Abstract
Yeast prions are protein-based genetic elements capable of self-perpetuation. One such prion, [RNQ+], requires the J-protein Sis1, an Ssa Hsp70 co-chaperone, as well as the AAA+ ATPase, Hsp104, for its propagation. We report that, upon depletion of Sis1, as well as upon inactivation of Hsp104, Rnq1 aggregates increased in size. Subsequently, cells having large aggregates, as well as an apparently soluble pool of Rnq1, became predominant in the cell population. Newly synthesized Rnq1 localized to both aggregates and bulk cytosol, suggesting that nascent Rnq1 partitioned into pools of prion and nonprion conformations, and implying that these large aggregates were still active as seeds. Ultimately, soluble Rnq1 predominated, and the prion was lost from the population. Our data suggest a model in which J-protein:Hsp70 machinery functions in prion propagation, in conjunction with Hsp104. Together, these chaperones facilitate fragmentation of prion polymers, generating a sufficient number of seeds to allow efficient conversion of newly synthesized Rnq1 into the prion conformation.
Keywords: AAA+ ATPase, Hsp40, Hsp70, Hsp104, [PIN+]
Introduction
Prion proteins exist in distinct conformational states. Once attained, maintenance of the prion conformation requires not only the conversion of newly synthesized protein into the prion conformation, but also a mechanism that allows distribution of prion ‘seeds' to progeny such that the prion is propagated from generation to generation. The yeast, Saccharomyces cerevisiae, is a useful model for studying this unique phenomenon of protein conversion and transmission. [PSI+], a prion formed by the Sup35 translation termination factor, is arguably the best-understood yeast prion. A second yeast prion, [RNQ+], the prion form of the Rnq1 protein is also known as [PIN+], because it is required for the de novo formation of [PSI+], and was first identified as a Psi-inducibility factor (Derkatch et al, 2001; Osherovich and Weissman, 2001). Prion-forming domain-containing fragments of both Sup35 and Rnq1 can form amyloid fibers in vitro that are able to recruit other conformers to the self-replicating conformation (Glover et al, 1997; King et al, 1997; Patel and Liebman, 2007).
Molecular chaperones, proteins that function to facilitate protein folding, dispersal of protein aggregates and the remodeling of multimeric protein complexes, have been linked to prion biogenesis. Both [RNQ+] and [PSI+] require Hsp104, a homohexameric AAA+ ATPase molecular chaperone, for propagation (Chernoff et al, 1995; Sondheimer and Lindquist, 2000). Inhibition of Hsp104 function leads to a reduction in the number of [PSI+] seeds (Ness et al, 2002). The currently favored model of prion propagation purports that fragmentation of prion fibers generates additional seeds that are critical for efficient prion transmission (Tanaka et al, 2006; Satpute-Krishnan et al, 2007). Indeed, Hsp104 has been reported to fragment Sup35 fibers in vitro, providing additional templates for conversion of Sup35 to the prion conformation (Shorter and Lindquist, 2004). Consistent with such fragmentation activity, inactivation of Hsp104 results in an increase in the size of Sup35 particles in [PSI+] cells (Paushkin et al, 1996; Wegrzyn et al, 2001; Kryndushkin et al, 2003; Borchsenius et al, 2006).
Sis1, an essential J-protein co-chaperone of the yeast cytosol, is the only factor other than Hsp104 known to be required for [RNQ+] maintenance (Sondheimer and Lindquist, 2000). Consistent with this observation, Sis1 can be efficiently immunoprecipitated with Rnq1 prion aggregates (Sondheimer et al, 2001; Lopez et al, 2003). Sis1, like other J-proteins, partners with an Hsp70 chaperone, serving to stimulate its ATPase activity, thus increasing the binding of Hsp70 to client proteins. Sis1 is one of several J-protein partners of Ssa, an abundant cytosolic Hsp70 involved in protein folding, translocation of proteins across membranes and assembly/disassembly of protein complexes (Craig et al, 2006).
Whereas Hsp104 activity is required for propagation of all known yeast prions, this stringent requirement for a J-protein is unparalleled. To better understand the role of Sis1 in [RNQ+] maintenance, we established a system to repress Sis1 synthesis and followed the effect of lowering Sis1 levels on the conformational state of Rnq1 over time. The process of prion loss, often called curing, due to reduced Sis1 activity correlated with an increase in [RNQ+] polymer size, and subsequent increase in soluble Rnq1 monomers. Our results are consistent with the model that Sis1 is critical for the fragmentation of Rnq1 fibrils, generating seed particles required for prion propagation.
Results
Curing of [RNQ+] upon depletion of Sis1
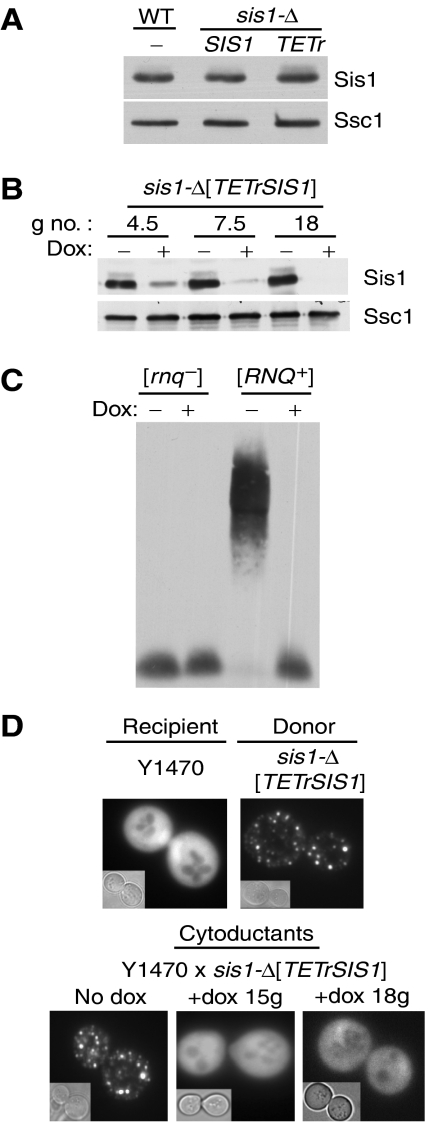
The initial observations establishing the requirement of Sis1 for maintenance of [RNQ+] utilized a mutant protein, Sis1ΔG/F, having a deletion of a small internal glycine/phenylalanine-rich region (Sondheimer et al, 2001). Advantageously, Sis1ΔG/F was able to support cell viability (Yan and Craig, 1999), but not the [RNQ+] prion. However, prion loss was very rapid, being virtually complete by the time cells expressing only the mutant Sis1 protein were obtained. Thus, analysis of the mutant provided no insight into the mechanism of prion loss. We therefore established a system that would allow study of the progression of a cell population from [RNQ+] to [rnq−] over time by placing SIS1 under the control of a tetracycline-repressible promoter. A sis1-Δ strain carrying this construct (referred to as sis1-Δ[TETrSIS1] throughout) expressed Sis1 at levels similar to wild-type cells (Figure 1A). On addition of the tetracycline analog, doxycycline, SIS1 expression was repressed, with levels being reduced to about 20% of normal within 4–5 generations (Figure 1B). The growth rate of cells in the presence and absence of drug was indistinguishable from wild-type cells for approximately 10 generations. Whereas growth slowed somewhat in subsequent generations, high cell viability was maintained for at least 30 generations, past the point relevant for experiments presented here (data not shown).
