Abstract
Incretins, endogenous polypeptide hormones released in response to food intake, potentiate insulin secretion from pancreatic β cells after oral glucose ingestion (the incretin effect). This response is signaled by the two peptide hormones glucose-dependent insulinotropic polypeptide (GIP) (also known as gastric inhibitory polypeptide) and glucagon-like peptide 1 through binding and activation of their cognate class 2 G protein-coupled receptors (GPCRs). Because the incretin effect is lost or significantly reduced in patients with type 2 diabetes mellitus, glucagon-like peptide 1 and GIP have attracted considerable attention for their potential in antidiabetic therapy. A paucity of structural information precludes a detailed understanding of the processes of hormone binding and receptor activation, hampering efforts to develop novel pharmaceuticals. Here we report the crystal structure of the complex of human GIP receptor extracellular domain (ECD) with its agonist, the incretin GIP1–42. The hormone binds in an α-helical conformation in a surface groove of the ECD largely through hydrophobic interactions. The N-terminal ligand residues would remain free to interact with other parts of the receptor. Thermodynamic data suggest that binding is concomitant with structural organization of the hormone, resulting in a complex mode of receptor–ligand recognition. The presentation of a well structured, α-helical ligand by the ECD is expected to be conserved among other hormone receptors of this class.
Keywords: glucose-dependent insulinotropic polypeptide, hormone binding, diabetes mellitus, x-ray
Incretins, intestinal peptide hormones released in response to food intake, potentiate insulin secretion from pancreatic β cells after oral glucose ingestion. This so-called incretin effect is strictly glucose-dependent and an essential factor in maintaining glucose homeostasis. In humans, this response is signaled by the two peptide hormones the 42-residue glucose-dependent insulinotropic polypeptide (GIP) (also known as gastric inhibitory polypeptide) and glucagon-like peptide (GLP)-1 (31 aa). Each hormone binds to a specific cognate class 2 G protein-coupled receptor (GPCR) to mediate signal transduction by activation of adenylate cyclase via a stimulatory G protein, leading to enhanced exocytosis of insulin-containing granules, β cell proliferation, and inhibition of apoptosis (1, 2).
The incretin effect is lost or significantly reduced in patients with type 2 diabetes mellitus, with severe consequences for glucose homeostasis (3). In addition to the insulinotropic activity of GIP, a number of concomitant extrapancreatic effects have been described, e.g., stimulation of glucose uptake in muscle (4), osteotropic activity (5), and regulation of lipid metabolism in adipocytes (6), thereby providing a link between GIP signaling and obesity (7).
GLP-1 and GIP have attracted considerable attention in light of their potential use in the treatment of type 2 diabetes (8). The rapid degradation (circulating half-life of 2–5 min) of incretin hormones in vivo by the enzyme dipeptidyl-peptidase IV (DP4) (9, 10), a significant obstacle in their therapeutic administration, has been addressed by applying natural or synthetic DP4 resistant analogues of GLP-1 (11) and GIP (12, 13) instead of the native peptides. The inhibition of DP4 represents a promising approach to prolong the half-life of circulating incretin hormones (14). Nevertheless, reliance on peptide analogues restricts possibilities for generating completely synthetic agonist and antagonist molecules that could prevent potential side-effects of the enzyme inhibitors.
The GIP receptor (GIPR) belongs to the glucagon receptor subfamily of class 2 GPCRs, a class that also includes the receptors for GLP-1, glucagon, parathyroid hormone, calcitonin, corticotropin-releasing factor (CRF), and other therapeutically important peptide hormones (15). As with other GPCRs of this class, the GIPR comprises an N-terminal extracellular domain (ECD), a central domain consisting of seven transmembrane α-helices, and a C-terminal, cytoplasmic domain that mediates intracellular signal transduction by physical association with the G protein (16–18). The current understanding of ligand recognition and receptor activation mechanisms of class 2 GPCRs is largely restricted to in vitro analysis of ligand binding and cAMP stimulation by using ligand and receptor variants, chimeric constructs, and isolated receptor fragments. These studies have suggested a two-step mechanism for class 2 GPCR activation: (i) the C-terminal region of the peptide ligand binds the N-terminal ECD of the receptor, after which (ii) the N-terminal portion of the ligand interacts with the central transmembrane domain, causing a conformational rearrangement that activates the receptor [reviewed elsewhere (19)]. A paucity of structural information, however, precludes a detailed understanding of these fundamental physiological processes, hampering efforts to develop pharmaceuticals for this therapeutically important class of molecules.
A number of structural studies have been carried out on isolated peptide ligands of class 2 GPCRs, including GIP (20, 21). All such hormones studied so far display a propensity to form α-helices, although the degree of helicity depends on the experimental conditions. The only known structure of a full-length GPCR is that of bovine rhodopsin, a class 1 GPCR (22). Rhodopsin is activated by light-induced isomerization of the covalently bound photosensor retinal, so that inferences from this structure are of limited value for ligand-activated GPCRs. Insights into ligand recognition of class 3 GPCRs have been obtained through x-ray studies on the ECDs of the metabotropic glutamate receptor (23). The structures of the isolated ECDs from class 2 GPCRs murine CRF receptor (CRFR)-2β (24) and Methuselah from Drosophila (25) reveal strongly diverging folds. Very recently, the solution structure of an N-terminally truncated form of the CRFR-2β ECD has been described in the presence of the peptide antagonist astressin (26).
