Abstract
The TodS/TodT two-component system controls expression of the toluene dioxygenase (TOD) pathway for the metabolism of toluene in Pseudomonas putida DOT-T1E. TodS is a sensor kinase that ultimately controls tod gene expression through its cognate response regulator, TodT. We used isothermal titration calorimetry to study the binding of different compounds to TodS and related these findings to their capacity to induce gene expression in vivo. Agonistic compounds bound to TodS and induced gene expression in vivo. Toluene was a powerful agonist, but ortho-substitutions of toluene reduced or abolished in vivo responses, although TodS recognized o-xylene with high affinity. These compounds were called antagonists. We show that agonists and antagonists compete for binding to TodS both in vitro and in vivo. The failure of antagonists to induce gene expression in vivo correlated with their inability to stimulate TodS autophosphorylation in vitro. We propose intramolecular TodS signal transmission, not molecular recognition of compounds by TodS, to be the phenomenon that determines whether a given compound will lead to activation of expression of the tod genes. Molecular modeling identified residues F46, I74, F79, and I114 as being potentially involved in the binding of effector molecules. Alanine substitution mutants of these residues reduced affinities (2- to 345-fold) for both agonistic and antagonistic compounds. Our data indicate that determining the inhibitory activity of antagonists is a potentially fruitful alternative to design specific two-component system inhibitors for the development of new drugs to inhibit processes regulated by two-component systems.
Keywords: histidine kinases, isothermal titration calorimetry, Pseudomonas, two-component systems, aromatic hydrocarbons
The most widely distributed type of transcriptional control in prokaryotic microorganisms exposed to environmental cues is the two-component regulatory system (TCS) (1). In fact, genes encoding TCSs are present in almost all bacteria and typically represent ≤1% of their genomes (1, 2). TCSs are also present, although to a lesser extent, in archaea and eukaryotes such as fungi, slime molds, and plants (3, 4). These systems are often made up of a histidine protein kinase (HPK) and a response regulator (RR). The recognition of physical or chemical signals at the input domain of HPKs typically initiates modulation of its autophosphorylation activity. The phosphate of the HPK is transferred to the RR, triggering alterations in the functional properties of its output domain and eventually leading to the stimulation of transcription. The HPK and their RR together comprise a large and diverse group. Their diversity is particularly pronounced in the input domain of HPKs and the output domain of RR, which were shown to belong to many different protein families (1). This variety ensures that HPKs recognize many different signals and RRs are involved in the regulation of different cellular processes (5–7).
Although many TCSs have been studied so far, the primary environmental signals recognized by HPKs remain unknown for most TCSs (8). This lack of information is often a consequence of difficulties in expressing and purifying sensor kinases, which are membrane-bound in most cases (8). Furthermore, in recent years, an increasing number of TCSs have been able to recognize and respond to structurally different agonists. This finding is exemplified by PhoQ, which was initially reported to recognize bivalent cations (9), although recent studies showed that PhoQ also binds cationic antimicrobial peptides (10, 11) by the same binding site for both types of agonists (12). Signal-recognition HPKs are generally assumed to trigger a regulatory response. However, in the light of data showing that some HPKs recognize a wide range of chemical signals, it is of interest to establish whether the binding of a ligand at the sensor domain of an HPK is enough to trigger this kind of response.
We have attempted to shed light on these issues by investigating the TodS HPK in Pseudomonas putida, which forms a TCS with the TodT RR (13, 14). This TCS controls the expression of the toluene dioxygenase (TOD) pathway (15) responsible for the metabolism of toluene into Krebs cycle intermediates (16). The catabolic genes of the TOD pathway form an operon that is transcribed from the PtodX promotor (13–16).