Figure 1.
Loss of [RNQ+] upon reduction of Sis1 levels. (A) Expression of Sis1 from the TETr promoter. Extracts were prepared from wild-type (wt) cells of sis1-Δ cells expressing Sis1 under the control of either its native promoter (SIS1) or the TETr promoter (TETr). Protein was separated by SDS–PAGE and subjected to immunoblot analysis using antibodies specific for Sis1 or, as a loading control, Ssc1. (B) sis1-Δ[TETrSIS1] cells cultured for the indicated number of generations (g no.) in the absence (−) or presence (+) of doxycycline (dox). Lysates were prepared and subjected to SDS–PAGE and immunoblot analysis using antibodies specific for Sis1 or, as a loading control, Ssc1. (C, D) Effect of reduced SIS1 expression by repression of the TETr promoter. (C) sis1-Δ[TETrSIS1] [rnq−] and [RNQ+] strains subcultured in the absence (−) or presence (+) of doxycycline were harvested after 16 generations. Prepared lysates were resolved by SDD–AGE and immunoblot analysis with α-Rnq1 antibodies. (D) Rnq1-GFP visualization by fluorescent microscopy of cytoduction recipient (Y1470) and donor sis1-Δ[TETrSIS1]) strains transiently overexpressing CUP1-Rnq1-GFP (top). sis1-Δ[TETrSIS1] cells subcultured in the absence (no dox) or presence of doxycycline (+dox) for up to 15–18 generations (g) cytoduced with Y1470, and transiently overexpressing CUP1-Rnq1-GFP, visualized by fluorescence microscopy (bottom). In each case, eight or more individual cytoductants were analyzed and >200 cells were observed; >99% of cells in each sample showed either diffuse or punctate fluorescence. Representative cells are shown.
To assess the effect of depletion of Sis1 on [RNQ+] prion maintenance analyses, cells were analyzed before and 15–35 generations after doxycycline addition. Rnq1, like Sup35, is present in detergent-resistant subparticles when in the prion form (Bagriantsev and Liebman, 2004; Kryndushkin et al, 2003). After treatment of cell extracts with sodium dodecyl sulfate (SDS), these subparticles can be separated by electrophoresis, in semi-denaturing detergent agarose gels (SDD–AGE). As expected from these previous reports, Rnq1 in extracts from untreated sis1-Δ[TETrSIS1] cells migrated as high molecular weight particles (Figure 1C) averaging approximately 750 kDa in molecular weight (Supplementary Figure 1). Rnq1 in extracts from drug-treated cells co-migrated with Rnq1 from [rnq−] cells, that is, at the position of monomers.
Whereas the disappearance of large aggregates and the concomitant increase in protein migrating as monomer in cells having reduced levels of Sis1 strongly suggests prion loss, the continued presence of small numbers of seeds cannot be ruled out by such biochemical assays. A definitive genetic test for maintenance (or loss) of the [RNQ+] prion is the ability to transmit (or not) the [RNQ+] prion to [rnq−] cells through cytoplasmic contact (Conde and Fink, 1976; Sondheimer and Lindquist, 2000). Typical for cytoduction analyses, we utilized a recipient strain having a dominant mutation that prevents karyogamy, allowing fusion of cytosols, but not nuclei, and thus not nuclear genetic material, upon mating. After cell division, haploids having only the recipient nucleus, but a mixture of donor and recipient cytosols, were selected.
Such haploids, resulting from cytoductants obtained before and after addition of doxycycline sis1-Δ[TETrSIS1] cells, as well as the donor and recipient strains, were transformed with a plasmid capable of transiently driving expression of a Rnq1-green fluorescent protein fusion (Rnq1-GFP). As expected, the donor [RNQ+] cells uniformly displayed distinct foci of fluorescence, indicative of Rnq1 being in prion aggregates (Figure 1D). Consistently, the fluorescence of recipient [rnq−] cells was dispersed throughout the cytosol. Haploids derived from cytoductants formed before addition of doxycycline to the culture displayed a punctate pattern, whereas those from cells treated with doxycycline for 15 or 18 generations displayed diffuse fluorescence, indicative of prion loss.
We also tested the ability of extracts from sis1-Δ[TETrSIS1] cells to ‘infect' [rnq−] cells, that is to act as seeds to facilitate the conversion to [RNQ+] (King and Diaz-Avalos, 2004; Tanaka et al, 2004; Patel and Liebman, 2007). [rnq−] spheroplasts were co-transformed with lysates from sis1-Δ[TETrSIS1] cells and a plasmid carrying the inducible CUP1-Rnq1-GFP plasmid. Transformants, selected by virtue of the marker on the plasmid, were then tested for their prion state by fluorescence microscopy. When extract made from cells harvested before doxycycline addition were tested, 64 of 100 (64%) transformants tested were [RNQ+]. Only 1 out of 50 transformants obtained using lysates from cells harvested 20 generations after doxycycline addition were [RNQ+]. In sum, we conclude that depletion of Sis1 results in loss of the [RNQ+] prion.
Increase in Rnq1 polymer size upon depletion of Sis1
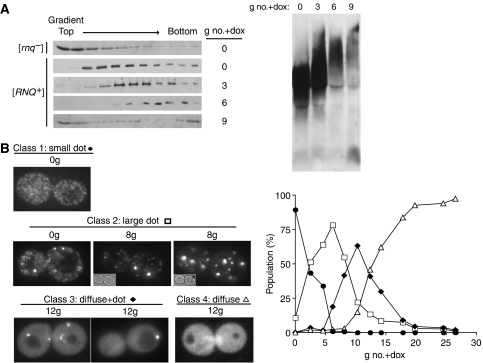
To better understand the effect of Sis1 depletion on the dynamics of Rnq1 structure, we followed the size of Rnq1 particles with time after repression of Sis1 expression. Cells were harvested 0, 3, 6 or 9 generations after doxycycline was added to log-phase cultures, and the resulting extracts analyzed by sucrose gradient centrifugation and SDD–AGE. The majority of Rnq1 in extracts from [rnq−] cells remained near the top of the gradient (Figure 2A, left panel), consistent with previous results (Bagriantsev and Liebman, 2004). Rnq1 from [RNQ+] cells migrated into the gradient, with the average sized aggregate migrating slightly further than an 80S marker (Figure 2A and Supplementary Figure 2). Rnq1 from cells harvested three and six generations after initiation of repression of Sis1 synthesis migrated progressively further into the gradient, whereas the majority of the Rnq1 in the nine-generation samples remained near the top of the gradient. The average size of the detergent-resistant particles also increased with time, before accumulation of low molecular weight Rnq1 (Figure 2A, right panel). Thus, over several generations after initiation of Sis1 repression, the size of Rnq1 polymers increased.