Here we report the crystal structure of the complex of human GIPR ECD (residues 24–138) with its agonist, the incretin hormone GIP1–42 [nomenclature: residues of the ECD are numbered according to the full-length sequence of the GIPR; residue numbers of the hormone are given a prime (′) suffix]. Accompanying thermodynamic measurements reveal determinants for ligand recognition and underlying mechanistic principles of complex formation.
Results
Overall Structure of the Ligand-Bound ECD.
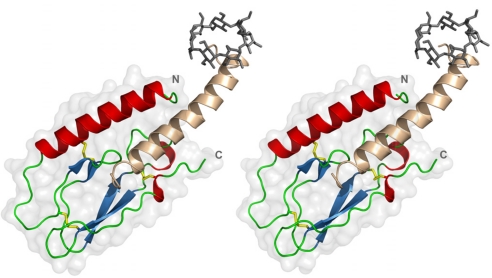
The crystal structure of the ECD of the human GIPR in complex with the ligand GIP1–42, (Fig. 1) reveals an N-terminal α-helix (spanning residues Ala-32 to Ala-52) that packs against a core with a sandwich-like arrangement of two antiparallel β-sheets, each comprising two short β strands (β1a: Ser-64–Phe-65; β1b: Cys-70–Trp-71; β2a: Ala-78–Ser-83; β2b: Phe-98–Cys-103), as well as two short helical C-terminal segments (His-91–Val-94 and Thr-116–Cys-118). The GIPR ECD displays only a moderate amount of secondary structure elements for a domain of this size, and is stabilized by a set of three disulfide bonds: the first (Cys-46–Cys-70) links the N-terminal α-helix to the first β-sheet, the second (Cys-61–Cys-103) connects the two β-sheets, whereas the third disulfide bond (Cys-84–Cys-118) holds the C terminus of the domain in close proximity to the central β-sheets. The topology imposed by the disulfide-linked β-sheets resembles the short consensus repeat, a small module commonly found in proteins of the complement system (24, 27). Because short consensus repeat domains, however, lack the N-terminal α-helix and hence the first disulfide bond, we suggest the structure of the GIPR ECD represents a prototype of a related yet distinct fold that we term the “glucagon hormone family recognition fold”.
Fig. 1.
Overall structure of the GIPR ECD in complex with GIP1–42. Stereoview of the peptide hormone GIP1–42 (residues 1′–32′ beige; residues 33′–42′ were not visible in the electron density) bound to the ECD of GIPR [colored according to secondary structure, residues 29(N)–122(C) visible]. Disulfide bridges in the GIPR ECD are shown as yellow sticks. The N terminus of the ligand is bound by a methyl-β-cyclodextrin molecule (gray sticks).
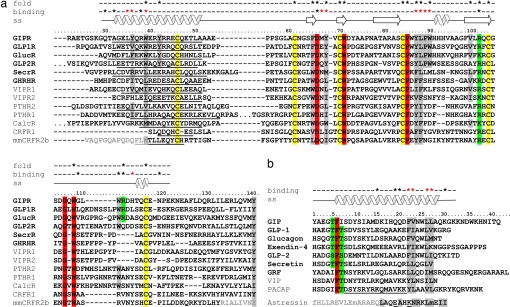
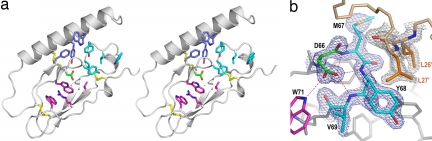
In addition to the disulfide bond pattern, the structure is defined by a set of key intramolecular interactions involving residues that are conserved within this receptor class (Fig. 2a), e.g., residues Trp-71 and Trp-109, the aromatic indol ring systems of which sandwich the basic residue Arg-101 (Fig. 3a). This central cluster (cluster 1) is further framed by a hydrophobic residue (Val-99) supporting Trp-71 on one side and the disulfide bond Cys-61–Cys-103 adjacent to Trp-109 on the opposite side. The absolutely conserved residue Asp-66 plays a central role in stabilizing the structure, making hydrogen bonds to the backbone amides of Met-67, Tyr-68, and Val-69 (stabilizing the β-turn at the base of the ligand-binding site; Fig. 3b), to the indole nitrogen of Trp-71, and to the guanidinium moiety of Arg-113. Although it has been suggested that the equivalent Asp-65 in CRFR-2β ECD forms a salt bridge to Arg-101 (25), we found no evidence for such an interaction. Mutation of Asp-66 to Gly in the murine growth hormone-releasing factor receptor leads to inactivation, resulting in a severe growth defect (28). The orientation of the N-terminal helix toward the core region of the GIPR ECD is held by a second cluster of hydrophobic interactions involving less conserved residues at positions 39 (Trp), 42 (Tyr), and 65 (Phe) and the adjacent first disulfide bond (Cys-46–Cys-70). The GIPR ECD possesses a further hydrophobic cluster near the third disulfide bridge (Cys-84–Cys-118) formed by the absolutely conserved Pro-85 and hydrophobic residues Tyr-68, Tyr-87, Leu-88, and Trp-90. Mutation of the proline residue corresponding to Pro-85 to Leu in the human parathyroid hormone receptor 1 results in an embryonic lethal disorder (29). These residues are located in loop regions between the β strands 1a–1b and 2a–2b and contribute in part to the ligand-binding surface of the domain (Fig. 4).