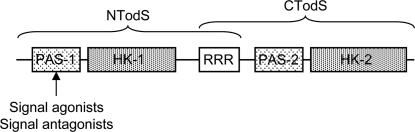
The architecture of the 108-kDa HPK TodS is atypical and complex. TodS has two supradomains, each containing a periodic circadian-Ah receptor single-minded protein (PAS) sensor domain and a histidine kinase domain (Fig. 1), which are separated by an RR receiver domain. TodS lacks transmembrane regions and is thus likely to be located in the cytosol (8, 13). The N-terminal PAS domain of TodS binds toluene with high affinity (KD ≈ 700 nM) (14). This binding increases the basal autophosphorylation rate of TodS, leading to transphosphorylation of TodT and transcription stimulation from PtodX (14). TodS seems to belong to a subfamily of HPKs involved in the control of catabolic pathways for the degradation of solvents. For example, TmoS (82% identity with TodS) controls toluene degradation by the T4MO pathway in Pseudomonas mendocina (17), TutC (49% identity) regulates the anaerobic degradation of toluene in Thaurea sp. strain T1 (18), and StyS (41% identity) in Pseudomonas sp. strain Y2 is involved in styrene degradation (19).
Fig. 1.
Domain organization of TodS. The NTodS and CTodS recombinant proteins are indicated. Agonists and antagonists bind to the PAS-1 domain. PAS, PAS-type sensor domain; HK, histidine kinase domain; RRR, response regulator receiver domain.
In the present study, we used isothermal titration calorimetry (ITC) to measure the thermodynamic parameters for the binding of a wide range of different compounds to purified TodS. We then related these data to the capacity of these compounds to induce gene expression in vivo and to their ability to stimulate TodS autophosphorylation activity in vitro. Almost all of the mono- and bicyclic aromatics we analyzed bound to TodS. However, only some of these molecules induced gene expression in vivo, and this ability was related to their capacity to increase TodS autophosphorylation in vitro.
Results
Comparison of the in vitro Ligand Affinities of TodS and the Capacity of the Compounds to Induce Gene Expression in vivo.
Purified TodS was subjected to microcalorimetric titration with different compounds to determine the effector profile in vitro. In parallel, the potential of these compounds to stimulate gene expression in vivo was determined by measuring the β-gal activity with a PtodX::lacZ fusion. The in vitro binding parameters and β-gal measurements are listed in Table 1.
Table 1.
in vitro thermodynamic parameters for the binding of different hydrocarbons to TodS and their capacity to induce expression from PtodX in vivo
| Compound | Binding parameters to TodS in vitro |
β-Gal expression, Miller units | |
|---|---|---|---|
| KD, μM | ΔH, kcal/mol | ||
| Benzene and singly substituted benzene derivatives | |||
| Benzene | 0.76 ± 0.1 | −11.0 ± 0.2 | 79 ± 3* |
| Toluene | 0.69 ± 0.1 | −5.5 ± 0.1 | 333 ± 55* |
| Ethylbenzene | 3.1 ± 0.1 | −3.6 ± 0.1 | 15 ± 2* |
| Propylbenzene | 18 ± 2 | −2.7 ± 0.4 | 2 ± 1* |
| Butylbenzene | 81 ± 4 | −5.4 ± 0.1 | 2 ± 1* |
| Styrene | 0.58 ± 0.1 | −12.4 ± 0.3 | 129 ± 42* |
| Nitrobenzene | 6.6 ± 0.1 | −7.3 ± 0.9 | 141 ± 20* |
| Chlorobenzene | 1.2 ± 0.1 | −9.9 ± 0.4 | 92 ± 5* |
| Fluorobenzene | 1.2 ± 0.1 | −5.3 ± 0.1 | 111 ± 22 |
| Disubstituted benzene derivatives | |||
| o-Xylene | 0.58 ± 0.1 | −9.4 ± 0.1 | 2 ± 1* |
| m-Xylene | 1.2 ± 0.1 | −9.0 ± 0.1 | 34 ± 2* |
| p-Xylene | 0.76 ± 0.1 | −10.1 ± 0.1 | 88 ± 18* |
| o-Chlorotoluene | 0.73 ± 0.1 | −6.7 ± 0.2 | 2 ± 1† |
| m-Chlorotoluene | 8.3 ± 0.1 | −11.0 ± 2 | 186 ± 12* |
| p-Chlorotoluene | 0.29 ± 0.1 | −8.9 ± 0.1 | 146 ± 32* |
| o-Toluidine | 3.2 ± 0.2 | −28.7 ± 2.5 | 17 ± 4* |
| m-Toluidine | 8.6 ± 0.7 | −7.3 ± 0.8 | 52 ± 16* |
| p-Toluidine | 11 ± 1 | −20.7 ± 3 | 63 ± 14* |
| Catechol | 16 ± 4 | −1.6 ± 0.6 | 13 ± 2 |
| TMB isomers | |||
| 1,2,3-TMB | 0.58 ± 0.1 | −4.2 ± 0.1 | 0* |
| 1,2,4-TMB | 1.9 ± 0.2 | −2.6 ± 0.1 | 0* |
| 1,3,5-TMB | 133 ± 20 | −3.2 ± 1.6 | 0* |
| Biaromatics | |||
| 1-naphthol | 2.1 ± 0.1 | −12.4 ± 0,3 | 0 |
| 2,3-dihydroxynaphthalene | 7.5 ± 0.3 | −15.1 ± 0.3 | 0 |
*Data were initially reported in ref. 13.