Figure 2.
SIS1 repression alters state of [RNQ+] before curing cells of prion. (A) Lysates from sis1-Δ[TETrSIS1] [rnq−] or [RNQ+] cells treated with doxycycline for the indicated number of generations (g no.+dox) were (left panel) separated by centrifugation through sucrose gradients, and fractions were isolated and analyzed by SDS–PAGE and immunoblotted with α-Rnq1, or (right panel) resolved by SDD–AGE and immunoblot analysis with α-Rnq1. (B) Cells expressing Rnq1-GFP from the RNQ1 promoter were visualized by fluorescence microscopy. Representative images of classes Rnq1-GFP phenotypes: small dots (•), large dot (□), diffuse+dot (⧫), diffuse (▵) (left panel). The percentage of cells that exhibited the indicated Rnq1-GFP phenotype, out of >800 cells per sample, plotted against the number of generations cells were subcultured in doxycycline (g no.+dox) (right panel). See Supplementary Table 1.
Cell population analysis of prion loss
Biochemical analyses are instructive concerning the effect of Sis1 on prion maintenance, but provide an averaged view of changes in Rnq1 structure taking place in the culture. To assess changes occurring at the cellular level, we utilized an Rnq1-GFP fusion whose expression was driven by the RNQ1 promoter. sis1-Δ[TETrSIS1] carrying this Rnq1-GFP fusion construct constitutively expressed Rnq1-GFP at approximately twice the level of endogenous Rnq1 (data not shown). When observed under the microscope, cells displayed a punctate pattern of fluorescence in a nonfluorescent background, consistent with the cells being [RNQ+] (Figure 2B).
Doxycycline was added to repress Sis1 synthesis and samples taken at intervals, up to 30 generations after doxycycline addition. With time, the percentage of cells showing a diffuse pattern of fluorescence increased (Figure 2B), reaching 15% at 10 generations, 84% at 18 generations and greater than 90% by 20 generations, consistent with the predominance of Rnq1 in the monomer form 16 generations after Sis1 repression as assayed by SDD–AGE (Figure 1C). Closer observations allowed us to place the cells having punctate fluorescence into several categories. Class 1 consisted of cells displaying multiple small foci of uniform size that moved very rapidly around the cytosol, which we refer to as the ‘small dot' phenotype. Class 2, which we term ‘large dot', are cells having one or more focus of fluorescence larger than those observed in the first class that either appeared to move throughout the cytosol more slowly than the small dots or remained relatively stationary. In the starting culture, the vast majority of cells, on the order of 90%, were ‘small dot' and 10% ‘large dot'.
Over time after drug addition, the proportion of ‘small dot' and ‘large dot' cells changed. At 4 generations, 64% of the cells were ‘large dot' and 34% ‘small dot'. In addition, it should be pointed out that the character of the ‘large dot' phenotype changed with time after initiation of repression of Sis1 synthesis. At the early time points, those classified as ‘large dot' typically contained many rapidly moving small foci and relatively few larger foci. At later time points, the ‘large dots' were bigger and fewer rapidly moving small foci were present, as indicated by the samples of the classes presented in Figure 2B.
Both Class 1 and Class 2 cells displayed punctate fluorescence against a dark background. In the intermediate time points, cells exhibited punctate fluorescence against a diffuse fluorescent background. Such cells were placed in Class 3, termed ‘diffuse+dot'. Greater than 60% of the cells at generation 10 had the ‘diffuse+dot' phenotype, as the proportion of cells having ‘large dots' rapidly decreased. The number of cells in the ‘diffuse+dot' category also decreased with time, as the majority of the population progressed to displaying only diffuse fluorescence (Class 4).
Accumulation of soluble and aggregated forms of newly synthesized Rnq1 after SIS1 repression
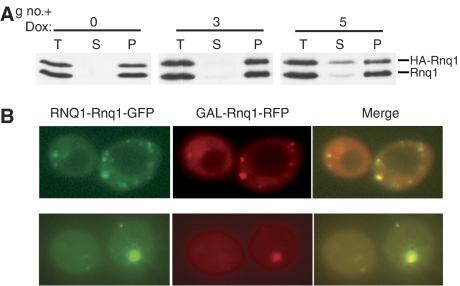
For a prion to be maintained, newly synthesized protein must be efficiently converted to the prion conformation. To directly assess the conversion of Rnq1 synthesized after initiation of Sis1 repression, we needed to distinguish newly synthesized Rnq1 from that present at earlier time points. To achieve that goal, we expressed Rnq1 with a HA tag (HA-Rnq1) under control of the GAL1 promoter to allow upregulation of the tagged Rnq1 upon transfer to galactose-based medium. To determine if HA-Rnq1 could be efficiently incorporated into prion particles, sis1-Δ[TETrSIS1] cells carrying the GAL-HA-RNQ1 construct were cultured in glucose media, and then shifted to galactose-based media for 3 h, at which point the level of HA-Rnq1 was approximately equivalent to that of untagged Rnq1. Extracts from these cells were subjected to high-speed centrifugation to separate prion aggregates from soluble protein. All of the tagged and untagged Rnq1 pelleted (Figure 3A), indicating efficient incorporation of Rnq1-HA into prion aggregates.
Figure 3.
Accumulation of newly synthesized Rnq1 after SIS1 repression. (A) sis1-Δ[TETrSIS1] cells transformed with GAL-HA-Rnq1 were subcultured in rich media in the presence of doxycycline for 0, 3 or 5 generations (g no.+dox). Cells were harvested, washed and transferred to doxycycline-containing rich, galactose-based media to induce HA-Rnq1 expression. After 3 h in galactose, extracts were prepared and subjected to high-speed centrifugation. Total (T), supernatant (S) and pellet (P) fractions were resolved by SDS–PAGE and analyzed by immunoblotting with α-Rnq1. (B) sis1-Δ[TETrSIS1] cells constitutively expressing Rnq1-GFP and harboring the GAL-Rnq1-RFP plasmid were cultured in doxycycline-containing glucose-based medium for 8 (upper panel) or 12 (lower panel) generations, and then cultured in the galactose-based medium in the presence of doxycycline for 3 h. Shown are two examples of diffuse+dot phenotype filtered for GFP, RFP or the overlay of the two images with yellow indicating colocalization.
Next, we wanted to compare the distribution of ‘new' and constitutively expressed Rnq1 after Sis1 repression. At three or five generations after addition of doxycycline, cells were transferred to doxycycline-containing galactose-based medium, and incubation continued for an additional 3 h. HA-Rnq1 from cells grown in the presence of doxycycline for three generations before induction of the tagged protein was nearly quantitatively found in the pellet fraction, along with endogenous Rnq1, indicating that incorporation of Rnq1 synthesized 3 to ∼4 generations after Sis1 repression was efficiently incorporated into aggregates. However, the distribution of tagged and untagged Rnq1 differed in the five-generation sample. Approximately 32% of the HA-Rnq1 was in the supernatant. However, only ∼7% of the constitutively expressed Rnq1, which is a mixture of pre-existing and ‘newly synthesized' Rnq1, was found in the supernatant. Thus, a portion of Rnq1, synthesized between 5 and ∼6 generations after Sis1 repression, was not incorporated into aggregates and accumulated in the soluble fraction.