Fig. 2.
Hormone receptor ECDs and ligands. (a) Sequence alignment of the ECDs of human hormone receptors (CRFR-2β: murine sequence). The numbering is according to GIPR residues, and the names of the glucagon receptor family members are in bold. The conserved cysteines are marked in yellow, other absolutely conserved residues are in red, and less conserved amino acids are highlighted in gray (hydrophobic) and green (basic), respectively. The dots in the sequence represent additional residues that are not shown. Amino acids of the predicted N-terminal α-helix are underlined; residues missing from the construct of CRFR-2β used for structure determination (26) are in gray. Amino acids involved in stabilizing the GIPR ECD core structure are marked by asterisks in the line “fold.” (b) Sequence alignment of human peptide hormones (exendin-4 from Heloderma suspectum), with numbering according to GIP; the names of glucagon receptor family ligands are in bold. The absolutely conserved Phe-6′ is marked in red, and the positions of hydrophobic amino acids corresponding to the GIP residues interacting with the ECD are highlighted in gray. Potential N-terminal helix-capping residues are marked in green, and PACAP residues forming an α-helix when bound to its receptor are underlined (47). The sequence of astressin used in the NMR structure determination (26) is aligned according to superposition of the ECDs of GIPR and CRFR-2β; f denotes DPhe12, m indicates norleucine residues 21 and 38, and underlined residues Glu-30 and Lys-33 are chemically linked through a lactam bridge. Residues not observed in the NMR structure are in gray, and those involved in ECD binding are highlighted. The GIPR ECD and GIP1–42 residues comprising the binding interface are marked with asterisks in the line “binding”; red asterisks are assigned to residues involved in hydrophobic interactions. The secondary structure found in the structure of the complex is depicted in the line “ss.”
Fig. 3.
The glucagon hormone family recognition fold. (a) Stereoview of the GIPR ECD with the three clusters of residues characteristic for the domain structure, consisting of the following: W71, W109, R101, and V99 (cluster 1, magenta); residues W39, Y42, and F65 (cluster 2, dark blue); and residues P85, Y68, Y87, L88, and W90 (cluster 3, cyan). The absolutely conserved D66 is shown in green, and disulfide bridges are in yellow. (b) Stick representation of the GIPR ECD loop region around D66 (green), with a 2Fo − Fc map at 1.5σ contour (blue mesh), and of GIP residues L26′ and L27′ (orange), with corresponding 2Fo − Fc map (gray mesh). The side chain of D66 is involved in hydrogen bonds (dashed lines) to W71 (magenta) and residues M67, Y67, and V69 (shown in cyan) and thereby fixes the loop that is involved in ligand binding.
Fig. 4.
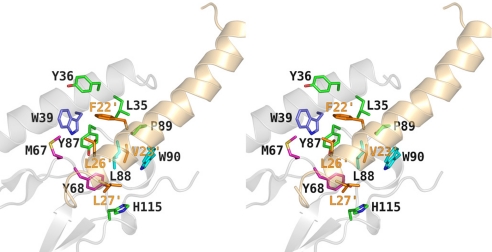
Hydrophobic interactions involved in ligand binding. Stereoview of GIP1–42 (beige) bound to the GIPR ECD (white) with key ligand residues (stick representation, orange) that occupy the hydrophobic binding groove of the GIPR ECD formed by residues from cluster 2 (dark blue), cluster 3 (cyan), the D66-loop region (magenta), or other residues at the ECD surface (green).
Structural Basis of Ligand Binding to the ECD.
Electron density is observed for GIP1–42 ligand residues Tyr-1′ to Lys-32′; residues 33′ to 42′ are disordered. The peptide hormone adopts a slightly curved α-helix between residues Phe-6′ and Ala-28′. The N terminus of the ligand is bound by a molecule of methyl-β-cyclodextrin, a cyclic carbohydrate added to the receptor–ligand complex that proved essential for crystallization. In the crystallized complex, the aromatic ligand residue Tyr-1′ and hydrophobic Ile-7′ are buried in a host–guest-like manner inside the ring of the cyclodextrin, resulting in a well defined loop structure for residues Tyr-1′ to Thr-5′ [supporting information (SI) Fig. 5a].
