 5 of them: the HDAC5, Tribbles-1, Bcl2, Scotin, and CCR8 genes (data not shown). Of these, HDAC5 is the only gene differentially expressed only during positive selection, according to our data and another published report (30). HDAC5 is a class IIa HDAC, like HDAC7, and is strongly upregulated by MEF2D-VP16. We also find that HDAC5 is upregulated after PMA-ionomycin stimulation of thymocytes (Fig. 8A). The putative MEF2 site in the HDAC5 promoter perfectly matches the MEF2 recognition site consensus (CTATTTTTAG), is 59 base pairs upstream of the transcriptional start site of the mouse HDAC5 gene, and is perfectly conserved between the human and mouse HDAC5 sequences. We therefore chose to focus on HDAC5, as a gene differentially expressed only during positive selection, to directly demonstrate the association of HDAC7 with its promoter. For a gene upregulated during negative selection, we chose Nur77. Nur77 is upregulated during both positive and negative thymic selection, and the Nur77 promoter contains two MEF2D sites that are regulated by HDAC7 (17). However, a direct interaction between HDAC7 and the Nur77 promoter has not been demonstrated. We prepared chromatin from unstimulated thymocytes and PMA-ionomycin-treated thymocytes and subjected it to immunoprecipitation with antibodies to HDAC7, MEF2D, or the nonspecific influenza hemagglutinin epitope. The immunoprecipitated material was analyzed by PCR with primers surrounding the putative MEF2 site in the HDAC5 promoter, two known MEF2 sites in the Nur77 promoter (97), and a nonspecific intergenic region near the BCL2 gene. In unstimulated thymocytes, both MEF2D and HDAC7 were associated with the promoters of Nur77 and HDAC5, but not with the nonspecific intergenic region (Fig. 8B, left panel). After PMA-ionomycin treatment, MEF2D remained associated with both the HDAC5 and Nur77 promoters, but HDAC7 was no longer present (Fig. 8B). These results directly demonstrate the signal-dependent dissociation of HDAC7 from the HDAC5 and Nur77 promoters.
FIG. 8.
Association of MEF2D and HDAC7 with HDAC5 and Nur77 promoters. (A) Whole cell extracts prepared from untreated wild-type mouse thymocytes or thymocytes treated with PMA plus ionomycin for 2.5 or 5 h were probed with antibodies to HDAC5, Nur77, or β-actin. (B) Chromatin extracts were prepared from wild-type thymocytes that were either untreated or treated for 1 h with PMA plus ionomycin. The extracts were immunoprecipitated with control antibodies (Ab), antibodies to MEF2D, or antibodies to HDAC7, as indicated. Immunoprecipitated chromatin (left) and serial dilutions of input chromatin (right) were amplified with primers specific for putative MEF2 sites in the promoters of Nur77 or HDAC5, 60 and 242 bp upstream of the respective genes, or in an intergenic region 30 kb upstream of the BCL2 gene (bottom).
Effects of HDAC7 mutants on T-cell differentiation in vitro.
Our results support the model that HDAC7 regulates genes that respond transcriptionally to TCR signaling during both positive and negative thymic selection. Thus, HDAC7 may be functionally important in both processes. HDAC7 appears to regulate activation-induced death in T cells (17), but there is as yet no evidence that HDAC7 plays a functional role in thymocyte differentiation. To examine the functional role of HDAC7 in these processes, we used the DPK cell line model. DPK is a CD4+ CD8+ cell line expressing the AND TCR (35), which recognizes a pigeon cytochrome c peptide on the I-Ek class II MHC molecule. When plated on a layer of APCs expressing I-Ek and ICAM (DCEK-ICAM cells), DPK cells differentiate from DP CD4+ CD8+ to single-positive CD4+ cells only in the presence of the antigenic peptide (34).
We infected DPK cells with empty virus or viral vectors encoding HDAC7-VP16 or HDAC7-ΔP and examined their ability to differentiate into single-positive CD4+ cells in the presence of APCs (DCEK-ICAM cells) with or without antigenic peptide. In the absence of exogenous stimulation, all of the infected DPK lines remained CD4+ CD8+ (Fig. 8A). When the same cell populations were plated for 3 days on DCEK-ICAM cells without peptide, the cells infected with empty virus and HDAC7-ΔP remained unchanged. Strikingly, a substantial fraction of the cells expressing HDAC7-VP16 had become CD4 single positive (Fig. 8B). When cultured for 3 days on DCEK-ICAM cells in the presence of antigenic peptide, cells infected with empty virus or HDAC7-VP16 differentiated into CD4 single-positive cells, but cells transduced with HDAC7-ΔP nearly all remained CD4+/CD8+ and appeared blocked in their differentiation. In addition to losing CD8 expression, we observed that DPK cells transduced with HDAC7-VP16 and plated on DCEK-ICAM cells without antigenic peptide upregulate CD5, a putative HDAC7 target, to a much greater extent than do untransduced cells or cells transduced with empty virus or HDAC7-ΔP (Fig. 9D). An effect on the differentiation of DPK cells can also be observed if the expression of HDAC7 is abrogated. We transfected DPK cells with siRNA oligonucleotides specific for HDAC7 or a mismatched control sequence. HDAC7 expression was markedly reduced in the cells that had received the HDAC7-specific siRNA, but not in those that had received the control siRNA (Fig. 9E). The cells were then plated on DCEK-ICAM cell layers in the presence of different concentrations of antigenic peptide and assayed after 2 days for differentiation into CD4 single-positive cells. Compared to untransfected cells or cells transfected with the control siRNA, the DPK cells that had reduced HDAC7 expression showed significantly higher percentages of CD4 single-positive cells at all peptide concentrations.
