Abstract
Eukaryotic genomes encode a zinc finger protein (ZPR1) with tandem ZPR1 domains. In response to growth stimuli, ZPR1 assembles into complexes with eukaryotic translation elongation factor 1A (eEF1A) and the survival motor neurons protein. To gain insight into the structural mechanisms underlying the essential function of ZPR1 in diverse organisms, we determined the crystal structure of a ZPR1 domain tandem and characterized the interaction with eEF1A. The ZPR1 domain consists of an elongation initiation factor 2-like zinc finger and a double-stranded β helix with a helical hairpin insertion. ZPR1 binds preferentially to GDP-bound eEF1A but does not directly influence the kinetics of nucleotide exchange or GTP hydrolysis. However, ZPR1 efficiently displaces the exchange factor eEF1Bα from preformed nucleotide-free complexes, suggesting that it may function as a negative regulator of eEF1A activation. Structure-based mutational and complementation analyses reveal a conserved binding epitope for eEF1A that is required for normal cell growth, proliferation, and cell cycle progression. Structural differences between the ZPR1 domains contribute to the observed functional divergence and provide evidence for distinct modalities of interaction with eEF1A and survival motor neuron complexes.
Keywords: growth factor receptor, structure, neurodegeneration, spinal muscular atrophy, cell cycle
ZPR1 was identified as a zinc finger (ZnF) protein that binds to the inactive form of the EGF receptor in quiescent cells and is released after activation (1). In proliferating cells treated with mitogens or other growth stimuli, ZPR1 binds directly to eukaryotic translation elongation factor 1A (eEF1A) (2), assembles into multiprotein complexes with the survival motor neurons (SMN) protein (3), and accumulates in subnuclear structures (gems and Cajal bodies) (1, 3, 4). Targeted disruption of the ZPR1 gene in yeast (2) and mice (5) indicates that ZPR1 is essential for viability in diverse eukaryotic organisms. Reduction of ZPR1 expression in mammalian cells by antisense or siRNA knockdown causes defects in transcription, prevents DNA synthesis, and results in an accumulation of cells in the G1 and G2 phases of the cell cycle (6).
The interaction of ZPR1 with SMN complexes is disrupted in cells derived from patients with spinal muscular atrophy (SMA) (3), an autosomal recessive disease linked to mutations in the telomeric copy of the SMN1 gene and characterized by progressive loss of spinal cord motor neurons during early childhood (7, 8). Interestingly, ZPR1 is expressed at low levels in patients with severe forms of SMA and at significantly higher levels in unaffected siblings with an identical SMN1 mutation (9). Consistent with this observation, Zpr1−/+ mice exhibit axonal defects and degeneration of spinal cord motor neurons (10). Recent studies suggest that the neurodegenerative phenotype of Zpr1−/+ mice, and potentially the severity of SMA also may be related to the interaction with eEF1A. Mammalian genomes contain two eEF1A genes, eEF1A1 and eEF1A2, which encode proteins that are 92% identical (11). Expression of eEF1A2 is restricted to terminally differentiated cells of skeletal muscle, heart, and brain and dominates in the postnatal period (12). Loss of eEF1A2 expression because of mutation of the eEF1A2 gene in wasted (wst) mice results in progressive motor neuron degeneration, muscle atrophy, paralysis, and death within 30 days after birth (12–14). The neurodegenerative phenotype of wst mice is similar to that of Zpr1−/+ mice (10) as well as mice with reduced expression of SMN (15, 16).
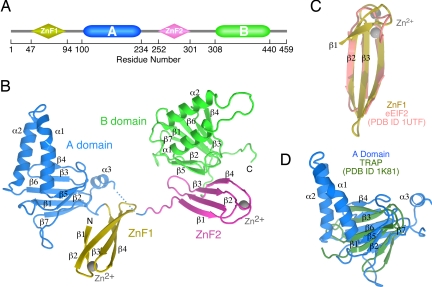
ZPR1 has a broadly conserved tandem architecture [Fig. 1A and supporting information (SI) Fig. 6] consisting of a duplicated module designated the “ZPR1 domain” in the SMART database (17). The ZPR1 domain is in fact a composite of two apparently modular domains: a C4-type Zn2+ finger and an ≈130-residue domain of unknown tertiary structure. Hereafter, we refer to the N- and C-terminal ZPR1 domains as the ZnF1-A domain and ZnF2-B domain modules, respectively. A homologous protein containing a single ZPR1 domain is present in Archae bacteria, suggesting that the tandem architecture arose early in the evolution of eukaryotic organisms through successive gene duplication and fusion events. Complementation analyses in yeast demonstrate that the ZnF2-B domain module is required for viability, whereas the ZnF1-A domain module is required for normal cell growth and proliferation (2). The interaction with the inactive EGF receptor is mediated by the ZnFs, whereas the binding sites for eEF1A and the SMN complex are located within the ZnF1-A domain and ZnF2-B domain modules, respectively (1–3).