The crystal structure clearly shows that the N-terminal portion of the ligand (Tyr-1′–Met-14′) makes no contacts to the extracellular receptor domain. The binding interface of GIP1–42 spans an almost exclusively α-helical ligand C-terminal region comprising residues Asp-15′ to Lys-30′. This helix is, in part, amphipathic (Gln-20′–Ala-28′), exposing hydrophobic ligand residues (Phe-22′, Val-23′, Leu-26′, Leu-27′) toward the ECD, suggesting that binding is dominated by hydrophobic interactions. These residues occupy a complementary binding groove, consisting of residues of the conserved third hydrophobic cluster of the GIPR ECD (Tyr-68, Tyr-87, and Leu-88), the adjacent amino acids Pro-89 and Trp-90 (present also in GLP1R, GLP2R, and GlucR), two nonconserved residues (His-115 and Met-67) in the same region, and three additional residues of the N-terminal α-helix (Leu-35, Tyr-36, and Trp-39, partially conserved within the receptor class; Figs. 4 and 2a).
The structure of the GIPR ECD–GIP1–42 complex also reveals a distinct pattern of intermolecular hydrogen bonds, mediated, in part, by water molecules that augment ligand binding. Two regions of the ligand are involved: (i) the central part of the ligand α-helix (Asp-15′, Gln-19′, and Gln-20′) and (ii) the C-terminal residues (Leu-26′, Leu-27′, and Lys-30′). Lys-30′, in particular, is involved in an extensive hydrogen-bonding network with several residues in the C-terminal part of the GIPR ECD (Arg-101, Gly-110, and Trp-112) and also with the conserved residue Asp-66. The carbonyl groups of Leu-26′ and Leu-27′ act as hydrogen bond acceptors from Arg-114 and Asp-66, either directly or mediated by water molecules at the binding interface. A second cluster of hydrogen bonds between GIP1–42 and the GIPR ECD involves three central ligand residues and several N- and C-terminal residues of the ECD. Whereas Asp-15′ and Gln-19′ form hydrogen bonds to Ala-32, Gln-30, and Arg-38, located in the N-terminal helix of the receptor domain, the side chain of Gln-20′ participates in two hydrogen bonds to Asn-120 at the C terminus of the GIPR ECD (SI Fig. 5b).
Analysis of Ligand Binding.
Complex formation of the GIPR ECD with different ligands was assessed by isothermal titration calorimetry and far-/near-UV circular dichroism (CD) spectroscopy (SI Text, SI Figs. 6–8, and SI Tables 2–6). The binding studies confirm the structural analysis, revealing the dominance of hydrophobic interactions in complex formation. Thermodynamic parameters and CD analysis of GIP1–42 binding to the GIPR ECD suggest structural rearrangements within the binding partners upon complex formation. This is seen in (i) the burial of additional solvent accessible surface with respect to the actual binding interface formed by the GIPR ECD and GIP1–42, (ii) an increase in α-helicity observed by far-UV CD upon ligand binding, and (iii) the binding parameters determined by isothermal titration calorimetry of successively truncated GIP variants lacking N-terminal amino acids, suggesting that these residues, even though they are not in contact with the ECD in the crystal structure, are indirectly involved in binding to the GIPR ECD, presumably through participation in a structural reorganization that provides additional binding enthalpy, as would be the case for α-helical structure formation. An ≈10-fold weaker binding to the GIPR ECD was observed for the incretin GLP-17–36, indicating that the interaction of GIP1–42 and GLP-17–36 with the ECDs of their closely related receptors involves certain common principles, although the significantly lower values for enthalpy and entropy of GLP-17–36 binding to the GIPR ECD compared with GIP1–42 reflect differences in their binding mode.
Discussion
The GIPR ECD exhibits the same overall fold as that of the CRFR-2β ECD (24). The similar arrangement of disulfide bonds of other class 2 GPCR ECDs (30–32) and conservation of key residues such as Trp-71, Trp-109, Arg-101, and Asp-66 (Fig. 1) indicate that this structure may be considered a prototype for other endogenous peptide hormone receptors. The most striking difference to the ligand-free CRFR-2β ECD NMR structure is found at the N terminus of the ECDs (SI Fig. 9a). The GIPR ECD exhibits a distinct N-terminal α-helix; this region is undetermined in the NMR structures of the CRFR-2β ECD (26), presumably because the construct used was truncated by 14 residues. Secondary structure predictions suggest that all of the peptide hormone receptors should be α-helical in this region (data not shown), although a shorter helix is predicted for the CRFR-1 ECD (Fig. 2a).