†No binding to TodS or transcription stimulation was observed for 1-hexanol, cyclohexane, benzamide, benzoate, anthracene, or naringenine.
ITC experiments revealed that TodS bound benzene with an affinity as high as that of toluene (Table 1 and Fig. 2A), but not cyclohexane (Fig. 2A) and aliphatic compounds such as 1-hexanol. We then investigated the influence of single substitutions on the aromatic ring [supporting information (SI) Fig. 6]. Styrene was found to be recognized by TodS with the highest affinity (KD = 580 ± 70 nM) among benzene derivatives carrying an aliphatic substitution. Ethyl, propyl, and butyl substitutions were recognized with lower affinity in comparison to benzene by a factor of 4, 23, and 110, respectively (Table 1). Ethylbenzene was found to be a weak inducer in vivo, and propyl- and butylbenzene did not induce expression from PtodX in vivo (Table 1). Nitro-, chloro-, and fluorobenzene bound to TodS with affinities in the low micromolar range and were found to be potent inducers of expression from PtodX (Table 1). Benzamide and benzoate were not bound by TodS, which is consistent with their failure to induce gene expression in vivo.
Fig. 2.
Microcalorimetric titration of TodS with different hydrocarbons. (A) (Upper) Heat changes for the titration of 12 to 13.2 μM TodS with 4.8-μl aliquots of 750 μM cyclohexane and 1.6-μl aliquots of 750 μM benzene and 500 μM 1-naphthol. (Lower) Integrated and corrected peak areas for the titration with benzene and 1-naphthol. (B) Heat changes (Upper) and integrated peak areas (Lower) for the titration of 10–12 μM TodS with 1.6-μl aliquots of the three xylenes. For clarity, raw titration data have been shifted arbitrarily on the y axis. Derived thermodynamic data are given in Table 1.
Taking into consideration that toluene is an efficient inducer in vivo, the next set of experiments was aimed at evaluating the impact of toluene substitutions on the binding parameters. Initial experiments were carried out with the three xylene isomers (methyl-substituted toluene). Surprisingly, all three xylenes bound to TodS with similar affinities (Fig. 2B and Table 1), but only m- and p-xylene were powerful inducers in vivo. In contrast, o-xylene failed to induce gene expression (Table 1).
To verify whether the data recorded for the three xylenes represented a general pattern in toluene derivatives, TodS was titrated with the three chlorotoluenes. Like xylenes, all three chlorotoluenes were bound by TodS, but only m- and p-chlorotoluene showed in vivo activity, whereas o-chlorotoluene was inactive in vivo (Table 1). To further verify these findings, we investigated the interaction of the three toluidines (amino toluenes). Again, o-toluidine was found to be a significantly weaker inducer in vivo than the other two isomers (Table 1), although it bound to TodS more tightly than m- or p-toluidine. Our interpretation of the combined data for xylenes, chlorotoluenes, and toluidines is that o-substitutions either fully abolished (xylene, chlorotoluene) or reduced (toluidine) the in vivo response without exerting a significant impact on binding affinity. This apparent lack of correlation between the affinity measured in vitro and expression studies in vivo was further confirmed by the fact that the second-best inducer in vivo, m-chlorotoluene, was recognized by TodS with modest in vitro affinity (KD = 8.3 ± 0.1 μM).