Fig. 1.
Domain organization and tertiary structure of ZPR1. (A) Schematic illustration of the modular architecture of ZPR1, which consists of two homologous Zn2+ finger-A/B domain modules. (B) Ribbon representation of the tertiary structure of ZPR1. (C) Structural alignment of the first ZnF in ZPR1 with the ZnF in eEIF2. (D) Structural alignment of the β-helix in the ZPR1 A domain with the β-helix in the Trp RNA-binding protein TRAP.
Despite the essential role of ZPR1 in diverse cellular processes, including growth, proliferation, and neurodegeneration, little is known about the structural bases for interaction with receptors, eEF1A, and SMN complexes. Furthermore, it is unclear how ZPR1 binding depends on and/or regulates the GTPase cycle of eEF1A or whether the interaction with eEF1A is required for any of the cellular functions previously associated with the ZnF1-A domain region of ZPR1. To address these questions and gain insight into the underlying structural bases, we determined the crystal structure of a nearly full-length construct of ZPR1; identified conserved surfaces that likely mediate interactions with receptors and SMN complexes; characterized the nucleotide dependence of ZPR1 binding to eEF1A as well as the effect on the GTPase cycle; mapped the eEF1A binding epitope by mutational analysis; and assessed the effect of disrupting the interaction with eEF1A on cell growth, proliferation, and cell cycle progression.
Results
Tertiary Structure of a ZPR1 Domain Tandem.
Crystals of a ZPR1 domain tandem were obtained for a construct corresponding to residues 46–440 of mouse ZPR1. The structure of the selenomethionine-substituted form was determined by single-wavelength anomalous diffraction (SAD) at the f ″ maximum of the selenium edge and refined to 2.0 Å (SI Table 1 and SI Fig. 7). Continuous density was observed throughout the ZnF1-A domain and ZnF2-B domain modules; however, residues 235–244 in the variable linker region between the A domain and ZnF2 were disordered. One possibility depicted in Fig. 1B is that the disordered residues connect nearest neighbors, which are separated by distance of 13 Å. In this case, the relative orientations of ZnF1, the A domain, and the ZnF2-B domain module would be constrained by intermolecular contacts within a crystallographic dimer (SI Fig. 7B Left) as well as lattice contacts between crystallographic dimers. The only other possibility would require that the disordered linker region adopt a largely extended configuration to span the longer distance of 29 Å between next nearest neighbors (SI Fig. 7B Right). In this case, the relative orientation of the various domains would be constrained as described above, with the exception that the contact between the A domain and ZnF2 would be intramolecular, giving rise to a more organized arrangement of the A domain with respect to the ZnF2-B domain tandem. Given that full-length ZPR1 as well as the construct used for crystallization run as monomers on gel filtration even at concentrations approaching 1 mM (data not shown), the physiological significance of the crystallographic dimer, if any, is unclear. Likewise, we find no evidence to support an interaction between the A domain and the ZnF2-B domain module in solution by either coprecipitation or isothermal titration calorimetry (data not shown). On the other hand, as discussed below, residues in the A domain that contact ZnF2 in the crystallographic dimer are important for binding to eEF1A.
The ZnFs consist of a four-stranded antiparallel β-sheet stabilized by a zinc-binding site located at one end (Fig. 1B). A search for structural homologues by tertiary structural comparison using secondary structure matching (18) revealed a high degree of structural similarity with the three-stranded antiparallel ZnF of elongation initiation factor 2 (eIF2; rmsd, 1.5 Å) (Fig. 1C). The A and B domains share a common composite fold consisting of a helical hairpin inserted within a double-stranded, antiparallel β-helix (Fig. 1B). Apart from the hairpin insertion and differences in loops, the β-helix in the ZPR1 A and B domains is most similar to that in the Trp RNA-binding attenuation protein (rmsd, 3.2 Å) (Fig. 1D). In contrast to the Trp RNA-binding attenuation protein, for which the β-helix facilitates formation of a stable elevenfold symmetric ring, ZPR1 runs as a monomer on gel filtration (data not shown).
Although ZnF1 and ZnF2 have similar tertiary structures (rmsd, 1.6 Å; SI Fig. 8A), variations occur in the NH2-terminal strand, which is longer in ZnF2, and in the Zn2+ binding site, which is well ordered in ZnF1 but exhibits relatively high B-factors in ZnF2 (SI Fig. 7C). The largest differences in the A and B domains occur in the helical hairpins and at the C terminus, which adopts a helical conformation in the A domain compared with an extended conformation in the B domain (SI Fig. 8A). The differences in the helical hairpins reflect dissimilar amino acid sequences encoding the α1 helices (SI Fig. 8B) and contribute to the functional divergence of the A and B domains.
Potential Conserved Binding Surfaces for Receptors and SMN Complexes.