These results suggest that the upregulation of HDAC7 target genes in T cells by HDAC7-VP16 or the suppression of HDAC7 expression by siRNA reduces the threshold of TCR signaling required for thymocyte differentiation, as assessed by the expression of two different cell surface markers associated with the activation of thymocytes in vivo. Conversely, blockade of the signal-dependent upregulation of HDAC7 target genes by the expression of HDAC7-ΔP raises the threshold of signaling required for thymocyte differentiation.
DISCUSSION
We have identified genes that are transcriptional targets of HDAC7 in T cells. Our prior observation that HDAC7 regulates Nur77, a mediator of programmed cell death (17), suggested that HDAC7 plays a role in thymic negative selection. By using the expression of two mutant HDAC7 proteins to perturb the transcriptional regulation of HDAC7 targets, we identified 96 target genes that are upregulated during positive or negative selection. Surprisingly, more of these putative HDAC7 targets were associated with differentiation and positive selection than with negative selection and deletion.
HDAC7 appears to regulate its targets in T cells through both MEF2-dependent and MEF2-independent mechanisms. Interestingly, Nur77, a direct target of HDAC7 via MEF2 sites in its promoter and a mediator of apoptosis in thymocytes, regulates genes that are associated with both positive and negative thymic selection. Based on its strong regulation by MEF2, but not Nur77, we identify HDAC5 as a potential direct target of HDAC7 that is upregulated during thymic positive, but not negative, selection. By using ChIP analysis, we demonstrated the signal-dependent association of HDAC7 with an MEF2 site in the promoter of HDAC5 and Nur77.
These observations provide evidence that HDAC7 directly regulates at least one gene associated with thymic differentiation and/or positive selection. However, the exact molecular mechanisms whereby HDAC7 regulates most of the targets identified in this screen remain unknown. In future studies, we intend to analyze HDAC7 recruitment in thymocytes at a genomic level by using promoter array hybridization (ChIP on chip) (62), as well as the generation and high-throughput sequencing of serial analysis of gene expression (SAGE) libraries (72, 73). We expect that the comparison of these data to the identification of transcriptional targets of HDAC7 presented in this report will be very informative in defining the full set of direct and indirect targets of HDAC7 in thymocytes.
We also show that perturbation of HDAC7 function in a model of thymic T-cell maturation, the DPK cell line, can alter activation-induced differentiation. Constitutive activation of HDAC7 transcriptional targets by the expression of HDAC7-VP16 or by knockdown of HDAC7 lowers the threshold of TCR signaling required to cause DPK cells to differentiate from CD4+ CD8+ into CD4+ single-positive cells. Conversely, blockade of the signal-dependent derepression of HDAC7 targets strongly suppresses the differentiation of DPK cells in response to an antigen receptor signal.
The results obtained in DPK cells, combined with the prior observation that blockade of signal-dependent HDAC7 release by the expression of HDAC7-ΔP in T-cell hybridomas is protective against activation-induced apoptosis (22), suggest that HDAC7 plays a role in the signal-dependent regulation of genes that are involved in both positive and negative thymic selection. Many identified HDAC7 transcriptional targets play critical roles in signal transduction. In particular, many of these proteins modulate the coupling between antigen receptor engagement and developmental responses in T cells.
Among the most strongly differentially expressed targets of HDAC7 were five cell surface receptors that have costimulatory functions in T cells. These include the immunoglobulin superfamily receptors CD28 and PD-1, the scavenger receptor cysteine-rich family members CD5 and CD6, and the integrin LFA-1. CD28 is important in the activation of naive T cells (reviewed in reference 42), and there is considerable evidence for a role for CD28 in negative selection (7, 61). PD-1 is expressed in thymocytes only after antigen receptor ligation (1, 59). Its engagement inhibits the proliferation of naive T cells (23, 40), and PD-1 deficiency can enhance either positive or negative selection, depending on the TCR background examined (6, 36). PD-1 was also previously identified as a target of Nur77 (69). CD5 and CD6 are both members of the scavenger receptor cysteine-rich family of cell surface receptors (76) and are upregulated upon antigen receptor engagement in DP thymocytes (4, 80). CD5 plays a negative role in the regulation of antigen receptor signaling, raising the signaling threshold for both positive and negative selection (3, 66), while CD6 plays a positive role, promoting positive selection and increasing resistance to apoptosis (80). Integrin β2, the CD18 subunit of LFA-1, is a major component of the outer adhesion zone of the immunological synapse (56). Its ligand, ICAM1, is highly expressed on thymic APCs (93). The ICAM-LFA-1 interaction has a potent positive modulatory effect on TCR signaling (2, 34, 81). Interestingly, this effect appears to enhance survival more than death in thymocytes, suggesting that the upregulation of LFA-1 on thymocytes might favor positive selection (48).