Functional studies using receptor chimera of the GIPR and GLP-1 receptor (17) indicate that the N-terminal domain, in conjunction with other parts of the receptor such as the first extracellular loop, confers affinity to the hormone. Similar approaches demonstrate this to be valid for other members of this receptor family and suggest further that the C-terminal part of the hormone ligand is recognized by the ECD (33–35). In agreement with these results, the crystal structure reveals that the ECD binds the 16 ligand residues Asp-15–Lys-30 in an α-helical conformation, corresponding to the C-terminal residues in most of the related peptide hormones, such as GLP-1, glucagon, and secretin (Fig. 2b). Residues 33–42 of GIP, not seen in our structure, have been linked to the somatotropic properties of GIP and do not contribute to the insulinotropic action of the incretin (36). Binding of GIP to the GIPR ECD is accomplished by hydrophobic interactions between a distinct set of residues in the ligand and ECD that are either conserved or of similar nature in the ligands and ECDs of the glucagon receptor family (Fig. 2a). Although a similar situation is seen in the astressin–CRFR-2β ECD complex (26), where the C-terminal 14 aa of the antagonist (corresponding to CRF residues 27′–41′) bind to the ECD in an α-helical conformation with the burial of hydrophobic surfaces, superposition of the ECDs reveals a relative displacement of the two ligands by 5Å perpendicular to the helix axis (SI Fig. 9b). Furthermore, the hydrophobic amino acids of astressin involved in binding show no correspondence to those in GIP (Fig. 2b). This suggests that primary ligand recognition may differ significantly between various families of the class 2 GPCRs (15).
The predominant structure of the bound ligand is a single α-helix, spanning also N-terminal residues (Phe-6′–Met-14′) that are not in contact with the ECD and thus free to interact with other regions of the GIPR. Several studies have shown that N-terminal GIP residues confer significant affinity toward the full-length receptor and are essential for activation of the GIPR (37–40). It has also been reported that the biological activity can be influenced by modifying the degree of α-helicity of the ligand (41). In the present study, thermodynamic and spectroscopic analysis of the GIPR ECD–GIP1–42 complex formation provide evidence for structural reorganization during binding (SI Text). These data, in part, can be explained by assuming that the ligand adopts the observed helical conformation upon binding to the ECD. Structural investigations on isolated ligands of class 2 hormone receptors, including GIP, GLP-1, parathyroid hormone, glucagon, and calcitonin, reveal in each case an α-helical propensity that depends on the ambient solvent conditions (21, 42–45). In addition, almost all of these ligands possess a potential helix-capping residue (46) at position 5′ or 7′ (Thr-5′ in GIP1–42; Fig. 2b), which could aid initiation of helix formation of the receptor-bound peptide hormone. In support of this conjecture, the NMR structure of pituitary adenylate cyclase-activating polypeptide PACAP1–21 elucidated in complex with its cognate receptor shows an α-helical conformation of residues 8–21 of the bound hormone (47). In the CRFR-2β ECD-astressin structure (26), helix formation of the ligand is also observed upon binding the receptor, together with a structuring of residues 87–97 that are disordered in the free ECD.
Members of the glucagon receptor family are thought to follow a common two-domain mechanism of activation, in which ligand binding by the ECD initiates a series of events that result in subsequent signal transduction at the receptor (19). Our results provide a view of the ligand-binding step: (i) the peptide hormone is captured by hydrophobic residues in the C-terminal part of the ligand; (ii) binding induces adoption of an α-helical conformation of the ligand, restricting the conformation of the N-terminal residues; and (iii) the ECD presents the N terminus to receptor domains responsible for subsequent activation and signal transmission.
The proposed concomitant folding and binding of the peptide hormone upon interaction with the receptor provides additional possibilities for achieving ligand selectivity. Affinity for the receptor would be governed by a combination of the inherent ability for the peptide to form an α-helix, together with presentation of the correct residues to the ECD-binding site. Such a complex interaction mechanism might explain the differences in thermodynamic parameters observed for GIP1–42 and GLP-17–36 binding to the GIPR ECD (SI Text), despite their high degree of sequence similarity (40, 48).
Clearly, many questions remain open concerning the precise mechanisms of GPCR activation. The data presented in this paper provide a detailed atomic understanding of part of this vital process. Complementary structural and dynamic information of the hormone ligand–receptor domain complexes, and ultimately of the full-length receptor, will pave the way for the development of tailor-made drugs conferring high potency and specificity in diabetes therapy.
Methods
Protein expression, refolding, purification, peptide synthesis, and binding studies are described in the SI Text.
Crystallization and Structure Determination of the GIPR ECD–GIP Complex.
Solutions of GIPR ECD and GIP1–42 in buffer [10 mM Tris (pH 7.4)/150 mM NaCl/1 mM EDTA] were mixed at a 1:1 molar ratio and subsequently concentrated to achieve a concentration of 20 mg/ml. After addition of 9% (wt/vol) methyl-β-cyclodextrin, crystals grew by hanging drop vapor diffusion at 15°C in the presence of 100 mM Hepes (pH 7.2)/1 M potassium–sodium tartrate over the course of 4 weeks. Before measurement, crystals were rapidly transferred to a cryobuffer consisting of mother liquor with an additional 30% (vol/vol) glycerol.