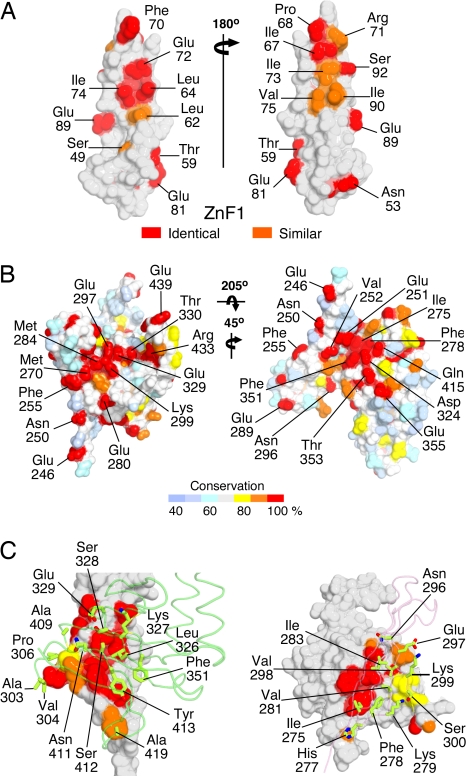
Biochemical studies have shown that: (i) both Zn2+ finger domains can bind independently to inactive tyrosine kinase receptors, including the EGF and PDGF receptors (1); (ii) the ZnF1-A domain module is sufficient for eEF1A binding (2); and (iii) the B domain is required for binding to the SMN complex (3). Although ZPR1 binds directly to receptors and eEF1A, the particular components of the SMN complex that mediate the interaction with ZPR1 have not yet been identified. Likewise, it is not yet known whether the A and B domains are sufficient for the interactions with eEF1A and the SMN complex or whether these interactions also require ZnF1 and ZnF2, respectively. In ZnF1 and ZnF2, the exposed residues that are conserved between the two ZnFs cluster in two patches (Fig. 2A and SI Fig. 9), one or both of which could contribute to the interaction with receptors. In the ZnF2-B domain module, the majority of exposed residues that are conserved among ZPR1 homologues cluster in two distinct patches located at the interface between ZnF2 and the B domain (Fig. 2B and SI Fig. 10). The conserved surface residues in both patches are distributed roughly evenly between the two domains, which adopt a specific organized tertiary structure with a substantial buried surface area of 1,000 Å2. The majority of residues that are buried at the interface between ZnF2 and the B domain are highly conserved (Fig. 2C).
Fig. 2.
Identification of putative conserved interaction epitopes in ZPR1 for receptors and SMN complexes. (A) Conservation of residues between ZnF1 and ZnF2 mapped to the surface of ZPR1. (B) Conservation of exposed residues in the ZnF2-B domain modules of ZPR1 homologues. (C) Conservation of residues buried in the interface between ZnF2 and the B domain of ZPR1 homologues.
ZPR1 Binds Preferentially to GDP-Bound eEF1A.
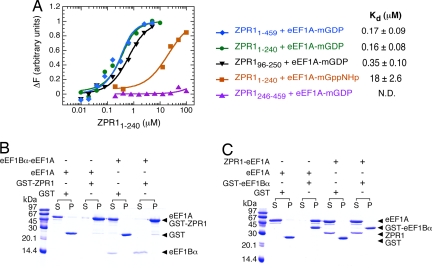
Although eEF1A binds directly to the ZnF1-A domain region of ZPR1 (2), the nucleotide dependence of the interaction and effect on the GTPase cycle of eEF1A have not been investigated. To further characterize the interaction of ZPR1 with eEF1A, we developed a quantitative binding assay that takes advantage of the environmentally sensitive emission of a methylanthraniloyl [2′-(3′)-bis-O-(N-methylanthraniloyl (mant)] fluorophore attached to the 2′/3′ hydroxyl groups of GDP or the nonhydrolyzeable GTP analogue guanyl-5′yl-imidodiphosphate. As shown in Fig. 3A, the emission intensity of mant–GDP-loaded eEF1A increases after titration with full-length ZPR1. The increase in emission intensity as a function of ZPR1 concentration has a hyperbolic shape characteristic of a binding isotherm and saturates in the high nanomolar range at a maximum increase of ≈10%. Titration with a truncated ZnF1-A domain construct (ZPR11–240) yields equivalent results, whereas a control titration with the corresponding ZnF2-B domain construct (ZPR1246–459), which does not coprecipitate eEF1A (2), has no significant effect on the emission intensity even at much higher concentrations. The data are well described by a simple 1:1 binding model, yielding indistinguishable dissociation constants (Kd) of 170 and 160 nM for full-length ZPR1 and ZPR11–240, respectively. The affinity of mant–GDP-loaded eEF1A for the A domain alone (ZPR196–250) is only 2-fold weaker, indicating that the binding site for eEF1A is located primarily on the A domain. Conversely, the affinity of mant–guanyl-5′yl-imidodiphosphate-loaded eEF1A for ZPR11–240 is two orders of magnitude lower. Finally, ZPR1 has only a modest effect (<2.5-fold decrease) on the affinity of eEF1A for nucleotides and no detectable effect on the kinetics of nucleotide exchange or GTP hydrolysis (SI Fig. 11). Thus, ZPR1 preferentially recognizes the GDP-bound form of eEF1A but does not directly influence the GTPase cycle.