In addition to these costimulatory receptors, HDAC7 regulates a number of modulators of intracellular signaling events directly connected to the membrane TCR complex. These include ZAP70, T-cell-specific adaptor protein (TSAd/SH2D2A), diacylglycerol kinase ζ (DGKZ), and CYBR (PSCDBP). The syk-family kinase ZAP70 is essential in both positive and negative thymic selection (58). TSAd plays an essential role in the activation of peripheral T cells, coupling TCR engagement to the phosphorylation of lck (51, 68). DGKZ negatively regulates TCR signaling by converting diacylglycerol to phosphatidic acid. DGKZ-deficient mice show enhanced upregulation of activation markers upon TCR engagement in both mature T cells and thymocytes (100). Finally, CYBR is a positive modulator of TCR signaling, enhancing the TCR-mediated activation of JNK/p38 MAP kinase activity by modulating the activity of the guanine nucleotide exchange factor VAV (12).
We also identify a number of modulators of intracellular signaling events not directly connected to the TCR, associated with MAP kinase activity, cytokine signaling, and apoptosis. Regulators of MAP kinase activity comprise the largest group, consisting of PAC-1 (DUSP2), MKP5 (DUSP10), GADD45β, and Tribbles-1 (Trib1). The phosphatases PAC-1 and MKP5 are both members of the MAPK phosphatase family which downregulate MAP kinase activity by dephosphorylating extracellular signal-regulated kinase and JNK/p38 kinases (87). GADD45β, in contrast, promotes apoptosis via the activation of JNK/p38 kinases (46) and is important in maintaining peripheral tolerance (44). Tribbles-1 also functions as a positive regulator of the MAPK cascade via its interaction with the MAPK kinase proteins (38). We also identified two members of the suppressor of cytokine signaling (SOCS) family, CIS (CISH) and SOCS1. SOCS1 plays a role in thymic selection, modulating proliferation in response to interleukin 7 and negatively regulating the positive selection of CD8 cells (14, 15). The regulators of apoptosis we identify include BCL2, homeodomain-interacting protein kinase (HIPK3), interferon regulatory factor 4 (IRF4), and Scotin. BCL2 is an antiapoptotic factor that can participate in both positive and negative selection (reviewed in reference 11). HIPK3 is a serine/threonine protein kinase that interacts with Fas (71). It is regulated via JNK MAPK activity, and inhibits apoptosis via FADD phosphorylation (16). IRF4 is a member of an extensive family of transcription factors that have diverse roles in lymphoid development (reviewed in reference 45).
Finally, we isolated a transcriptional corepressor, NGFI-A binding protein 2 (NAB2), which is of clear potential significance in thymic selection. It is a negative regulator of EGR1 target genes (29). EGR1 is important in positive selection, promoting both maturation to the single-positive stage and the survival of recently emigrated thymocytes (5, 78).
In summary, our screen has revealed that HDAC7 regulates an extensive functional cassette of molecules that are involved in the responses to antigen receptor and costimulatory signals in developing T cells, many with apparently opposed functions. The release of HDAC7 gene repression may thus allow thymocytes to respond to extracellular signals of several types. It is difficult to predict how this broad priming of the ability of a thymocyte to respond to multiple signals would affect its developmental fate in the complex environment of the thymus. In different cell culture model systems, we have now acquired evidence that HDAC7 can modulate both differentiation and death in response to TCR signaling (Fig. 9) (17). The release of HDAC7 may, therefore, be an important aspect of a developing T cell's ability to adopt any developmental fate in response to TCR signals. Our findings with respect to the differentiation and death of T-cell lines expressing HDAC7-ΔP support this model (Fig. 9) (17).
The definitive elucidation of the role of HDAC7 in thymic T-cell development will require the evaluation of its function in its normal physiologic context, using mice either deficient in HDAC7 expression in the thymus or expressing HDAC7 mutants employed in this study. Since the very broad range of potential avidities and specificities of the wild-type pool of antigen receptors can compensate for perturbations that have a quantitative rather than qualitative effect on the developmental response of thymocytes to antigen receptor signaling, we will evaluate the effect of the perturbation of HDAC7 function in a variety of defined antigen receptor systems. These studies are currently ongoing in our laboratory. We anticipate that the work described here will inform our analysis of any phenotypic effects we may observe.
Acknowledgments
We thank Yanxia Hao for extensive assistance with microarray hybridizations and Michael Salazar for assistance with the use of the Acuity software. We thank Christopher Barker, Andrea Barczak, and David Erle for helpful consultations about microarray data analysis. We also thank Warner Greene, Melanie Ott, and Maribel Parra-Bola for critical reading of the manuscript and helpful discussions.
This work was supported by the Gladstone Institutes.
Footnotes
Published ahead of print on 30 April 2007.