A highly redundant data set was collected in house from a single crystal at −180°C diffracting up to 1.9 Å with Cu Kα radiation (λ = 1.5418 Å) by using a rotating-anode source (RA Micro 007; Rigaku/MSC, Tokyo, Japan) and an image plate detector (R-AXIS IV++; Rigaku/MSC) (see Table 1). Oscillation photographs were integrated, merged, and scaled by using DENZO and SCALEPACK (49). Phase determination was accomplished by using sulfur single-wavelength anomalous diffraction. Six of the nine putative sulfur atom positions (corresponding to three disulfide bonds and three methionine residues in the asymmetric unit) were determined with the program SHELXD (50) by using diffraction data up to 3.5 Å, providing a starting point for density modification (solvent content, 60.2%) and phase extension to 1.9 Å by using SHELXE. The resulting electron density was used for automatic main-chain tracing and side-chain docking carried out with the ARP/wARP software (51) and resulted in a model comprising two chains (88 residues of the GIPR ECD and 29 residues of GIP ligand), with the three disulfide bridges assigned (C46/C70, C61/C103, and C84/C118). Subsequent manual building of missing fragments and maximum-likelihood refinement cycles were performed by using programs COOT and REFMAC5 from the CCP4 suite (52).
Table 1.
Data collection and refinement statistics
| Data collection | |
| Space group | R32 |
| Cell dimensions | |
| a, Å | 84.01 |
| b, Å | 84.01 |
| c, Å | 180.95 |
| α, ° | 90 |
| β, ° | 90 |
| γ, ° | 120 |
| Resolution, Å | 1.9 |
| Rmerge, % | 9.7 (55.5) |
| I/σI | 27.3 (8.2) |
| Completeness, % | 99.5 (98.4) |
| Redundancy | 26.4 (25.4) |
| Refinement | |
| Resolution, Å | 67.6–1.9 |
| No. of reflections (work/test) | 18,643/1,004 |
| Rwork/Rfree | 16.6/18.3 |
| No. of atoms | |
| GIPR ECD | 763 |
| GIP | 265 |
| Methyl-β-cyclodextrin | 84 |
| Tartrate | 5 |
| Water | 133 |
| B factors, Å2 | |
| GIPR–ECD | 25.78 |
| GIP | 26.23 |
| Methyl-β-cyclodextrin | 34.95 |
| Tartrate | 16.67 |
| Water | 38.60 |
| rmsd | |
| Bond lengths, Å | 0.010 |
| Bond angles, ° | 1.17 |
The data set was collected from a single crystal. The data collection statistics treat Friedel mates independently. The values in parentheses are for the highest-resolution shell (1.97–1.90 Å).
The final model, consisting of residues 29–122 of the GIPR ECD receptor fragment, residues 1–32 of the ligand GIP1–42, one molecule of methyl-β-cyclodextrin, one half tartrate molecule, and 133 solvent molecules in the asymmetric unit, was refined at 1.9 Å resolution to an R factor of 16.6% and Rfree of 18.3% (Table 1). For some residues (GIP Ser-11′ and the GIPR ECD Arg-43), the electron density indicated multiple side-chain conformations in the crystal. The stereochemistry of the final model was assessed by using PROCHECK (52). Ramachandran statistics showed 94.4% of side chains in favored regions, 4.7% in allowed regions, 0.9% generously allowed, and none in disallowed regions. Figures were prepared by using PyMOL (DeLano Scientific LLC, Palo Alto, CA).
Supplementary Material
Acknowledgments
We thank Michael Wermann and René Wetzel for excellent technical assistance and our colleagues Alison Blakey, Jim McCormack, Martin Procter, and Christine Reynet (Prosidion, Melville, NY) for helpful advice and discussion. This work was supported by a grant from the Federal State of Sachsen–Anhalt, Germany (Wertschöpfung durch Proteine als Wirkstoffe und Werkzeuge).
Abbreviations
- CRF
corticotropin-releasing factor
- CRFR
murine CRF receptor
- ECD
extracellular domain
- GIP
glucose-dependent insulinotropic polypeptide or gastric inhibitory polypeptide
- GIPR
GIP receptor
- GLP
glucagon-like peptide
- GPCR
G protein-coupled receptor.
Footnotes
The authors declare no conflict of interest.
The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2QKH).
This article contains supporting information online at www.pnas.org/cgi/content/full/0706404104/DC1.