Fig. 3.
Characterization of the interaction between ZPR1 and eEF1A. (A) Quantitative analysis of the binding of ZPR1 constructs to eEF1A loaded with mant–GDP or mant–guanyl-5′yl-imidodiphosphate. Solid lines represent fitted model functions for a hyperbolic binding isotherm. (B) Coprecipitation of eEF1A or the preformed eEF1A–eEF1Bα116–206 complex with GST–ZPR11–240. Note that eEF1A and GST-ZPR11–240 have identical mobility on SDS/PAGE. (C) Coprecipitation of eEF1A or the preformed eEF1A–His6 ZPR11–240 complex with GST–eEF1Bα116–206.
ZPR1 Competes with eEF1Bα for Binding to eEF1A.
GTP-bound eEF1A is required for recruitment of aminoacyl tRNAs to the A-site of the ribosome during translation elongation. GTP is loaded onto eEF1A through the catalytic action of eEF1B, an essential heterooligomeric complex that includes the GDP/GTP exchange factor eEF1Bα (19, 20). As shown in Fig. 3B, ZPR1 competes effectively with eEF1Bα for binding to eEF1A, displacing eEF1Bα from a preformed nucleotide-free eEF1A–eEF1Bα complex to form a stable eEF1A–ZPR1 complex. In contrast, eEF1Bα fails to displace ZPR1 from a preformed eEF1A (GDP)–ZPR1 complex (Fig. 3B), which is consistent with the ≈30-fold higher affinity of eEF1A (GDP) for ZPR1 compared with eEF1Bα. The simplest possibility is that ZPR1 binds to a site that overlaps with or occludes that for eEF1Bα. Alternatively, ZPR1 binding might cause a change in the conformation of eEF1A that would decrease the affinity for eEF1Bα. Although ZPR1 does not directly modulate the intrinsic nucleotide exchange kinetics of eEF1A (SI Fig. 11), the results presented here suggest that ZPR1–eEF1A complexes in cells would be resistant to nucleotide exchange catalyzed by eEF1B and might contribute to the regulation of translation elongation or otherwise play a distinct role in cell proliferation.
Mutational Analyses of eEF1A-Binding Epitope.
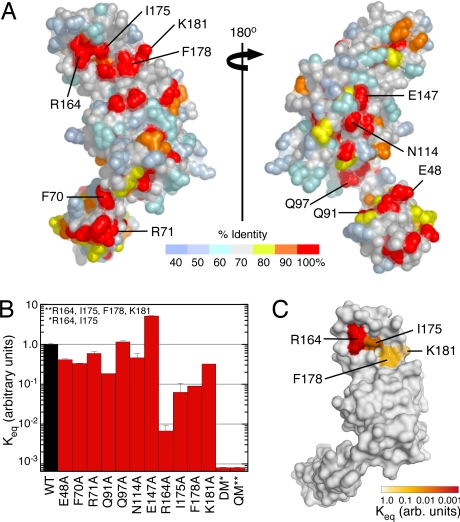
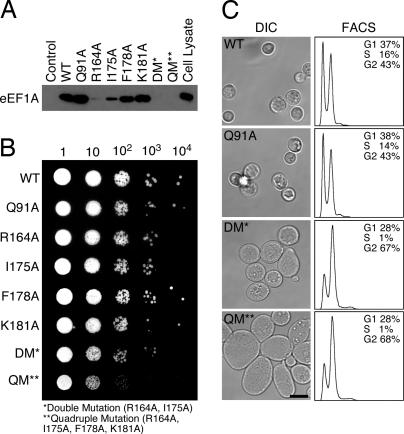
To locate the binding site for eEF1A and identify key interaction determinants, conserved surface residues in the ZnF1-A domain module were substituted with alanine, and the binding affinity for mant–GDP-loaded eEF1A was quantified as described above (Fig. 4A and SI Fig. 12). All of the mutants characterized expressed in a soluble form at levels comparable with the wild-type protein. As shown in Fig. 4B, the majority of substitutions had a relatively small effect (1- to 3-fold difference). Three single substitutions, R164A, I175A, and F178A, decreased the binding affinity by one to two orders of magnitude. Double (R164A, I175A) and quadruple (R164A, I175A, F178A, K181A) mutations reduced the binding affinity below the detection limit (>1,000-fold decrease). The four residues that are most critical for eEF1A binding (Arg-164, Ile-175, Phe-178, and Lys-181) are clustered on the surface of the helical hairpin in the A domain just above the β4-strand (Fig. 4C). It is likely that these residues are located within the binding epitope for eEF1A. Although not mutated, three other invariant resides (Glu-129 in the β3/β4 loop and Glu-133 and Pro-135 in β4) are clustered on a proximal surface that may also contribute to the eEF1A binding site. Between the adjacent clusters of conserved residues, the exposed edge of the β4-strand presents backbone NH and CO groups that could participate in polar interactions. Finally, as shown in Fig. 5A, the single, double, and quadruple alanine substitutions of Arg-164, Ile-175, Phe-178, and Lys-181 impair or disrupt the ability of 6xHis ZPR11–240 to coprecipitate endogenous eEF1A from HeLa cell lysates.