REFERENCES
- 1.Agata, Y., A. Kawasaki, H. Nishimura, Y. Ishida, T. Tsubata, H. Yagita, and T. Honjo. 1996. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int. Immunol. 8:765-772. [DOI] [PubMed] [Google Scholar]
- 2.Altmann, D. M., N. Hogg, J. Trowsdale, and D. Wilkinson. 1989. Cotransfection of ICAM-1 and HLA-DR reconstitutes human antigen-presenting cell function in mouse L cells. Nature 338:512-514. [DOI] [PubMed] [Google Scholar]
- 3.Azzam, H. S., J. B. DeJarnette, K. Huang, R. Emmons, C. S. Park, C. L. Sommers, D. El-Khoury, E. W. Shores, and P. E. Love. 2001. Fine tuning of TCR signaling by CD5. J. Immunol. 166:5464-5472. [DOI] [PubMed] [Google Scholar]
- 4.Azzam, H. S., A. Grinberg, K. Lui, H. Shen, E. W. Shores, and P. E. Love. 1998. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 188:2301-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bettini, M., H. Xi, J. Milbrandt, and G. J. Kersh. 2002. Thymocyte development in early growth response gene 1-deficient mice. J. Immunol. 169:1713-1720. [DOI] [PubMed] [Google Scholar]
- 6.Blank, C., I. Brown, R. Marks, H. Nishimura, T. Honjo, and T. F. Gajewski. 2003. Absence of programmed death receptor 1 alters thymic development and enhances generation of CD4/CD8 double-negative TCR-transgenic T cells. J. Immunol. 171:4574-4581. [DOI] [PubMed] [Google Scholar]
- 7.Buhlmann, J. E., S. K. Elkin, and A. H. Sharpe. 2003. A role for the B7-1/B7-2:CD28/CTLA-4 pathway during negative selection. J. Immunol. 170:5421-5428. [DOI] [PubMed] [Google Scholar]
- 8.Calnan, B. J., S. Szychowski, F. K. Chan, D. Cado, and A. Winoto. 1995. A role for the orphan steroid receptor Nur77 in apoptosis accompanying antigen-induced negative selection. Immunity 3:273-282. [DOI] [PubMed] [Google Scholar]
- 9.Chang, S., T. A. McKinsey, C. L. Zhang, J. A. Richardson, J. A. Hill, and E. N. Olson. 2004. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 24:8467-8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, S., B. D. Young, S. Li, X. Qi, J. A. Richardson, and E. N. Olson. 2006. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell 126:321-334. [DOI] [PubMed] [Google Scholar]
- 11.Chao, D. T., and S. J. Korsmeyer. 1998. BCL-2 family: regulators of cell death. Annu. Rev. Immunol. 16:395-419. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Q., A. Coffey, S. G. Bourgoin, and M. Gadina. 2006. Cytohesin binder and regulator augments T cell receptor-induced nuclear factor of activated T cells. AP-1 activation through regulation of the JNK pathway. J. Biol. Chem. 281:19985-19994. [DOI] [PubMed] [Google Scholar]
- 13.Cheng, L. E., F. K. Chan, D. Cado, and A. Winoto. 1997. Functional redundancy of the Nur77 and Nor-1 orphan steroid receptors in T-cell apoptosis. EMBO J. 16:1865-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Chomczynski, P., and N. Sacchi. 1987. Single step method of RNA isolation by acid guanidine thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 14.Cornish, A. L., M. M. Chong, G. M. Davey, R. Darwiche, N. A. Nicola, D. J. Hilton, T. W. Kay, R. Starr, and W. S. Alexander. 2003. Suppressor of cytokine signaling-1 regulates signaling in response to interleukin-2 and other gamma c-dependent cytokines in peripheral T cells. J. Biol. Chem. 278:22755-22761. [DOI] [PubMed] [Google Scholar]
- 15.Cornish, A. L., G. M. Davey, D. Metcalf, J. F. Purton, J. E. Corbin, C. J. Greenhalgh, R. Darwiche, L. Wu, N. A. Nicola, D. I. Godfrey, W. R. Heath, D. J. Hilton, W. S. Alexander, and R. Starr. 2003. Suppressor of cytokine signaling-1 has IFN-γ-independent actions in T cell homeostasis. J. Immunol. 170:878-886. [DOI] [PubMed] [Google Scholar]
- 16.Curtin, J. F., and T. G. Cotter. 2004. JNK regulates HIPK3 expression and promotes resistance to Fas-mediated apoptosis in DU 145 prostate carcinoma cells. J. Biol. Chem. 279:17090-17100. [DOI] [PubMed] [Google Scholar]
- 17.Dequiedt, F., H. Kasler, W. Fischle, V. Kiermer, M. Weinstein, B. G. Herndier, and E. Verdin. 2003. HDAC7, a thymus-specific class II histone deacetylase, regulates Nur77 transcription and TCR-mediated apoptosis. Immunity 18:687-698. [DOI] [PubMed] [Google Scholar]
- 18.Dequiedt, F., J. Van Lint, E. Lecomte, V. Van Duppen, T. Seufferlein, J. R. Vandenheede, R. Wattiez, and R. Kettmann. 2005. Phosphorylation of histone deacetylase 7 by protein kinase D mediates T cell receptor-induced Nur77 expression and apoptosis. J. Exp. Med. 201:793-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeRyckere, D., D. L. Mann, and J. DeGregori. 2003. Characterization of transcriptional regulation during negative selection in vivo. J. Immunol. 171:802-811. [DOI] [PubMed] [Google Scholar]
- 20.Dressel, U., P. J. Bailey, S. C. Wang, M. Downes, R. M. Evans, and G. E. Muscat. 2001. A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J. Biol. Chem. 276:17007-17013. [DOI] [PubMed] [Google Scholar]