References
- 1.Fehmann HC, Göke R, Göke B. Endocr Rev. 1995;16:390–410. doi: 10.1210/edrv-16-3-390. [DOI] [PubMed] [Google Scholar]
- 2.Drucker DJ. Cell Metab. 2006;3:153–165. doi: 10.1016/j.cmet.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Nauck MA, Baller B, Meier JJ. Diabetes. 2004;53(Suppl 3):S190–S196. doi: 10.2337/diabetes.53.suppl_3.s190. [DOI] [PubMed] [Google Scholar]
- 4.O'Harte FP, Gray AM, Flatt PR. J Endocrinol. 1998;156:237–243. doi: 10.1677/joe.0.1560237. [DOI] [PubMed] [Google Scholar]
- 5.Xie D, Cheng H, Hamrick M, Zhong Q, Ding KH, Correa D, Williams S, Mulloy A, Bollag W, Bollag RJ, et al. Bone. 2005;37:759–769. doi: 10.1016/j.bone.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 6.Yip RG, Wolfe MM. Life Sci. 2000;66:91–103. doi: 10.1016/s0024-3205(99)00314-8. [DOI] [PubMed] [Google Scholar]
- 7.Miyawaki K, Yamada Y, Ban N, Ihara Y, Tsukiyama K, Zhou H, Fujimoto S, Oku A, Tsuda K, Toyokuni S, et al. Nat Med. 2002;8:738–742. doi: 10.1038/nm727. [DOI] [PubMed] [Google Scholar]
- 8.Meier JJ, Gallwitz B, Nauck MA. BioDrugs. 2003;17:93–102. doi: 10.2165/00063030-200317020-00002. [DOI] [PubMed] [Google Scholar]
- 9.Kieffer TJ, McIntosh CH, Pederson RA. Endocrinology. 1995;136:3585–3596. doi: 10.1210/endo.136.8.7628397. [DOI] [PubMed] [Google Scholar]
- 10.Deacon CF, Nauck MA, Toft-Nielsen M, Pridal L, Willms B, Holst JJ. Diabetes. 1995;44:1126–1131. doi: 10.2337/diab.44.9.1126. [DOI] [PubMed] [Google Scholar]
- 11.Holst JJ, Orskov C. Diabetes. 2004;53(Suppl 3):S197–S204. doi: 10.2337/diabetes.53.suppl_3.s197. [DOI] [PubMed] [Google Scholar]
- 12.Hinke SA, Lynn F, Ehses J, Pamir N, Manhart S, Kuhn-Wache K, Rosche F, Demuth H-U, Pederson RA, McIntosh CH. Adv Exp Med Biol. 2003;524:293–301. doi: 10.1007/0-306-47920-6_35. [DOI] [PubMed] [Google Scholar]
- 13.Gault VA, Flatt PR, O'Harte FP. Biochem Biophys Res Commun. 2003;308:207–213. doi: 10.1016/s0006-291x(03)01361-5. [DOI] [PubMed] [Google Scholar]
- 14.Demuth H-U, McIntosh CH, Pederson RA. Biochim Biophys Acta. 2005;1751:33–44. doi: 10.1016/j.bbapap.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 15.Foord SM, Bonner TI, Neubig RR, Rosser EM, Pin JP, Davenport AP, Spedding M, Harmar AJ. Pharmacol Rev. 2005;57:279–288. doi: 10.1124/pr.57.2.5. [DOI] [PubMed] [Google Scholar]
- 16.Usdin TB, Mezey E, Button DC, Brownstein MJ, Bonner TI. Endocrinology. 1993;133:2861–2870. doi: 10.1210/endo.133.6.8243312. [DOI] [PubMed] [Google Scholar]
- 17.Gelling RW, Wheeler MB, Xue J, Gyomorey S, Nian C, Pederson RA, McIntosh CH. Endocrinology. 1997;138:2640–2643. doi: 10.1210/endo.138.6.9104. [DOI] [PubMed] [Google Scholar]
- 18.Wheeler MB, Gelling RW, Hinke SA, Tu B, Pederson RA, Lynn F, Ehses J, McIntosh CH. J Biol Chem. 1999;274:24593–24601. doi: 10.1074/jbc.274.35.24593. [DOI] [PubMed] [Google Scholar]
- 19.Hoare SR. Drug Discov Today. 2005;10:417–427. doi: 10.1016/S1359-6446(05)03370-2. [DOI] [PubMed] [Google Scholar]
- 20.Alana I, Hewage CM, Malthouse JP, Parker JC, Gault VA, O'Harte FP. Biochem Biophys Res Commun. 2004;325:281–286. doi: 10.1016/j.bbrc.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 21.Alana I, Parker JC, Gault VA, Flatt PR, O'Harte FP, Malthouse JP, Hewage CM. J Biol Chem. 2006;281:16370–16376. doi: 10.1074/jbc.M510414200. [DOI] [PubMed] [Google Scholar]
- 22.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, et al. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 23.Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 24.Grace CR, Perrin MH, DiGruccio MR, Miller CL, Rivier JE, Vale WW, Riek R. Proc Natl Acad Sci USA. 2004;101:12836–12841. doi: 10.1073/pnas.0404702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.West AP, Jr, Llamas LL, Snow PM, Benzer S, Bjorkman PJ. Proc Natl Acad Sci USA. 2001;98:3744–3749. doi: 10.1073/pnas.051625298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grace CR, Perrin MH, Gulyas J, DiGruccio MR, Cantle JP, Rivier JE, Vale WW, Riek R. Proc Natl Acad Sci USA. 2007;104:4858–4863. doi: 10.1073/pnas.0700682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norman DG, Barlow PN, Baron M, Day AJ, Sim RB, Campbell ID. J Mol Biol. 1991;219:717–725. doi: 10.1016/0022-2836(91)90666-t. [DOI] [PubMed] [Google Scholar]