Fig. 4.
Structure-based mutational analysis of the ZPR1–eEF1A interaction. (A) Distribution of conserved residues in the ZnF1-A domain region of ZPR1. (B) Effect of ZPR1 mutations on the interaction with eEF1A (mean ± SD, n = 2). The expression, solubility, and stability of the wild-type and mutant proteins were similar. (C) Binding affinity of ZPR1 mutants for eEF1A (relative to wild-type ZPR1) mapped to the surface of the ZnF1-A domain module of ZPR1.
Fig. 5.
Mutations that disrupt eEF1A binding cause growth, proliferation, and cell cycle defects in yeast. (A) Immunoblot analysis of the effect of ZPR1 mutations on the binding of human eEF1A. The His6-tagged fusion proteins were purified, immobilized on Ni2+-NTA-agarose, and used for binding assays using HeLa cell lysates. Bound eEF1A was detected with anti-eEF1A antibody. (B) Growth of haploid yeast strains (zpr1::LEU2) complemented with plasmid expression vectors encoding wild-type and mutated cZPR1 proteins. Liquid cultures (0.1 OD600) were serially diluted 10-fold, spotted onto yeast extract peptone dextrose plates, and incubated at 30°C. (C) (Left) The morphology of haploid yeast strains (zpr1::LEU2) complemented with plasmid expression vectors encoding wild-type and mutated cZPR1 proteins was examined by differential interference confocal (DIC) microscopy. (Scale bar, 6.0 μm.) (Right) Also shown are the results of flow cytometric analyses using FACS of the haploid yeast strains expressing wild-type and mutated cZPR1.
Disruption of ZPR1–eEF1A Interaction Causes Growth Defects.
Although the ZnF1-A domain region of ZPR1 is required for normal cell growth, proliferation, and cell cycle progression, the functional role of the ZPR1–eEF1A complex is unclear. To assess the functional consequences of substitutions that disrupt the interaction with eEF1A, we investigated the effect of ZPR1 mutations on the viability of a Saccharomyces cerevisiae ZPR1 (cZPR1)-null strain. Haploid yeast with disrupted zpr1::LEU2 were complemented with wild-type cZPR1 (strain MY7) (2) and were used to examine the effect of cZPR1 mutations by plasmid shuffling (21) using 5′-fluoroorotic acid (2). As shown in Fig. 5B, both the wild-type and mutated cZPR1 proteins complement the viability of the cZPR1-null strain. This result is consistent with our earlier finding that the N-terminal region is dispensable for viability but is required for normal growth (2). Nevertheless, the small colony size observed for yeast complemented with cZPR1 containing double and in particular quadruple mutations suggests that the interaction with eEF1A likely plays an important role in cell growth and proliferation. Indeed, the interaction of ZPR1 with eEF1A is induced by extracellular growth stimuli (2). To further investigate this possibility, we compared the growth phenotype of the cZPR1 null strain complemented with plasmids encoding wild-type or mutated cZPR1 proteins. Control experiments demonstrated that cZPR1 mutations corresponding to substitutions that had only a small effect on eEF1A binding, such as Q91A, exhibited normal growth (Fig. 5B). In contrast, mutations corresponding to the R164A and I175A substitutions that reduced the affinity for eEF1A by 10- to 100-fold exhibited decreased cell growth (data not shown), whereas mutations corresponding to the double and quadruple amino acid substitutions that completely disrupted eEF1A binding caused a profound decrease (>20 fold) in growth compared with yeast expressing wild-type cZPR1 (Fig. 5B). Furthermore, yeast strains complemented with mutated cZPR1 containing double and quadruple amino acid substitutions grow in clusters and exhibit a marked (2- to 4-fold) increase in cell size (Fig. 5C Left).
ZPR1-eEF1A Interaction Is Required for Normal Cell Cycle Progression.