These results are consistent with pre-existing Rnq1 protein being stably maintained in aggregates, whereas newly synthesized protein is inefficiently incorporated into Rnq1 polymers as early as six generations after repression of Sis1 synthesis. It was intriguing to us that this was approximately the time at which cells with ‘diffuse+dot' phenotype, those that contained visible aggregates, but also had substantial diffuse fluorescence, were beginning to comprise a significant proportion of the population. To allow evaluation of the fate of newly synthesized Rnq1 in such cells, we constructed a chimeric gene allowing expression of red fluorescent protein (RFP) fused to Rnq1 (Rnq1-RFP), under the control of a galactose-regulatable promoter. Doxycycline was added to sis1-Δ[TETrSIS1] cells carrying both the constitutive GFP and the regulatable RFP expression plasmids and then cells were shifted to galactose-based media to induce expression of Rnq1-RFP. The GFP and RFP signals colocalized in cells having ‘diffuse+dot' phenotype (Figure 3B), suggesting that newly synthesized Rnq1 was partitioning, some being incorporated into aggregates, and the remainder not, but rather joining the soluble pool. The localization of newly synthesized Rnq1 to large dots in these cells suggested that large aggregates in Class 3 cells remained active as seeds and were able to incorporate newly synthesized Rnq1.
Timing of loss of prion conversion capacity upon Sis1 repression
The biochemical and cellular analyses provided insight into the change in structure of Rnq1 upon Sis1 depletion. To correlate these data with the ability of cells to transmit the prion, two types of assays were performed. First, cytoduction assays were carried out as described above to test for the ability of the cytosol of individual cells to convert a recipient [rnq−] strain to [RNQ+]. Haploid strains were derived from eight independent cytoductants obtained from cells taken at 0, 5, 10 or 15 generations after addition of doxycycline. The prion states of the strains were monitored by visualization after transient expression of Rnq1-GFP (Table I). All eight cytoductants from the 5-generation time point were [RNQ+], as indicated by a punctate pattern of fluorescence; seven of the eight cytoductants from the 10-generation time point were [rnq−], with the progeny of one cytoductant showing a combination of cells having either diffuse or punctate fluorescence. All cytoductants from the 15-generation time point were [rnq−].
Table 1.
Frequency of [rnq−] strain conversion to [RNQ+] by introduction of doxycyline-treated sis1-Δ[TETrSIS1]cytoplasm by cytoduction, or lysates by transformation
| g no.+dox | No. of colonies tested | |
|---|---|---|
| Cytoduction | Yeast lysate transformation | |
| [rnq−]/[RNQ+] | [rnq−]/[RNQ+] | |
| 0 | 0/8 | 6/21 |
| 5 | 0/8 | 21/9 |
| 10 | 7/1 | 49/1 |
| 15 | 8/0 | 49/1 |
We also tested the ability of extracts from cells taken at various times after Sis1 repression to convert [rnq−] to [RNQ+]. When extract made from cells harvested before doxycycline addition were tested, 21 of 27 (78%) transformants tested were [RNQ+], as indicated by a punctate pattern of fluorescence (Table I). By five generations after drug addition, this fraction was reduced, with 9 of 30 (30%) transformants testing [RNQ+] positive. At the 10- and 15-generation time points, the number of transformants that tested [RNQ+] was dramatically lower, only 1 of 50.
Results of both these biological assays indicate that the capacity for prion transmission was dramatically reduced by 10 generations after Sis1 repression. By this point, significant soluble Rnq1 was detected in the biochemical assay and the observation of fluorescence generated by Rnq1-GFP indicated that the vast majority of cells in the population had either a ‘diffuse+dot' or ‘diffuse' phenotype. The infectivity analyses also revealed that by five generations, at which point significant enhancement in the size of Rnq1 polymers had occurred, a reduction in the number of seeds in the population was beginning to occur, as efficiency of infectivity was reduced from 78 to 30% in our assay.
Evidence for requirement of J-domain function for prion maintenance
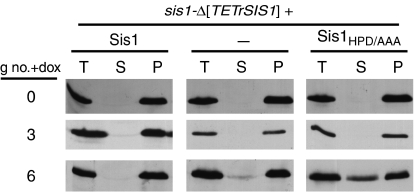
Sis1 is a J-protein co-chaperone known to function with the Hsp70 Ssa. Although J-proteins have not been observed to function independently of Hsp70s, it is possible that Sis1's function in prion maintenance does not depend on its interaction with an Hsp70. However, because Ssa has multiple essential functions, it was difficult to directly critically test its role in the maintenance of the [RNQ+] prion. Therefore, to assess whether prion loss during Sis1 repression is due to loss of Sis1's J-protein function, we devised a test utilizing a Sis1 mutant protein, Sis1HPD/AAA (Yan and Craig, 1999), containing a triple alanine substitution of the conserved HPD motif of the J-domain, which is required for ATPase stimulation of Hsp70s by J-proteins. We set up conditions such that expression of Sis1HPD/AAA was maintained at normal levels, whereas expression of wild-type Sis1 was repressed. The logic behind this test was that, as higher levels of Sis1 function are required for prion maintenance than for cell viability, if the prion maintenance function was independent of Sis1's co-chaperone activity, expression of Sis1HPD/AAA at wild-type levels should be able to substitute for wild-type Sis1 in prion maintenance.
Plasmids capable of expressing Sis1 or Sis1HPD/AAA under the native SIS1 promoter, as well as a vector control, were transformed into sis1-Δ[TETrSIS1]. Prion maintenance was monitored at three and six generations after the addition of doxycycline. Over the time course of the experiment, Rnq1 was maintained in the aggregated state in cells expressing wild-type Sis1 from the endogenous promoter, as all of the Rnq1 was found in the pellet fraction as assessed by centrifugation analysis (Figure 4). In contrast, a substantial portion of Rnq1 partitioned to the supernatant upon centrifugation of extracts from the six-generation time point of cells expressing Sis1HPD/AAA. These results are consistent with a role of Sis1 as an Hsp70 co-chaperone in prion maintenance. The accumulation of soluble Rnq1 occurred more rapidly than in the absence of Sis1HPD/AAA, likely because of interference of the mutant Sis1 with the function of the wild-type protein.
Figure 4.
Sis1 is an Hsp70 co-chaperone in prion maintenance. sis1-Δ[TETrSIS1] cells expressing wild-type Sis1, empty vector (−), or Sis1HPD/AAA were cultured in the presence of doxycycline for the indicated number of generations (g no.+dox). Total (T), supernatant (S) and pellet (P) fractions of lysates following centrifugation were resolved by SDS–PAGE and immunoblot analysis with α-Rnq1.