- 28.Lin SC, Lin CR, Gukovsky I, Lusis AJ, Sawchenko PE, Rosenfeld MG. Nature. 1993;364:208–213. doi: 10.1038/364208a0. [DOI] [PubMed] [Google Scholar]
- 29.Karaplis AC, He B, Nguyen MT, Young ID, Semeraro D, Ozawa H, Amizuka N. Endocrinology. 1998;139:5255–5258. doi: 10.1210/endo.139.12.6522. [DOI] [PubMed] [Google Scholar]
- 30.Grauschopf U, Lilie H, Honold K, Wozny M, Reusch D, Esswein A, Schafer W, Rucknagel KP, Rudolph R. Biochemistry. 2000;39:8878–8887. doi: 10.1021/bi0001426. [DOI] [PubMed] [Google Scholar]
- 31.Perrin MH, Fischer WH, Kunitake KS, Craig AG, Koerber SC, Cervini LA, Rivier JE, Groppe JC, Greenwald J, Moller NS, et al. J Biol Chem. 2001;276:31528–31534. doi: 10.1074/jbc.M101838200. [DOI] [PubMed] [Google Scholar]
- 32.Bazarsuren A, Grauschopf U, Wozny M, Reusch D, Hoffmann E, Schaefer W, Panzner S, Rudolph R. Biophys Chem. 2002;96:305–318. doi: 10.1016/s0301-4622(02)00023-6. [DOI] [PubMed] [Google Scholar]
- 33.Holtmann MH, Hadac EM, Miller LJ. J Biol Chem. 1995;270:14394–14398. doi: 10.1074/jbc.270.24.14394. [DOI] [PubMed] [Google Scholar]
- 34.Bergwitz C, Gardella TJ, Flannery MR, Potts JT, Jr, Kronenberg HM, Goldring SR, Juppner H. J Biol Chem. 1996;271:26469–26472. doi: 10.1074/jbc.271.43.26469. [DOI] [PubMed] [Google Scholar]
- 35.Runge S, Gram C, Brauner-Osborne H, Madsen K, Knudsen LB, Wulff BS. J Biol Chem. 2003;278:28005–28010. doi: 10.1074/jbc.M301085200. [DOI] [PubMed] [Google Scholar]
- 36.Morrow GW, Kieffer TJ, McIntosh CH, MacGillivray RT, Brown JC, St Pierre S, Pederson RA. Can J Physiol Pharmacol. 1996;74:65–72. [PubMed] [Google Scholar]
- 37.Hinke SA, Manhart S, Pamir N, Demuth H-U, Gelling W, Pederson RA, McIntosh CH. Biochim Biophys Acta. 2001;1547:143–155. doi: 10.1016/s0167-4838(01)00181-9. [DOI] [PubMed] [Google Scholar]
- 38.Gault VA, Parker JC, Harriott P, Flatt PR, O'Harte FP. J Endocrinol. 2002;175:525–533. doi: 10.1677/joe.0.1750525. [DOI] [PubMed] [Google Scholar]
- 39.Gault VA, Harriott P, Flatt PR, O'Harte FP. Biosci Rep. 2002;22:523–528. doi: 10.1023/a:1022073819618. [DOI] [PubMed] [Google Scholar]
- 40.Hinke SA, Manhart S, Speck M, Pederson RA, Demuth HU, McIntosh CH. Life Sci. 2004;75:1857–1870. doi: 10.1016/j.lfs.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 41.Manhart S, Hinke SA, McIntosh CH, Pederson RA, Demuth H-U. Biochemistry. 2003;42:3081–3088. doi: 10.1021/bi026868e. [DOI] [PubMed] [Google Scholar]
- 42.Thornton K, Gorenstein DG. Biochemistry. 1994;33:3532–3539. doi: 10.1021/bi00178a009. [DOI] [PubMed] [Google Scholar]
- 43.Jin L, Briggs SL, Chandrasekhar S, Chirgadze NY, Clawson DK, Schevitz RW, Smiley DL, Tashjian AH, Zhang F. J Biol Chem. 2000;275:27238–27244. doi: 10.1074/jbc.M001134200. [DOI] [PubMed] [Google Scholar]
- 44.Sasaki K, Dockerill S, Adamiak DA, Tickle IJ, Blundell T. Nature. 1975;257:751–757. doi: 10.1038/257751a0. [DOI] [PubMed] [Google Scholar]
- 45.Motta A, Andreotti G, Amodeo P, Strazzullo G, Castiglione Morelli MA. Proteins. 1998;32:314–323. [PubMed] [Google Scholar]
- 46.Dasgupta S, Bell JA. Int J Pept Protein Res. 1993;41:499–511. [PubMed] [Google Scholar]
- 47.Inooka H, Ohtaki T, Kitahara O, Ikegami T, Endo S, Kitada C, Ogi K, Onda H, Fujino M, Shirakawa M. Nat Struct Biol. 2001;8:161–165. doi: 10.1038/84159. [DOI] [PubMed] [Google Scholar]
- 48.Gallwitz B, Witt M, Morys-Wortmann C, Folsch UR, Schmidt WE. Regul Pept. 1996;63:17–22. doi: 10.1016/0167-0115(96)00019-5. [DOI] [PubMed] [Google Scholar]
- 49.Otwinowski Z, Minor W. Macromol Crystallogr A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 50.Schneider TR, Sheldrick GM. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 51.Perrakis A, Morris R, Lamzin VS. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 52.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.