To determine the effect of disrupting the interaction with eEF1A on cell cycle progression, we examined the distribution of yeast cells in different phases of the cell cycle. Analysis of the DNA content by flow cytometry indicates that cZPR1 null cells complemented with wild-type or mutated cZPR1 proteins (e.g., Q91A) that do not disrupt interaction with eEF1A are distributed among the G1, S, and G2 phases of the cell cycle, corresponding to 1N and 2N DNA content, respectively (Fig. 5C Right). In contrast, cZPR1-null cells complemented with cZPR1 containing the double and quadruple mutations were distributed between the G1 and G2 phases of the cell cycle. The distribution of cells (>65% contain 2N DNA in the G2 phase) and larger size are indicative of G2/M phase accumulation. These results are consistent with the recent finding that reduced ZPR1 expression causes accumulation of mammalian cells in the G1 and G2 phases of the cell cycle (6) and support the conclusion that the interaction of ZPR1 with eEF1A is required for normal cell cycle progression.
Discussion
The crystal structure of a ZPR1 domain tandem provides insight into the tertiary structure and organization of the individual ZPR1 domain modules. There are few intramolecular contacts between ZnF1 and the A domain, indicative of a modular organization. Earlier studies established that either Zn2+ finger is sufficient for receptor binding, whereas eEF1A binding is disrupted by deletions in the A domain. Here, we find that the A domain is sufficient as well as necessary for eEF1A binding. Thus, both the structural and biochemical data support the conclusion that ZnF1 and the A domain function as independent binding modules. In contrast, ZnF2 and the B domain adopt a specific organized tertiary structure with a substantial interface composed of conserved residues. Furthermore, the conserved surface-exposed residues in the ZnF2-B domain module are clustered within two distinct patches that extend over both domains. These observations suggest the possibility of a concerted binding modality. Given that ZnF2 is sufficient for receptor binding, such a concerted binding modality would evidently involve the SMN complexes or other unknown partners. Further investigation awaits identification of the specific components of the SMN complex and/or other factors that mediate interactions with the ZnF2-B domain module. Finally, as a consequence of the disordered linker, we cannot unambiguously resolve whether the observed contact between the A domain and ZnF2 is crystallographic or intramolecular.
The apparently organized tertiary structure of the ZnF2-B domain module may also be related to the observation that cZPR1 nuclear export requires the cis–trans prolyl isomerase activity of cyclophilin A (22). Although not an essential gene, cyclophilin A exhibits specific synthetic lethality with three cZPR1 alleles that belong to the same complementation group. Two of the alleles involve a common single amino acid substitution in which an invariant aspartic acid is replaced by asparagine. The third allele encodes a single substitution in which an invariant glycine residue is replaced by aspartic acid. The corresponding aspartic acid and glycine residues in the ZPR1 structure are located at the C terminus of β7 in the B domain and at the C terminus of ZnF2, respectively. Although all of the proline residues in the ZPR1 crystal structure adopt the trans configuration, cis isomers might occur in vivo. Indeed, it has been proposed (23) that the highly conserved proline following the invariant aspartic acid might undergo a native state cis–trans isomerization switch analogous to the proline isomerization switch that modulates ligand recognition in the SH2 domain of the IL-2 tyrosine kinase (24). The proline residue in ZPR1 is located at the N terminus of the β6/β7-loop, the conformation of which would necessarily be altered by cis–trans isomerization. Given that the β6/β7-loop occupies the core of the intramolecular interface between ZnF2 and the B domain, it is likely that the latter interface also would be affected. Finally, the only obvious connection between the invariant aspartic acid in the B domain and the invariant glycine in ZnF2 is their proximity to the ZnF2-B domain interface. Thus, local structural perturbations induced by the asparagine and aspartic acid substitutions may indirectly affect the cis–trans equilibrium of the conserved proline in the β6/β7-loop through destabilization of the ZnF2-B domain interface.
Consistent with broad evolutionary conservation in eukaryotic organisms, ZPR1 is essential for viability in yeast and mice (2, 5). Whereas the ZnF2-B domain module is essential for viability in yeast, the ZnF1-A domain is required for normal growth, proliferation, and cell cycle progression (2). It also had been observed that eEF1A binding is disrupted by deletion of a highly conserved 20-residue region in the A domain. This deletion, which eliminates the β6/β7-loop as well as most of the β6- and β7-strands, would be expected to compromise the tertiary structure of the β-helix. Therefore, the loss of eEF1A binding is likely to be a secondary consequence of a structural defect. In the present study, we have used a structure-based mutational analysis of conserved surface residues to identify critical determinants in the eEF1A binding epitope. Individual substitutions involving four conserved residues in the helical hairpin substantially reduced the affinity for eEF1A without affecting the expression levels or solubility of the protein. Furthermore, complementation experiments demonstrated that double and quadruple substitutions that abolish eEF1A binding restore the viability of the ZPR1-null strain but exhibit growth, proliferation, and cell cycle defects similar to those previously reported for constructs that delete the ZnF1-A domain module (2).