- 21.Fiorini, E., I. Schmitz, W. E. Marissen, S. L. Osborn, M. Touma, T. Sasada, P. A. Reche, E. V. Tibaldi, R. E. Hussey, A. M. Kruisbeek, E. L. Reinherz, and L. K. Clayton. 2002. Peptide-induced negative selection of thymocytes activates transcription of an NF-kappa B inhibitor. Mol. Cell 9:637-648. [DOI] [PubMed] [Google Scholar]
- 22.Fischle, W., F. Dequiedt, M. Fillion, M. J. Hendzel, W. Voelter, and E. Verdin. 2001. Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo. J. Biol. Chem. 276:35826-35835. [DOI] [PubMed] [Google Scholar]
- 23.Freeman, G. J., A. J. Long, Y. Iwai, K. Bourque, T. Chernova, H. Nishimura, L. J. Fitz, N. Malenkovich, T. Okazaki, M. C. Byrne, H. F. Horton, L. Fouser, L. Carter, V. Ling, M. R. Bowman, B. M. Carreno, M. Collins, C. R. Wood, and T. Honjo. 2000. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 192:1027-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groettrup, M., K. Ungewiss, O. Azogui, R. Palacios, M. J. Owen, A. C. Hayday, and H. von Boehmer. 1993. A novel disulfide-linked heterodimer on pre-T cells consists of the T cell receptor beta chain and a 33 kd glycoprotein. Cell 75:283-294. [DOI] [PubMed] [Google Scholar]
- 25.Grozinger, C. M., and S. L. Schreiber. 2000. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. USA 97:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruffat, H., E. Manet, and A. Sergeant. 2002. MEF2-mediated recruitment of class II HDAC at the EBV immediate early gene BZLF1 links latency and chromatin remodeling. EMBO Rep. 3:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hildeman, D. A., Y. Zhu, T. C. Mitchell, P. Bouillet, A. Strasser, J. Kappler, and P. Marrack. 2002. Activated T cell death in vivo mediated by proapoptotic bcl-2 family member bim. Immunity 16:759-767. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann, R., L. Bruno, T. Seidl, A. Rolink, and F. Melchers. 2003. Rules for gene usage inferred from a comparison of large-scale gene expression profiles of T and B lymphocyte development. J. Immunol. 170:1339-1353. [DOI] [PubMed] [Google Scholar]
- 29.Houston, P., C. J. Campbell, J. Svaren, J. Milbrandt, and M. Braddock. 2001. The transcriptional corepressor NAB2 blocks Egr-1-mediated growth factor activation and angiogenesis. Biochem. Biophys. Res. Commun. 283:480-486. [DOI] [PubMed] [Google Scholar]
- 30.Huang, Y. H., D. Li, A. Winoto, and E. A. Robey. 2004. Distinct transcriptional programs in thymocytes responding to T cell receptor, Notch, and positive selection signals. Proc. Natl. Acad. Sci. USA 101:4936-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kao, H. Y., M. Downes, P. Ordentlich, and R. M. Evans. 2000. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 14:55-66. [PMC free article] [PubMed] [Google Scholar]
- 32.Kao, H. Y., A. Verdel, C. C. Tsai, C. Simon, H. Juguilon, and S. Khochbin. 2001. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 276:47496-47507. [DOI] [PubMed] [Google Scholar]
- 33.Kasler, H. G., J. Victoria, O. Duramad, and A. Winoto. 2000. ERK5 is a novel type of mitogen-activated protein kinase containing a transcriptional activation domain. Mol. Cell. Biol. 20:8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaye, J., and D. L. Ellenberger. 1992. Differentiation of an immature T cell line: a model of thymic positive selection. Cell 71:423-435. [DOI] [PubMed] [Google Scholar]
- 35.Kaye, J., M. L. Hsu, M. E. Sauron, S. C. Jameson, N. R. Gascoigne, and S. M. Hedrick. 1989. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature 341:746-749. [DOI] [PubMed] [Google Scholar]
- 36.Keir, M. E., Y. E. Latchman, G. J. Freeman, and A. H. Sharpe. 2005. Programmed death-1 (PD-1):PD-ligand 1 interactions inhibit TCR-mediated positive selection of thymocytes. J. Immunol. 175:7372-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kishimoto, H., and J. Sprent. 1999. Several different cell surface molecules control negative selection of medullary thymocytes. J. Exp. Med. 190:65-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiss-Toth, E., S. M. Bagstaff, H. Y. Sung, V. Jozsa, C. Dempsey, J. C. Caunt, K. M. Oxley, D. H. Wyllie, T. Polgar, M. Harte, L. A. O'Neill, E. E. Qwarnstrom, and S. K. Dower. 2004. Human tribbles, a protein family controlling mitogen-activated protein kinase cascades. J. Biol. Chem. 279:42703-42708. [DOI] [PubMed] [Google Scholar]
- 39.Kurobe, H., C. Liu, T. Ueno, F. Saito, I. Ohigashi, N. Seach, R. Arakaki, Y. Hayashi, T. Kitagawa, M. Lipp, R. L. Boyd, and Y. Takahama. 2006. CCR7-dependent cortex-to-medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity 24:165-177. [DOI] [PubMed] [Google Scholar]