Because of the unexpected results for ortho-substituted compounds, we explored the influence of polysubstituted benzene derivatives on the binding parameters. The three trimethylbenzene (TMB) isomers bound to TodS but did not induce expression in vivo, which supports the notion that binding in vitro does not automatically translate into induction by a compound in vivo.
Because only monoaromatic compounds have been reported thus far to induce the TOD pathway (14), we carried out experiments to define the upper size limit of TodS ligands. The biaromatic compounds 1-naphthol (Fig. 2A) and 2,3-dihydroxynaphthalene bound to TodS with KD values of 2.1 ± 0.1 and 7.5 ± 0.3 μM (Table 1), respectively, but were equally inactive in vivo. No binding of the polyaromatic hydrocarbon anthracene to TodS was observed.
In short, the compounds under study can be classified into three groups. The first group is made up of compounds such as cyclohexane or benzoate, which do not bind to TodS in vitro and do not activate gene expression in vivo. The second group, referred to here as agonists, consists of compounds that bind to TodS and induce expression from PtodX. The third group, called antagonists, includes chemicals that bind to TodS in vitro but exhibit no in vivo activity.
Agonists and Antagonists Bind to the Same PAS Domain.
We then studied the mode of action of antagonists, among which o-xylene, o-chlorotoluene, and 1,2,3-TMB were chosen as representatives because they were recognized by TodS with similar affinities to toluene and in the range of 580–730 nM (Table 1). TodS is predicted to contain two sensor domains of the PAS type (20). One domain is located at the N-terminal end of TodS (amino acids 31–168 according to Pfam) (21), whereas the second domain is found at the C-terminal half (amino acids 613–725). We recently showed that toluene only binds to the N-terminal PAS sensor domain (14), a finding that raised the possibility that agonists and antagonists bind to different sensor domains, causing the differences observed in vivo.
To test this possibility, we conducted sequential ITC experiments. In an initial series of experiments, TodS was saturated with toluene, and this complex was titrated with o-xylene, o-chlorotoluene, or 1,2,3-TMB. Binding of the second ligand would suggest that the binding sites of both ligands did not overlap. If the second ligand failed to bind to the TodS–toluene complex, binding sites would either be the same or overlap. In all cases, none of the three antagonists bound to the TodS–toluene complex (data not shown). When these experiments were repeated in reverse order (i.e., the titration of TodS complexed to o-xylene, o-chlorotoluene, or 1,2,3-TMB with toluene), the same results were obtained (data not shown).
To corroborate these findings, the NTodS and CTodS recombinant fragments (Fig. 1) were titrated with the three antagonists. No binding of o-xylene, o-chlorotoluene, or 1,2,3-TMB to CTodS was observed, whereas all three antagonists bound to NTodS with a 2- to 7-fold lower affinity compared with the binding to full TodS (SI Fig. 7 and SI Table 3). This reduction in affinity was similar to that observed for toluene (14).
Antagonists Inhibit Toluene-Mediated Up-Regulation of Gene Expression in vivo.
ITC data suggested that agonists and antagonists most likely compete for the same binding site in vitro. Gene expression studies with PtodX::lacZ and the todST genes in pMIR66 were carried out to determine whether this competition was observed in vivo (Fig. 3). In parallel experiments, the β-gal activity in cultures induced with toluene was compared with cultures to which o-xylene, o-chlorotoluene, or 1,2,3-TMB was added before toluene (Fig. 3). When antagonists and toluene were added at equimolar concentrations, gene expression was reduced by approximately half (Fig. 3). When a 5-fold molar excess of antagonist was used, gene expression was reduced by a factor of ≈4 (Fig. 3). When antagonists recognized by TodS with low affinity were used, higher concentrations of these compounds (≈10-fold) were necessary to observe similar inhibition effects. Therefore, these results are consistent with competition between toluene and each of the antagonists for binding to TodS.