Increase in Rnq1 polymer size upon inhibition of Hsp104
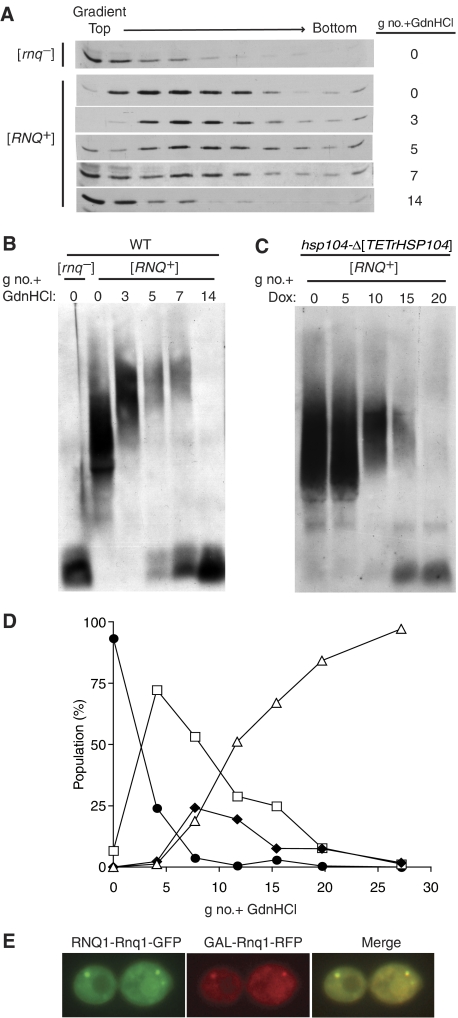
Hsp104 is required for maintenance of both [PSI+] and [RNQ+] in cell populations (Chernoff et al, 1995; Sondheimer and Lindquist, 2000). Reduced Hsp104 activity causes an increase in Sup35 polymer size in cells before detectable accumulation of monomers (Kryndushkin et al, 2003). To test the effect of inhibition of Hsp104 activity on the state of Rnq1 protein over time, we carried out two tests. First, we treated a culture of [RNQ+] cells with guanidine hydrochloride (GdnHCl), which is known to inhibit Hsp104's ATPase activity (Ferreira et al, 2001; Jung and Masison, 2001; Grimminger et al, 2004). Lysates prepared from [RNQ+] cells at various intervals after GdnHCl addition, as well as those from untreated [RNQ+] and [rnq−] cells, were subjected to centrifugation through sucrose gradients (Figure 5A). Rnq1 from cells harvested three generations after the addition of GdnHCl migrated even further into the gradient, whereas after longer times, increasing amounts of Rnq1 were found at the top of the gradient. Lysates were also subjected to SDD–AGE (Figure 5B). The average size of these particles increased with time after GdnHCl addition and the presence of monomers became more predominant, eventually accounting for the entire pool of Rnq1.
Figure 5.
Hsp104 and Sis1 inhibition result in similar pattern of distribution of Rnq1 over time. (A, B) GdnHCl treatment. Lysates prepared from wild-type [rnq−] and [RNQ+] cells subcultured in the presence of GdnHCl for 0, 3, 5, 7 or 14 generations (g no.) were separated by (A) sucrose gradient centrifugation, followed by SDS–PAGE or (B) SDD–AGE. Rnq1 was detected by immunoblotting with α-Rnq1 antibodies. (C) Depletion of Hsp104. Lysates prepared from hsp104-Δ[TETrHSP104] cells treated with 10 μg/ml of doxycycline were harvested 0, 5,10, 15 and 20 generations (g no.) were detected by immunoblotting with α-Rnq1 antibodies. (D) Time course of GdnHCl treatment of wild-type cells carrying the RNQ1-Rnq1-GFP plasmid. Cells were harvested after the indicated number of generations (g no.+GdnHCl), and visualized by fluorescence microscopy. Plots represent the percentage of cells, out of >500 per sample, that exhibited the indicated Rnq1-GFP classification (small dots (•), large dot (□), diffuse+dot (⧫), diffuse (▵)) plotted against number of generations cells were treated with GdnHCl (g no.+GdnHCl). See Supplementary Table 2. (E) Wild-type cells constitutively expressing Rnq1-GFP from the RNQ1 promoter and carrying the plasmid capable of expressing Rnq1-RFP in galactose-based medium were grown in glucose-based medium in the presence of GdnHCl for eight generations, then washed and resuspended in galactose-based medium for an additional 3 h. Shown is an example of diffuse+dot phenotype filtered for GFP, RFP or the overlay of the two images with yellow indicating colocalization.
As GdnHCl may have effects on cells in addition to inhibition of Hsp104 activity (Bradley et al, 2003), we also tested the effect of depletion of Hsp104, utilizing a construct in which Hsp104 expression was driven by the tetracycline-repressible promoter. Analysis by SDD–AGE revealed an increase in the average size of Rnq1 aggregates with time after addition of doxycycline (Figure 5C and Supplementary Figure 3). It should be noted that the timing of the increase in average aggregate size and accumulation of monomer cannot be directly compared between GndHCl additon and repression of Hsp104 synthesis, as the rate and extent of decrease of cellular Hsp104 activity is not comparable. However, together these results indicate that the size of Rnq1-containing aggregates become larger upon inactivation of Hsp104 before accumulation of monomeric Rnq1, similar to what has been observed for Sup35.
Inactivation of Hsp104 results in similar distribution of Rnq1-GFP as depletion of Sis1
Large aggregates accumulate upon lowering the activity of either Sis1 or Hsp104 (Figures 2A and 5A–C). Also, as described above (Figure 2B), the distribution of fluorescence in cells depleted of Sis1 followed a pattern in which the size of fluorescent foci increased and then a population of cells having dots against a diffuse background arose. This transient presence of cells displaying a ‘diffuse+dot' phenotype upon depletion of Sis1 raised the question of whether a similar class of cells exists during the process of prion loss due to inactivation of Hsp104.
To allow a comparison at the cellular level of the effect of lowering the cellular activity of the two chaperones, GdnHCl was added to wild-type cells carrying the plasmid driving Rnq1-GFP expression from the endogenous RNQ1 promoter (RNQ1-Rnq1-GFP) and the pattern of fluorescence monitored over time. By 28 generations, 97% of cells displayed diffuse fluorescence (Figure 5D). Similar to the case of depletion of Sis1, the percentage of cells having a ‘small dot' phenotype decreased, with a concomitant increase in ‘large dot' cells, followed by a significant portion of the population having a ‘diffuse+dot' pattern. Thus, initiation of prion loss by Hsp104 inactivation resulted in a similar sequence of events, very reminiscent of Sis1 repression.
As reduction in activity of either Hsp104 or Sis1 resulted in accumulation of cells having the ‘diffuse+dot' phenotype, we decided to assess the fate of newly synthesized Rnq1 in GdnHCl-treated cells. We again utilized cells that expressed Rnq1-GFP constitutively and were capable of regulated expression of Rnq1-RFP. Wild-type cells harboring the two plasmids were cultured in glucose-based medium for eight generations after the addition of GdnHCl, and then transferred to GdnHCl-containing, galactose-based medium for 3 h to allow expression of Rnq1-RFP. As was the case during Sis1 repression (Figure 3B), Rnq1-GFP and the Rnq1-RFP colocalized, indicating that newly synthesized Rnq1 partitioned into both aggregates and the soluble pool (Figure 5E). In addition, depletion of Hsp104 showed a similar sequence of events and colocalization (data not shown).