- 40.Latchman, Y., C. R. Wood, T. Chernova, D. Chaudhary, M. Borde, I. Chernova, Y. Iwai, A. J. Long, J. A. Brown, R. Nunes, E. A. Greenfield, K. Bourque, V. A. Boussiotis, L. L. Carter, B. M. Carreno, N. Malenkovich, H. Nishimura, T. Okazaki, T. Honjo, A. H. Sharpe, and G. J. Freeman. 2001. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol. 2:261-268. [DOI] [PubMed] [Google Scholar]
- 41.Lemercier, C., A. Verdel, B. Galloo, S. Curtet, M. P. Brocard, and S. Khochbin. 2000. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J. Biol. Chem. 275:15594-15599. [DOI] [PubMed] [Google Scholar]
- 42.Lenschow, D. J., T. L. Walunas, and J. A. Bluestone. 1996. CD28/B7 system of T cell costimulation. Annu. Rev. Immunol. 14:233-258. [DOI] [PubMed] [Google Scholar]
- 43.Linette, G. P., M. J. Grusby, S. M. Hedrick, T. H. Hansen, L. H. Glimcher, and S. J. Korsmeyer. 1994. Bcl-2 is upregulated at the CD4+ CD8+ stage during positive selection and promotes thymocyte differentiation at several control points. Immunity 1:197-205. [DOI] [PubMed] [Google Scholar]
- 44.Liu, L., E. Tran, Y. Zhao, Y. Huang, R. Flavell, and B. Lu. 2005. Gadd45 beta and Gadd45 gamma are critical for regulating autoimmunity. J. Exp. Med. 202:1341-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lohoff, M., and T. W. Mak. 2005. Roles of interferon-regulatory factors in T-helper-cell differentiation. Nat. Rev. Immunol. 5:125-135. [DOI] [PubMed] [Google Scholar]
- 46.Lu, B., H. Yu, C. Chow, B. Li, W. Zheng, R. J. Davis, and R. A. Flavell. 2001. GADD45gamma mediates the activation of the p38 and JNK MAP kinase pathways and cytokine production in effector TH1 cells. Immunity 14:583-590. [DOI] [PubMed] [Google Scholar]
- 47.Lu, J., T. A. McKinsey, C. L. Zhang, and E. N. Olson. 2000. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell 6:233-244. [DOI] [PubMed] [Google Scholar]
- 48.Lucas, B., and R. N. Germain. 2000. Opening a window on thymic positive selection: developmental changes in the influence of cosignaling by integrins and CD28 on selection events induced by TCR engagement. J. Immunol. 165:1889-1895. [DOI] [PubMed] [Google Scholar]
- 49.Ma, A., J. C. Pena, B. Chang, E. Margosian, L. Davidson, F. W. Alt, and C. B. Thompson. 1995. Bclx regulates the survival of double-positive thymocytes. Proc. Natl. Acad. Sci. USA 92:4763-4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marklund, U., K. Lightfoot, and D. Cantrell. 2003. Intracellular location and cell context-dependent function of protein kinase D. Immunity 19:491-501. [DOI] [PubMed] [Google Scholar]
- 51.Marti, F., G. G. Garcia, P. E. Lapinski, J. N. MacGregor, and P. D. King. 2006. Essential role of the T cell-specific adapter protein in the activation of LCK in peripheral T cells. J. Exp. Med. 203:281-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2001. Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev. 11:497-504. [DOI] [PubMed] [Google Scholar]
- 53.Mick, V. E., T. K. Starr, T. M. McCaughtry, L. K. McNeil, and K. A. Hogquist. 2004. The regulated expression of a diverse set of genes during thymocyte positive selection in vivo. J. Immunol. 173:5434-5444. [DOI] [PubMed] [Google Scholar]
- 54.Miska, E. A., E. Langley, D. Wolf, C. Karlsson, J. Pines, and T. Kouzarides. 2001. Differential localization of HDAC4 orchestrates muscle differentiation. Nucleic Acids Res. 29:3439-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mombaerts, P., A. R. Clarke, M. A. Rudnicki, J. Iacomini, S. Itohara, J. J. Lafaille, L. Wang, Y. Ichikawa, R. Jaenisch, M. L. Hooper, et al. 1992. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature 360:225-231. [DOI] [PubMed] [Google Scholar]
- 56.Monks, C. R., B. A. Freiberg, H. Kupfer, N. Sciaky, and A. Kupfer. 1998. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature 395:82-86. [DOI] [PubMed] [Google Scholar]
- 57.Murphy, K. M., A. B. Heimberger, and D. Y. Loh. 1990. Induction by antigen of intrathymic apoptosis of CD4+CD8+TCRlo thymocytes in vivo. Science 250:1720-1723. [DOI] [PubMed] [Google Scholar]
- 58.Negishi, I., N. Motoyama, K. Nakayama, S. Senju, S. Hatakeyama, Q. Zhang, A. C. Chan, and D. Y. Loh. 1995. Essential role for ZAP-70 in both positive and negative selection of thymocytes. Nature 376:435-438. [DOI] [PubMed] [Google Scholar]
- 59.Nishimura, H., Y. Agata, A. Kawasaki, M. Sato, S. Imamura, N. Minato, H. Yagita, T. Nakano, and T. Honjo. 1996. Developmentally regulated expression of the PD-1 protein on the surface of double-negative (CD4-CD8-) thymocytes. Int. Immunol. 8:773-780. [DOI] [PubMed] [Google Scholar]
- 60.Nishimura, H., T. Honjo, and N. Minato. 2000. Facilitation of beta selection and modification of positive selection in the thymus of PD-1-deficient mice. J. Exp. Med. 191:891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noel, P. J., M. L. Alegre, S. L. Reiner, and C. B. Thompson. 1998. Impaired negative selection in CD28-deficient mice. Cell. Immunol. 187:131-138. [DOI] [PubMed] [Google Scholar]