Fig. 3.
Inhibition of toluene-mediated induction of PtodX by o-xylene, o-chlorotoluene, and 1,2,3-TMB. Eleven 25-ml cultures of P. putida DOT-T1E harboring pMIR66 (containing todST) and pMIR77 (containing a PtodX::lacZ fusion) were grown in LB to a turbidity of 0.2 at 660 nm. Then, six cultures were exposed to o-xylene, o-chlorotoluene, or 1,2,3-TMB (asterisk) at 0.3 mM (hatched bars) or 1.5 mM (dotted bars). When the cultures reached a turbidity of 0.5, buffer (control culture), o-xylene, o-chlorotoluene, or 1,2,3-TMB (all at a final concentration of 0.3 mM) was added to four cultures without addition, and 0.3 mM toluene was added to the seven remaining cultures. The β-gal activity was measured 2 h later. Data are the means and corresponding standard errors derived from at least three independent assays, each done in triplicate.
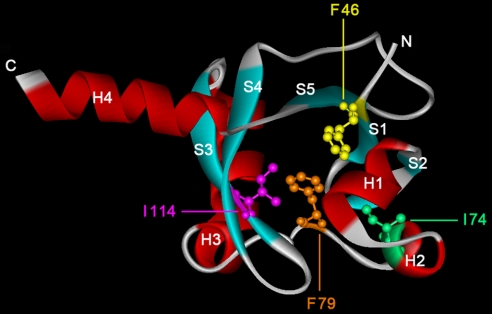
Identification of the Agonist/Antagonist Binding Site of TodS.
To identify the amino acids involved in ligand binding at the N-terminal PAS domain of TodS, we generated a 3D model of this domain. With the help of the DALI algorithm (22), we aligned this model to the structure of the effector-binding domain of the HPK CitA (Protein Data Bank ID code 1POZ), which has been solved in complex with its agonist citrate (23). The model showed a hydrophobic cavity at a position analogous to the citrate-binding site of CitA, which we hypothesized to be the ligand-binding pocket of TodS. Four amino acids located in this pocket (F46, I74, F79, and I114) were postulated to be involved in effector binding (Fig. 4). All four amino acids were conserved in an alignment of the HPKs TodS, TmoS (17), StyS (19), and TutC (18). These three regulators responded to similar agonists such as toluene (TodS, TmoS, and TutC) and styrene (StyS) (14, 17–19). To investigate the potential role of these residues in effector binding, we prepared and characterized the corresponding alanine replacement mutants. All four purified mutants were subjected to ITC assays with agonists (toluene and benzene), as well as with the o-xylene and o-chlorotoluene antagonists (Table 2). The affinity of mutants F46A, I74A, and I114A for agonists and antagonists was found to be reduced 2- to 12-fold compared with the wild-type protein (Table 2), which supports the hypothesis that agonists and antagonists probably bind to the same site at TodS. The titration of mutant F79A with a 4- to 5-mM ligand solution only gave rise to small heat changes for binding, suggesting that this mutant had lost the ability to bind agonist and antagonist molecules. We then tested the mutants in vivo by replacing the wild-type todS allele in pMIR66 with the mutant variants and measuring induction from PtodX::lacZ as β-gal. As expected, TodSF79A did not stimulate transcription in vivo with any of the tested agonists and antagonists. Also, as expected, none of the other three TodS mutant variants responded to o-xylene and o-chlorotoluene, although they did induce transcription with toluene and benzene (data not shown).
Fig. 4.
The 3D model of the N-terminal signal sensor domain of TodS. Secondary structure elements are indicated by S (strand) and H (helix). The amino acids in the proposed effector-binding site, which were replaced with alanine residues, are shown in ball-and-stick mode. This model contains amino acids 43–164 of TodS.
Table 2.