Discussion
The J-protein Sis1 functions in the propagation of the [RNQ+] prion of S. cerevisiae. The data presented here are most consistent with a model in which Sis1 functions as a J-protein partner of Hsp70, and in conjunction with Hsp104 generates new prion seeds by facilitating fragmentation of high molecular weight Rnq1 aggregates.
Sis1 function in [RNQ+] propagation
Rnq1 aggregates increase in size upon depletion of Sis1, as indicated by results of biochemical analysis of Rnq1 in cellular extracts, as well as direct in vivo visualization utilizing an Rnq1-GFP fusion. Within three generations of initiation of Sis1 repression, an increase in Rnq1 particle size was evident. Within five generations, reduction in the number of prion seeds was measurable. The decrease in number of seeds coupled with an increase in aggregate size points to a role of Sis1 in the fragmentation of Rnq1 prion aggregates. Several years ago, similar observations obtained during analysis of [PSI+] loss upon Hsp104 inactivation (Paushkin et al, 1996; Wegrzyn et al, 2001; Kryndushkin et al, 2003; Borchsenius et al, 2006) led to the currently favored idea that this chaperone functions in the fragmentation of Sup35 prion aggregates. Hsp104 is also essential for propagation of [RNQ+]. Indeed, the patterns of change of Rnq1 aggregate size upon decrease in Sis1 and Hsp104 activity we observed were very similar, supporting the idea that the Sis1:Hsp70 machinery and Hsp104 function together to maintain a population of prion seeds by fragmentation of Rnq1 aggregates. There is a precedent of J-protein:Hsp70 pairs functioning together with AAA+ ATPases in the dissolution of protein aggregates in vitro (Glover and Lindquist, 1998; Goloubinoff et al, 1999; Motohashi et al, 1999; Zolkiewski, 1999). In fact, Sis1 with its Hsp70 partner Ssa1 was shown to facilitate the solubilization and reactivation of denatured luciferase in an Hsp104-dependent reaction (Krzewska et al, 2001).
The mode of cooperation between J-protein:Hsp70 and Hsp104 machineries has been a matter of debate (Bosl et al, 2006), with proposals of both upstream and downstream action of the J-protein:Hsp70 machinery relative to AAA+ ATPases such as Hsp104. Sis1 is stably associated with Rnq1 in [RNQ+] strains (Lopez et al, 2003), whereas a physical interaction between Hsp104 and Rnq1 has not been reported. These observations support the idea that the J-protein:Hsp70 system functions upstream of Hsp104, which is consistent with recent results indicating that, in the bacterial system, the J-protein:Hsp70 system of DnaJ:DnaK acts before the AAA+ ATPase ClpB (Weibezahn et al, 2004; Zietkiewicz et al, 2004). Thus, whereas the exact mechanism of how such fragmentation occurs remains an enigma, we favor the following scenario in the case of [RNQ+]: the J-protein:Hsp70 machinery of Sis1:Ssa acts before Hsp104 in fiber fragmentation, perhaps facilitating a conformational change of an internal monomer, allowing the engagement of Hsp104 and ultimately removal of the monomer, resulting in fragmentation of the fiber into two.
Model of [RNQ+] prion propagation
The data presented here for [RNQ+], and elsewhere for [PSI+] as mentioned throughout the text, strongly support the idea that aggregates become larger, and therefore, the number of seeds in the cell population becomes fewer during the process of prion loss. But what occurs as at the cellular level as the cell population proceeds on the path to becoming [prion−]? Our data suggest that it is a combination of factors that result in the prion being lost from the population. First, a decrease in activity of either Sis1 or Hsp104 results in accumulation of a population of cells having large aggregates, and thus on average fewer seeds. At some point, the number of seeds reaches a critical point such that rate of conversion to the prion conformation cannot keep up with the rate of protein synthesis, visualized in our studies by the appearance of a class of cells having large aggregates in a background of diffuse fluorescence. Second, daughter cells fail to inherit seeds, both because they are fewer in number and because of they have reduced mobility due to their large size. Thus, these newly budded cells, as well as their progeny, are [prion−]. Such cells are represented in our assays as the late appearing cells having only diffuse fluorescence that proceed to dominate the population.
The colocalization of newly synthesized Rnq1with large aggregates indicates that even these large structures are active as seeds and capable of incorporating newly synthesized protein into prion fibers. However, it is likely that they are ‘inefficient seeds', having fewer ends available per monomer present in the aggregate for conversion of soluble monomers compared to smaller particles. This idea that larger aggregates are less efficient as seeds is consistent with a recent report in which different strains of [PSI+] were analyzed (Tanaka et al, 2006). A positive correlation was found between the size of the aggregates and the amount of monomeric Sup35 present in cell lysates, with those having the smallest polymers having the least monomeric Sup35.
Specificity of Sis1 requirement in [RNQ+] propagation
Although there are 13 J-proteins in the yeast cytosol, Sis1 is specifically required for propagation of [RNQ+]. Intriguingly, this specificity has been maintained in evolution, as the ortholog of Sis1 in mammalian cells, Hdj1, is able to maintain [RNQ+] when expressed in yeast, but the ortholog of the closely related cytosolic J-protein Ydj1, called Hdj2, cannot (Lopez et al, 2003). In addition to this high degree of specificity, the exquisite sensitivity of [RNQ+] prion propagation to the levels of Sis1 is also surprising. Whereas only a small fraction of the amount of Sis1 normally present in a cell is required for robust growth, even a 50% reduction in Sis1 levels destabilizes [RNQ+], even though it is between 18- and 65-fold more abundant than Rnq1 (Ghaemmaghami et al, 2003; Lopez et al, 2003). Neither why such high activity of Sis1 is required for prion maintenance, nor the mechanistic basis for this specificity is understood. However, it does not appear to lie in the ability of Sis1 to specifically bind to Rnq1, as Sis1 lacking its client protein-binding domain maintains its function in prion maintenance.
In summary, results presented here establish the requirement of Sis1 and Hsp104 for efficient prion seed generation needed for propagation of [RNQ+]. Further analysis will be required to elucidate their mechanism of cooperative action, and the basis of the specificity of Sis1 in prion maintenance.
Materials and methods
Strains and growth media
All strains, with the exception of the one used for the cytoduction assay, used in this study have the W303 genetic background. Wild-type W303 (PJ513a): [RNQ+] [psi−] MAT a trp1-1 ura3-1 leu2-3,112 his3-11,15 ade2-1 can1-100 GAL2 met2-1 lys2-2 (James P and Craig EA, unpublished data). Strains having Sis1 expression under the control of the tetracycline repressible (TETr) promoter (sis1-Δ[TETrSIS1]) were created by transforming sis1∷HIS3 or sis1∷LEU2 strains harboring a URA3-marked expression vector having the wild-type SIS1 gene (p316-SIS1) with the TRP1-marked TETrSIS1 expression vector, and counterselecting for p316-SIS1 on medium containing 5-fluororotic acid. The [RNQ+] hsp104-Δ[TETrHSP104] strain was constructed by crossing [rnq−] hsp104-Δ[TETrHSP104] containing the TETrHSP104 plasmid with [RNQ+] W303. Haploid progeny were recovered and their [RNQ+] phenotype confirmed.