- 62.Odom, D. T., N. Zizlsperger, D. B. Gordon, G. W. Bell, N. J. Rinaldi, H. L. Murray, T. L. Volkert, J. Schreiber, P. A. Rolfe, D. K. Gifford, E. Fraenkel, G. I. Bell, and R. A. Young. 2004. Control of pancreas and liver gene expression by HNF transcription factors. Science 303:1378-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Page, D. M. 1999. Cutting edge: thymic selection and autoreactivity are regulated by multiple coreceptors involved in T cell activation. J. Immunol. 163:3577-3581. [PubMed] [Google Scholar]
- 64.Parra, M., H. Kasler, T. A. McKinsey, E. N. Olson, and E. Verdin. 2005. Protein kinase D1 phosphorylates HDAC7 and induces its nuclear export after T-cell receptor activation. J. Biol. Chem. 280:13762-13770. [DOI] [PubMed] [Google Scholar]
- 65.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pena-Rossi, C., L. A. Zuckerman, J. Strong, J. Kwan, W. Ferris, S. Chan, A. Tarakhovsky, A. D. Beyers, and N. Killeen. 1999. Negative regulation of CD4 lineage development and responses by CD5. J. Immunol. 163:6494-6501. [PubMed] [Google Scholar]
- 67.Punt, J. A., B. A. Osborne, Y. Takahama, S. O. Sharrow, and A. Singer. 1994. Negative selection of CD4+CD8+ thymocytes by T cell receptor-induced apoptosis requires a costimulatory signal that can be provided by CD28. J. Exp. Med. 179:709-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rajagopal, K., C. L. Sommers, D. C. Decker, E. O. Mitchell, U. Korthauer, A. I. Sperling, C. A. Kozak, P. E. Love, and J. A. Bluestone. 1999. RIBP, a novel Rlk/Txk- and itk-binding adaptor protein that regulates T cell activation. J. Exp. Med. 190:1657-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rajpal, A., Y. A. Cho, B. Yelent, P. H. Koza-Taylor, D. Li, E. Chen, M. Whang, C. Kang, T. G. Turi, and A. Winoto. 2003. Transcriptional activation of known and novel apoptotic pathways by Nur77 orphan steroid receptor. EMBO J. 22:6526-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rincon, M., A. Whitmarsh, D. D. Yang, L. Weiss, B. Derijard, P. Jayaraj, R. J. Davis, and R. A. Flavell. 1998. The JNK pathway regulates the in vivo deletion of immature CD4(+)CD8(+) thymocytes. J. Exp. Med. 188:1817-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rochat-Steiner, V., K. Becker, O. Micheau, P. Schneider, K. Burns, and J. Tschopp. 2000. FIST/HIPK3: a Fas/FADD-interacting serine/threonine kinase that induces FADD phosphorylation and inhibits fas-mediated Jun NH(2)-terminal kinase activation. J. Exp. Med. 192:1165-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roh, T. Y., S. Cuddapah, K. Cui, and K. Zhao. 2006. The genomic landscape of histone modifications in human T cells. Proc. Natl. Acad. Sci. USA 103:15782-15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roh, T. Y., S. Cuddapah, and K. Zhao. 2005. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 19:542-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sabapathy, K., T. Kallunki, J. P. David, I. Graef, M. Karin, and E. F. Wagner. 2001. c-Jun NH2-terminal kinase (JNK)1 and JNK2 have similar and stage-dependent roles in regulating T cell apoptosis and proliferation. J. Exp. Med. 193:317-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saint-Ruf, C., K. Ungewiss, M. Groettrup, L. Bruno, H. J. Fehling, and H. von Boehmer. 1994. Analysis and expression of a cloned pre-T cell receptor gene. Science 266:1208-1212. [DOI] [PubMed] [Google Scholar]
- 76.Sarrias, M. R., J. Gronlund, O. Padilla, J. Madsen, U. Holmskov, and F. Lozano. 2004. The scavenger receptor cysteine-rich (SRCR) domain: an ancient and highly conserved protein module of the innate immune system. Crit. Rev. Immunol. 24:1-37. [DOI] [PubMed] [Google Scholar]
- 77.Schmitz, I., L. K. Clayton, and E. L. Reinherz. 2003. Gene expression analysis of thymocyte selection in vivo. Int. Immunol. 15:1237-1248. [DOI] [PubMed] [Google Scholar]
- 78.Schnell, F. J., and G. J. Kersh. 2005. Control of recent thymic emigrant survival by positive selection signals and early growth response gene 1. J. Immunol. 175:2270-2277. [DOI] [PubMed] [Google Scholar]
- 79.Simon, A. K., N. Auphan, M. Pophillat, C. Boyer, S. Ghosh, M. Rincon, R. A. Flavell, and A. M. Schmitt-Verhulst. 2000. The lack of NF-kappa B transactivation and PKC epsilon expression in CD4(+)CD8(+) thymocytes correlates with negative selection. Cell Death Differ. 7:1253-1262. [DOI] [PubMed] [Google Scholar]
- 80.Singer, N. G., D. A. Fox, T. M. Haqqi, L. Beretta, J. S. Endres, S. Prohaska, J. R. Parnes, J. Bromberg, and R. M. Sramkoski. 2002. CD6: expression during development, apoptosis and selection of human and mouse thymocytes. Int. Immunol. 14:585-597. [DOI] [PubMed] [Google Scholar]
- 81.Siu, G., S. M. Hedrick, and A. A. Brian. 1989. Isolation of the murine intercellular adhesion molecule 1 (ICAM-1) gene. ICAM-1 enhances antigen-specific T cell activation. J. Immunol. 143:3813-3820. [PubMed] [Google Scholar]