Thermodynamic parameters for the titration of TodS mutants with agonists and antagonists
| Mutants* and compounds | KD, μM | ΔH, kcal/mol | KDmut/KDwt |
|---|---|---|---|
| F46A | |||
| Benzene | 10.0 ± 2 | −8.20 ± 2.2 | 12 |
| Toluene | 3.03 ± 0.2 | −14.6 ± 0.7 | 4.3 |
| o-Chlorotoluene | 3.48 ± 0.3 | −13.5 ± 1 | 4.5 |
| o-Xylene | 2.46 ± 0.1 | −5.66 ± 0.1 | 4.2 |
| I74A | |||
| Benzene | 5.81 ± 1.3 | −2.52 ± 0.8 | 7.7 |
| Toluene | 8.62 ± 1.0 | −5.49 ± 1.3 | 12 |
| o-Chlorotoluene | 2.29 ± 0.7 | −2.02 ± 0.5 | 3.1 |
| o-Xylene | 3.20 ± 0.6 | −3.52 ± 0.8 | 12 |
| F79A | |||
| Benzene | >200† | — | >263 |
| Toluene | >200† | — | >290 |
| o-Chlorotoluene | >200† | — | >274 |
| o-Xylene | >200† | — | >345 |
| I114A | |||
| Benzene | 2.32 ± 0.2 | −5.14 ± 0.3 | 3.0 |
| Toluene | 1.63 ± 0.3 | −9.75 ± 1.2 | 2.4 |
| o-Chlorotoluene | 2.33 ± 0.1 | −5.14 ± 0.3 | 3.2 |
| o-Xylene | 1.27 ± 0.2 | −20.0 ± 9.3 | 2.2 |
KDmut/KDwt shows the ratio of the determined KD to the corresponding value obtained for the wild-type (wt) protein.
*The location of the amino acids in the 3D model of the N-terminal PAS domain of TodS is shown in Fig. 4.
†A titration of this mutant with a series of 12-μl injections of 4–5 mM ligand solution gave rise to minor heats of binding, which did not permit data analysis. The KD is estimated to be >200 μM.
Antagonist Compounds Do Not Stimulate TodS Autophosphorylation in Vitro.
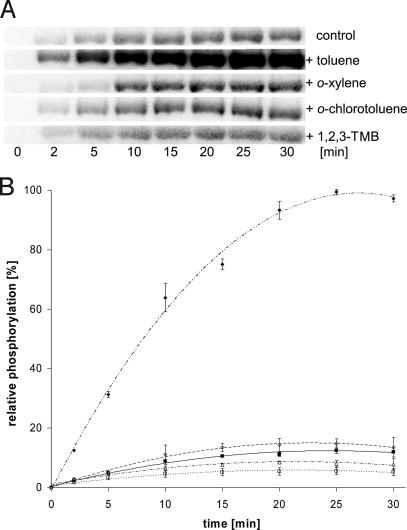
It often has been found that one of the critical steps in the mechanism of TCS's action is the autophosphorylation of HPK in response to ligands and the subsequent transphosphorylation of the RR. To shed further light on antagonists in vitro, we first performed autophosphorylation studies with TodS by using [γ-32P]ATP in the absence and presence of various concentrations of agonists and antagonists (SI Fig. 8). After electrophoresis, [32P]TodS was quantified densitometrically and plotted against time. From the linear curve fit of the data, we calculated the apparent rates of phosphorylation. In the presence of o-xylene, 1,2,3-TMB, or o-chlorotoluene, autophosphorylation rates were similar to the basal rate (0.8–1.4 ± 0.2) (Fig. 5). In contrast, the autophosphorylation rate in the presence of toluene was found to be 7.3 ± 0.1 times faster than in the absence of toluene. Other agonists such as fluorobenzene and ethylbenezene were also found to stimulate the rate of autophosphorylation between 3- and 5-fold (data not shown). This finding indicates that failure of the antagonists to induce gene expression in vivo correlates with their inability to promote autophosphorylation in vitro. We also tested autophosphorylation activity in mutant strain F79A in response to toluene. TodSF79A was found to have a relative basal autophosphorylation rate that did not increase in response to toluene (SI Fig. 9). We reasoned that antagonists could inhibit autophosphorylation if present in mixtures containing toluene. Similar assays to the ones described earlier were conducted with equimolecular concentrations of toluene and o-xylene. Under these conditions, the autophosphorylation rate was about half of that with toluene alone (data not shown).