Although these results provide strong evidence that the interaction with eEF1A is required for normal growth, proliferation, and cell cycle progression, the underlying mechanism of action remains to be determined. The observation that ZPR1 binds preferentially to the GDP-loaded form of eEF1A but has a negligible effect on nucleotide affinity, as well as the kinetics of GDP/GTP exchange and GTP hydrolysis, supports the conclusion that ZPR1 does not function within the context of the conventional GTPase paradigm as an effector, guanine nucleotide exchange factor, or GTPase activating protein. Nevertheless, the ability to efficiently displace eEF1Bα from preformed complexes with eEF1A suggests that ZPR1 might indirectly regulate the GTPase cycle of eEF1A by interfering with eEF1B-catalyzed activation. Thus, after release from receptors in mitogen/nutrient-stimulated cells, ZPR1 may act as a negative regulator of eEF1A. It also is possible that ZPR1–eEF1A complexes have an eEF1Bα-independent function in translation and/or other cellular processes that require eEF1A, such as regulation of the actin cytoskeleton (25, 26).
The studies presented here provide insights into the structural and mechanistic bases underlying the essential function of the evolutionarily conserved ZPR1 protein. We anticipate that the ZPR1 structure will continue to provide a valuable framework for structure–function analyses of the mechanisms through which ZPR1 regulates fundamental cellular processes in both dividing as well as terminally differentiated cells, including neurons.
Materials and Methods
Constructs.
Constructs of mouse ZPR1 (residues 1–459, 46–440, 1–240, 96–250, and 246–459) and S. cerevisiae eEF1Bα (residues 1–206 and 116–206) were amplified and subcloned into pGEX-6P1 (Amersham Pharmacia Biotech, Piscataway, NJ) for expression as GST fusions or into a modified pET15b vector, which incorporates an N-terminal His6 tag (MGHHHHHHGS). Site-specific mutations were generated with the QuikChange kit (Stratagene, La Jolla, CA). All constructs were verified by sequencing the entire coding region.
Crystallization and Structure Determination.
Selenomethionine-substituted mouse ZPR1 (residues 46–440) was crystallized at 18°C in hanging drops containing 20 mg/ml protein in 14% PEG 6000/0.2 M ammonium sulfate/0.1 M Tris, pH 8.0/10% glycerol/2 mM Tris(2-carboxyethyl)phosphine hydrochloride. Crystals appeared within 3–4 days and grew to maximum dimensions of 0.3 × 0.2 × 0.2 mm over 3 weeks. The crystals are in the C-centered monoclinic space group C2 with cell constants a = 55.7 Å, b = 142.6 Å, and c = 67.2 Å, with one molecule in the asymmetric unit. Crystals were transferred to a cryostabilizer solution (4.0 M lithium formate), flash-frozen in liquid propane, and maintained at 100 K in a nitrogen cryostream. Diffraction data at the peak wavelength of the selenium edge were collected at the National Synchrotron Light Source, beamline X25; processed with Denzo (HKL Research, Charlottesville, VA); and scaled with Scalepack (HKL Research) (27). The structure was determined by single-wavelength anomalous diffraction. Twelve Se sites were identified independently by Patterson methods with SOLVE (28) and direct methods with SnB (29) using the Bijvoet differences at the f ″ maximum. The heavy atom model was refined with SHARP (30). Single-wavelength anomalous diffraction phases were improved by solvent flipping with SOLOMON (31). A σA-weighted Fourier summation yielded an interpretable map with continuous density for the main chain and most side chains. An initial model generated by ARP/wARP (European Molecular Biology Laboratory, Heidelberg, Germany) (32) was completed by manual building using O (33). The model was refined in several iterative cycles by using ARP/wARP and Refmac5 (32). The refined model includes residues 47–234 and 245–440. Additional information related to the structure determination and refinement is compiled in SI Table 1. Structural Figures were rendered with PyMol.
For additional information, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Terri Goss Kinzy (University of Massachusetts Medical School, Worchester, MA) for a cDNA encoding eEF1Bα, Dr. Craig Peterson (Robert Wood Johnson Medical School, Piscataway, NJ) for the cy333 yeast strain, and Drs. C. Robert Matthews and Xiaojing Pan for assistance with stopped-flow measurements and GTP hydrolysis, respectively. This work was supported in part by research grants from the Families of SMA and the Muscular Dystrophy Association (to L.G.) and the National Institute of Neurological Diseases and Stroke (to R.J.D.). R.J.D. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- eEF1A
eukaryotic translation elongation factor 1A
- SMN
survival motor neurons
- SMA
spinal muscular atrophy
- ZnF
zinc finger
- mant
2′-(3′)-bis-O-(N-methylanthraniloyl).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2QKD).
This article contains supporting information online at www.pnas.org/cgi/content/full/0704915104/DC1.