[rnq−] strains for use as controls were constructed as follows. For the assays using GdnHCl treatment of wild-type cells, PJ513a was passaged on media containing 3 mM GdnHCl. To create the sis1-Δ[TETrSIS1] [rnq−] strain used in the Sis1 repression assays, sis1∷HIS3 [RNQ+] cells were cured of the Rnq1 prion by expression of Sis1ΔG/F, and after confirmation of prion loss, the TETrSIS1 plasmid was introduced into the strain by plasmid shuffling.
Plasmids
TETrSIS1 and TETrHSP104 were constructed by inserting the SIS1 or HSP104 open reading frame into the vector pCM184 (Gari et al, 1997), placing them under control of a tetracycline-repressible promoter (TETr). Unless otherwise indicated, all other plasmids used in this study were based on the pRS series of plasmids (Sikorski and Hieter, 1989). To regulate expression of a tagged Rnq1, a galactose inducible HA-tagged Rnq1 (GAL-HA-Rnq1) plasmid was constructed. Using PCR, a 3 × HA-tag was inserted at the 5′ end of RNQ1 in p316, followed by PCR amplification of the HA-RNQ1 fragment, which was then inserted into pYES2 vector (Invitrogen) placing it under control of the GAL1 promoter.
Rnq1-GFP fusions were constructed with a monomeric GFP to prevent nonspecific aggregation of Rnq1-GFP by GFP dimerization. A plasmid (RNQ1-Rnq1-GFP) encoding a monomeric Rnq1-GFP fusion under the control of the RNQ1 promoter was generated by inserting the RNQ1 promoter and coding sequence followed by a linker sequence (GSSGTSR) and a mutant Aequorea victoria GFP gene into p413 plasmid (Mumberg et al, 1995). The mutant GFP contains amino-acid alterations (S65T and V163A) to enhance fluorescence and facilitate folding (Heim et al, 1995; Tsien, 1998) and A206K to prevent dimerization (Zacharias et al, 2002). To generate monomeric Rnq1-GFP under the CUP1 promoter that is regulated by copper (CUP1-Rnq1-GFP), the Rnq1-GFP mutant described above was cloned into the p316-CUP1 plasmid (Sondheimer and Lindquist, 2000). To generate a galactose-inducible RFP construct (GAL-Rnq1-RFP), the Discosoma RFP gene, which expresses the monomeric form of RFP (Campbell et al, 2002), was ligated to the 3′ end of RNQ1 with the linker sequence as described above. The Rnq1-RFP fusion coding sequence was then inserted into p416-GALS plasmid (Mumberg et al, 1994).
Biochemical analysis of prion particle size
Lysates were prepared by vortexing cell pellets, resuspended in protein extraction buffer (PEB: 25 mM Tris (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 1 mM EDTA) supplemented with protease inhibitor cocktail (complete mini (Roche), PMSF, DTT and pepstatin), with glass beads. Separation of prion particles by sucrose gradient analysis was performed as described previously (Tanaka et al, 2006) with modifications. Lysate from each strain was precleared by centrifugation at 10 000 g for 5 min at 4°C to remove unlysed cells. Equivalent amounts of total protein extracts were layered onto 2 ml 15/40/60% sucrose cushions, and ultracentrifuged at 31 000 r.p.m. for 60 min in a SW55Ti (Beckman) rotor. Fractions were collected and resolved by SDS–PAGE, followed by immunoblotting with Rnq1-specific antibodies.
SDD–AGE was performed as described elsewhere (Kryndushkin et al, 2003; Bagriantsev and Liebman, 2004) with modifications. Lysates were precleared for 2 min at 600 g at 4°C and equivalent amounts of total protein were then incubated in nonreducing sample buffer for 7 min at 25°C, and resolved in a 1.6% Tris-glycine (0.1% SDS) agarose gel. The protein was transferred to nitrocellulose membrane at 10 V for 14 h at 4°C in a Tris-glycine/methanol buffer and probed with α-Rnq1 antibodies. A preparation of rat soleus extract was used as a marker to estimate the molecular weight of Rnq1 particles.
The centrifugation assay was performed as described previously (Sondheimer and Lindquist, 2000; Sondheimer et al, 2001; Lopez et al, 2003). Precleared lysates were centrifuged at 80 000 r.p.m. (Beckman rotor TLA120.1) for 30 min. Supernatant and pellet fractions were isolated and resolved by standard SDS–PAGE and immunoblot analysis.
Assays for prion curing
For the cytoduction assay, donor cells were mixed with the recipient [rnq−] cytoduction strain (Y1470) on glucose-based rich media and incubated overnight. Haploid cytoductants (containing recipient nucleus and donor cytoplasm) were selected by isolation of single colonies on glycerol-based medium containing G418. Isolated haploids were transformed with CUP1-Rnq1-GFP, subjected to colony purification and analyzed by fluorescence microscopy 4–6 h after addition of 50 μM CuSO4 (Sondheimer and Lindquist, 2000).
The procedure for yeast lysate transformations (adapted from King and Diaz-Avalos, 2004; Brachmann et al, 2005), was carried out by co-transforming spheroplasts of wild-type [rnq−] cells with cell extracts of the test strain containing equivalent amounts of total protein, and CUP1-Rnq1-GFP plasmid. Cells that took up the plasmid DNA were selected on medium lacking uracil, then patched onto media containing 50 μM CuSO4. After overnight incubation, the Rnq1 prion state was assessed by fluorescence microscopy. As controls, [rnq−] cells were transformed with CUP1-Rnq1-GFP plasmid in the absence or the presence of lysate from [rnq−] cells and the transformants observed before and after induction. In such controls, punctate fluorescence was only rarely observed in the cell population, and never observed in more than 1% of the cells. In addition, transformant were subjected to three rounds of colony purification. All transformants designated [rnq−] or [RNQ+] initially after isolation maintained their respective [rnq−] or [RNQ+] phenotypes.
Time course of prion loss assays
All cell cultures used for SIS1 repression assays and Hsp104 inactivation assays were maintained in log phase by continual subculturing into fresh liquid media. SIS1 repression time-course assays were carried out in the absence or presence of 5 μg/ml of the tetracycline analog doxycycline (Sigma). This concentration was chosen after determining that the level of Sis1 expression after doxycycline addition was similar over a large range of drug concentrations. Hsp104 inactivation was achieved by addition of GdnHCl to a concentration of 1–3 mM.
For the analysis of constitutively expressed Rnq1-GFP, sis1-Δ[TETrSIS1] cells containing RNQ1-Rnq1-GFP plasmid were subcultured in glucose-based complete minimal medium plus 5 μg/ml doxycycline, and visualized by fluorescence microscopy at the indicated number of generations. For the analysis of newly synthesized Rnq1 (HA-Rnq1 or Rnq1-RFP), cells were subcultured in glucose-based minimal plus drug for the indicated number of generations, washed, and transferred to galactose-based media plus drug for 3 h to induce expression of GAL-HA-Rnq1 or GAL-Rnq1-RFP. It is noted that the [RNQ+] phenotype of the sis1-Δ[TETrSIS1] cells carrying Rnq1-GFP was stable, being maintained over many months of propagation.