- 82.Song, K., J. Backs, J. McAnally, X. Qi, R. D. Gerard, J. A. Richardson, J. A. Hill, R. Bassel-Duby, and E. N. Olson. 2006. The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell 125:453-466. [DOI] [PubMed] [Google Scholar]
- 83.Sparrow, D. B., E. A. Miska, E. Langley, S. Reynaud-Deonauth, S. Kotecha, N. Towers, G. Spohr, T. Kouzarides, and T. J. Mohun. 1999. MEF-2 function is modified by a novel corepressor, MITR. EMBO J. 18:5085-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sugawara, T., T. Moriguchi, E. Nishida, and Y. Takahama. 1998. Differential roles of ERK and p38 MAP kinase pathways in positive and negative selection of T lymphocytes. Immunity 9:565-574. [DOI] [PubMed] [Google Scholar]
- 85.Sun, G., X. Liu, P. Mercado, S. R. Jenkinson, M. Kypriotou, L. Feigenbaum, P. Galera, and R. Bosselut. 2005. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat. Immunol. 6:373-381. [DOI] [PubMed] [Google Scholar]
- 86.Suzuki, A., M. T. Yamaguchi, T. Ohteki, T. Sasaki, T. Kaisho, Y. Kimura, R. Yoshida, A. Wakeham, T. Higuchi, M. Fukumoto, T. Tsubata, P. S. Ohashi, S. Koyasu, J. M. Penninger, T. Nakano, and T. W. Mak. 2001. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity 14:523-534. [DOI] [PubMed] [Google Scholar]
- 87.Theodosiou, A., A. Smith, C. Gillieron, S. Arkinstall, and A. Ashworth. 1999. MKP5, a new member of the MAP kinase phosphatase family, which selectively dephosphorylates stress-activated kinases. Oncogene 18:6981-6988. [DOI] [PubMed] [Google Scholar]
- 88.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ueno, T., F. Saito, D. H. Gray, S. Kuse, K. Hieshima, H. Nakano, T. Kakiuchi, M. Lipp, R. L. Boyd, and Y. Takahama. 2004. CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J. Exp. Med. 200:493-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vega, R. B., B. C. Harrison, E. Meadows, C. R. Roberts, P. J. Papst, E. N. Olson, and T. A. McKinsey. 2004. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 24:8374-8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vega, R. B., K. Matsuda, J. Oh, A. C. Barbosa, X. Yang, E. Meadows, J. McAnally, C. Pomajzl, J. M. Shelton, J. A. Richardson, G. Karsenty, and E. N. Olson. 2004. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 119:555-566. [DOI] [PubMed] [Google Scholar]
- 92.Verdin, E., F. Dequiedt, and H. G. Kasler. 2003. Class II histone deacetylases: versatile regulators. Trends Genet. 19:286-293. [DOI] [PubMed] [Google Scholar]
- 93.Volkmann, A., T. Zal, and B. Stockinger. 1997. Antigen-presenting cells in the thymus that can negatively select MHC class II-restricted T cells recognizing a circulating self antigen. J. Immunol. 158:693-706. [PubMed] [Google Scholar]
- 94.Wang, A. H., N. R. Bertos, M. Vezmar, N. Pelletier, M. Crosato, H. H. Heng, J. Th'ng, J. Han, and X. J. Yang. 1999. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell. Biol. 19:7816-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang, A. H., M. J. Kruhlak, J. Wu, N. R. Bertos, M. Vezmar, B. I. Posner, D. P. Bazett-Jones, and X. J. Yang. 2000. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell. Biol. 20:6904-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Woronicz, J. D., B. Calnan, V. Ngo, and A. Winoto. 1994. Requirement for the orphan steroid receptor Nur77 in apoptosis of T-cell hybridomas. Nature 367:277-281. [DOI] [PubMed] [Google Scholar]
- 97.Woronicz, J. D., A. Lina, B. J. Calnan, S. Szychowski, L. Cheng, and A. Winoto. 1995. Regulation of the Nur77 orphan steroid receptor in activation-induced apoptosis. Mol. Cell. Biol. 15:6364-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang, C. L., T. A. McKinsey, S. Chang, C. L. Antos, J. A. Hill, and E. N. Olson. 2002. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110:479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang, C. L., T. A. McKinsey, and E. N. Olson. 2001. The transcriptional corepressor MITR is a signal-responsive inhibitor of myogenesis. Proc. Natl. Acad. Sci. USA 98:7354-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhong, X. P., E. A. Hainey, B. A. Olenchock, M. S. Jordan, J. S. Maltzman, K. E. Nichols, H. Shen, and G. A. Koretzky. 2003. Enhanced T cell responses due to diacylglycerol kinase zeta deficiency. Nat. Immunol. 4:882-890. [DOI] [PubMed] [Google Scholar]
- 101.Zhou, X., V. M. Richon, R. A. Rifkind, and P. A. Marks. 2000. Identification of a transcriptional repressor related to the noncatalytic domain of histone deacetylases 4 and 5. Proc. Natl. Acad. Sci. USA 97:1056-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zucchelli, S., P. Holler, T. Yamagata, M. Roy, C. Benoist, and D. Mathis. 2005. Defective central tolerance induction in NOD mice: genomics and genetics. Immunity 22:385-396. [DOI] [PubMed] [Google Scholar]