References
- 1.Galcheva-Gargova Z, Konstantinov KN, Wu IH, Klier FG, Barrett T, Davis RJ. Science. 1996;272:1797–1802. doi: 10.1126/science.272.5269.1797. [DOI] [PubMed] [Google Scholar]
- 2.Gangwani L, Mikrut M, Galcheva-Gargova Z, Davis RJ. J Cell Biol. 1998;143:1471–1484. doi: 10.1083/jcb.143.6.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gangwani L, Mikrut M, Theroux S, Sharma M, Davis RJ. Nat Cell Biol. 2001;3:376–383. doi: 10.1038/35070059. [DOI] [PubMed] [Google Scholar]
- 4.Galcheva-Gargova Z, Gangwani L, Konstantinov KN, Mikrut M, Theroux SJ, Enoch T, Davis RJ. Mol Biol Cell. 1998;9:2963–2971. doi: 10.1091/mbc.9.10.2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gangwani L, Flavell RA, Davis RJ. Mol Cell Biol. 2005;25:2744–2756. doi: 10.1128/MCB.25.7.2744-2756.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gangwani L. J Biol Chem. 2006;281:40330–40340. doi: 10.1074/jbc.M608165200. [DOI] [PubMed] [Google Scholar]
- 7.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 8.Monani UR. Neuron. 2005;48:885–896. doi: 10.1016/j.neuron.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Helmken C, Hofmann Y, Schoenen F, Oprea G, Raschke H, Rudnik-Schoneborn S, Zerres K, Wirth B. Hum Genet. 2003;114:11–21. doi: 10.1007/s00439-003-1025-2. [DOI] [PubMed] [Google Scholar]
- 10.Doran B, Gherbesi N, Hendricks G, Flavell RA, Davis RJ, Gangwani L. Proc Natl Acad Sci USA. 2006;103:7471–7475. doi: 10.1073/pnas.0602057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lund A, Knudsen SM, Vissing H, Clark B, Tommerup N. Genomics. 1996;36:359–361. doi: 10.1006/geno.1996.0475. [DOI] [PubMed] [Google Scholar]
- 12.Chambers DM, Peters J, Abbott CM. Proc Natl Acad Sci USA. 1998;95:4463–4468. doi: 10.1073/pnas.95.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knudsen SM, Frydenberg J, Clark BF, Leffers H. Eur J Biochem. 1993;215:549–554. doi: 10.1111/j.1432-1033.1993.tb18064.x. [DOI] [PubMed] [Google Scholar]
- 14.Newbery HJ, Gillingwater TH, Dharmasaroja P, Peters J, Wharton SB, Thomson D, Ribchester RR, Abbott CM. J Neuropathol Exp Neurol. 2005;64:295–303. doi: 10.1093/jnen/64.4.295. [DOI] [PubMed] [Google Scholar]
- 15.Jablonka S, Schrank B, Kralewski M, Rossoll W, Sendtner M. Hum Mol Genet. 2000;9:341–346. doi: 10.1093/hmg/9.3.341. [DOI] [PubMed] [Google Scholar]
- 16.Le TT, Pham LT, Butchbach ME, Zhang HL, Monani UR, Coovert DD, Gavrilina TO, Xing L, Bassell GJ, Burghes AH. Hum Mol Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 17.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krissinel E, Henrick K. Acta Crystallogr D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 19.Janssen GM, Moller W. J Biol Chem. 1988;263:1773–1778. [PubMed] [Google Scholar]
- 20.Le Sourd F, Boulben S, Le Bouffant R, Cormier P, Morales J, Belle R, Mulner-Lorillon O. Biochim Biophys Acta. 2006;1759:13–31. doi: 10.1016/j.bbaexp.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Sikorski RS, Boeke JD. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 22.Ansari H, Greco G, Luban J. Mol Cell Biol. 2002;22:6993–7003. doi: 10.1128/MCB.22.20.6993-7003.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreotti AH. Biochemistry. 2003;42:9515–9524. doi: 10.1021/bi0350710. [DOI] [PubMed] [Google Scholar]
- 24.Mallis RJ, Brazin KN, Fulton DB, Andreotti AH. Nat Struct Biol. 2002;9:900–905. doi: 10.1038/nsb864. [DOI] [PubMed] [Google Scholar]
- 25.Gross SR, Kinzy TG. Nat Struct Mol Biol. 2005;12:772–778. doi: 10.1038/nsmb979. [DOI] [PubMed] [Google Scholar]
- 26.Gross SR, Kinzy TG. Mol Cell Biol. 2007;27:1974–1989. doi: 10.1128/MCB.00832-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Terwilliger TC, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weeks CM, Miller R. J Appl Crystallogr. 1999;32:120–124. [Google Scholar]
- 30.La Fortelle E, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 31.Abrahams JP, Leslie AG. Acta Crystallogr D. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 32.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 33.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47(2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.