Prion analysis by fluorescence microscopy
Images were obtained with a Nikon Eclipse E800 fluorescence microscope (× 60/1.4 NA plan apochromat objective), or a Zeiss inverted fluorescence microscope Axiovert 200 M (× 63/1.4 oil plan apochromat objective powered by × 1.6 tube lens (total magnification × 100.8)). Filter sets used are as follows: 17 (BP 485/20, FT 510, BP 515–565) for GFP; 20 (BP 546/12, FT 560, BP 575–640) for RFP. Images were captured by a photometrics CoolSnap HQ cooled CCD camera (Roper Scientific Inc.) or an AxioCam HRm CCD camera with the resolution of 1300 pixel × 1030 pixel and treated with AxioVison release 4.5 software, with the Nikon or Zeiss microscope, respectively.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Tables
Acknowledgments
We thank Marion Greaser, Roger Tsien and Susan Lindquist for rat soleus samples, RFP plasmids and the hsp104-Δ strain, respectively; Bill Sugden, Asuka Nanbo and Chris Wiese for use of microscopes; members of the Craig laboratory and Jaroslaw Marszalek for thoughtful comments on the manuscript. This work was supported by National Institutes of Health Grant GM53655 and the USDA Cooperative State Research, Education and Extension Service (CSREES) project WISO4769 (EAC); Uehara Memorial Foundation Postdoctoral and Human Frontier Science Program Long-Term Fellowships (TH).
References
- Bagriantsev S, Liebman SW (2004) Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J Biol Chem 279: 51042–51048 [DOI] [PubMed] [Google Scholar]
- Borchsenius AS, Muller S, Newnam GP, Inge-Vechtomov SG, Chernoff YO (2006) Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr Genet 49: 21–29 [DOI] [PubMed] [Google Scholar]
- Bosl B, Grimminger V, Walter S (2006) The molecular chaperone Hsp104—a molecular machine for protein disaggregation. J Struct Biol 156: 139–148 [DOI] [PubMed] [Google Scholar]
- Brachmann A, Baxa U, Wickner RB (2005) Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J 24: 3082–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, Bagriantsev S, Vishveshwara N, Liebman SW (2003) Guanidine reduces stop codon read-through caused by missense mutations in SUP35 or SUP45. Yeast 20: 625–632 [DOI] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY (2002) A monomeric red fluorescent protein. Proc Natl Acad Sci USA 99: 7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268: 880–884 [DOI] [PubMed] [Google Scholar]
- Conde J, Fink GR (1976) A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci USA 73: 3651–3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA, Huang P, Aron R, Andrew A (2006) The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev Physiol Biochem Pharmacol 156: 1–21 [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW (2001) Prions affect the appearance of other prions: the story of [PIN(+)]. Cell 106: 171–182 [DOI] [PubMed] [Google Scholar]
- Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF (2001) The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol 40: 1357–1369 [DOI] [PubMed] [Google Scholar]
- Gari E, Piedrafita L, Aldea M, Herrero E (1997) A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13: 837–848 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S (1997) Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89: 811–819 [DOI] [PubMed] [Google Scholar]
- Glover JR, Lindquist S (1998) Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82 [DOI] [PubMed] [Google Scholar]
- Goloubinoff P, Mogk A, Zvi AP, Tomoyasu T, Bukau B (1999) Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci USA 96: 13732–13737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimminger V, Richter K, Imhof A, Buchner J, Walter S (2004) The prion curing agent guanidinium chloride specifically inhibits ATP hydrolysis by Hsp104. J Biol Chem 279: 7378–7383 [DOI] [PubMed] [Google Scholar]
- Heim R, Cubitt AB, Tsien RY (1995) Improved green fluorescence. Nature 373: 663–664 [DOI] [PubMed] [Google Scholar]
- Jung G, Masison DC (2001) Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr Microbiol 43: 7–10 [DOI] [PubMed] [Google Scholar]
- King CY, Diaz-Avalos R (2004) Protein-only transmission of three yeast prion strains. Nature 428: 319–323 [DOI] [PubMed] [Google Scholar]
- King CY, Tittmann P, Gross H, Gebert R, Aebi M, Wuthrich K (1997) Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci USA 94: 6618–6622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV (2003) Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem 278: 49636–49643 [DOI] [PubMed] [Google Scholar]
- Krzewska J, Langer T, Liberek K (2001) Mitochondrial Hsp78, a member of the Clp/Hsp100 family in Saccharomyces cerevisiae, cooperates with Hsp70 in protein refolding. FEBS Lett 489: 92–96 [DOI] [PubMed] [Google Scholar]
- Lopez N, Aron R, Craig EA (2003) Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+]. Mol Biol Cell 14: 1172–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Watanabe Y, Yohda M, Yoshida M (1999) Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc Natl Acad Sci USA 96: 7184–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22: 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122 [DOI] [PubMed] [Google Scholar]
- Ness F, Ferreira P, Cox BS, Tuite MF (2002) Guanidine hydrochloride inhibits the generation of prion ‘seeds' but not prion protein aggregation in yeast. Mol Cell Biol 22: 5593–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherovich LZ, Weissman JS (2001) Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell 106: 183–194 [DOI] [PubMed] [Google Scholar]
- Patel BK, Liebman SW (2007) ‘Prion-proof' for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132-405) induces [PIN+]. J Mol Biol 365: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD (1996) Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J 15: 3127–3134 [PMC free article] [PubMed] [Google Scholar]
- Satpute-Krishnan P, Langseth SX, Serio TR (2007) Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol 5: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S (2004) Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304: 1793–1797 [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer N, Lindquist S (2000) Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell 5: 163–172 [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lopez N, Craig EA, Lindquist S (2001) The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J 20: 2435–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS (2004) Conformational variations in an infectious protein determine prion strain differences. Nature 428: 323–328 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Collins SR, Toyama BH, Weissman JS (2006) The physical basis of how prion conformations determine strain phenotypes. Nature 442: 585–589 [DOI] [PubMed] [Google Scholar]
- Tsien RY (1998) The green fluorescent protein. Annu Rev Biochem 67: 509–544 [DOI] [PubMed] [Google Scholar]
- Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO (2001) Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol 21: 4656–4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, Mogk A, Bukau B (2004) Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119: 653–665 [DOI] [PubMed] [Google Scholar]
- Yan W, Craig EA (1999) The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol Cell Biol 19: 7751–7758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY (2002) Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296: 913–916 [DOI] [PubMed] [Google Scholar]
- Zietkiewicz S, Krzewska J, Liberek K (2004) Successive and synergistic action of the Hsp70 and Hsp100 chaperones in protein disaggregation. J Biol Chem 279: 44376–44383 [DOI] [PubMed] [Google Scholar]
- Zolkiewski M (1999) ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J Biol Chem 274: 28083–28086 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Tables