Fig. 5.
Modulation of TodS autophosphorylation activity in vitro. TodS autophosphorylation activity with [32P]ATP was measured in the absence of signal (control) and in the presence of 100 μM toluene, o-xylene, 1,2,3-TMB, or o-chlorotoluene. (A) SDS/PAGE of TodS in the absence and presence of different ligands. (B) Densitometric analysis of the data in A. Data are the means of three independent assays. Linear fit of the points at 2.5 and 10 min was used to calculate relative rates of autophosphorylation. Filled square, control; open triangle, o-chlorotoluene; open square, trimethylbenzene; open diamond, o-xylene; filled diamond, toluene.
Discussion
Both the Recognition of Agonists by TodS and Intra-TodS Signal Transmission Determine Whether a Compound Stimulates Expression from PtodX.
This study was designed to answer the following question: Is ligand binding to HPKs enough to set up the regulatory cascade in response to environmental cues? To answer this question, we used the TCS TodS/TodT involved in toluene metabolism, and we analyzed recognition by TodS of molecules with different structural features. TodS only bound compounds containing at least one aromatic ring, although the presence of a benzene ring did not guarantee binding, as evidenced by the lack of recognition of benzamide and benzoate by TodS. Among the compounds recognized by TodS, we distinguished agonist and antagonist molecules, and three lines of evidence are consistent with the notion that agonists and antagonists share the same binding site: (i) agonists and antagonists compete for binding at TodS in vitro and in vivo, (ii) agonists and antagonists bind to NTodS but not to CTodS, and (iii) amino acid substitutions at the N-terminal PAS-binding domain of TodS reduce binding of agonists and antagonists in a similar fashion. Although all molecules appear to bind to the same site, the binding of agonists is likely to generate a signal that is transmitted through conformational changes to the kinase domain of the HKP, which in turn stimulates autophosphorylation activity. In contrast, the binding of antagonists seems to keep TodS in an inactive conformational state and thus has no significant effect on TodS autophosphorylation (Fig. 5).
Of the 26 mono- and biaromatic compounds analyzed here, 24 were found to bind to TodS in vitro (SI Fig. 6). Surprisingly, only 14 of these 24 compounds were able to increase gene expression in vivo. It is generally assumed that binding of ligands at the sensor domain of a histidine kinase is the dominant prerequisite for the regulatory activity of a TCS. However, the situation in TodS is different because the capacity of a molecule to stimulate gene expression from PtodX is not primarily determined by molecular recognition of the chemical by TodS, but rather by its ability to trigger the phosphorylation cascade.
Our findings with regard to the TodS sensor kinase system show clear parallels with the repressor TtgV (24, 25), a one-component regulator system that, like TodS, exerts its action in response to mono- and biaromatic compounds. We have shown that both types of compounds activate TtgV-mediated transcription with different efficiencies (26), and it was concluded that the effect of mono- and bicyclic compounds on TtgV intramolecular signal transmission is different from one to the other. It remains to be established whether such differences in the mechanism of intramolecular signal transmission are a general feature of regulatory proteins with a broad effector profile.
Identification of Amino Acids Involved in Signal Binding.
A 3D model of the N-terminal sensor domain was generated and aligned to the structure of the sensor domain of CitA. A hydrophobic pocket in an analogous position to the citrate-binding site of CitA was hypothesized to be the effector-binding site of TodS. We generated alanine replacement mutants of F46, I74, I114, and F79 located in this pocket. Three of these mutants (F46A, I74A, and I114A) had a 2- to 12-fold lower affinity for agonists and antagonists, whereas the affinity of the F79A protein was reduced by a factor of >250 with the tested ligands (Table 2). These data suggest that the proposed hydrophobic cavity is the common binding pocket for agonists and antagonists in TodS. The aromatic side chain of F79 in TodS appears to play a central role in the recognition of a broad series of ligands, and we propose effector recognition to be dominated by pi-pi stacking interactions of effectors and protein residues, a common feature in the molecular recognition of aromatic ligands by proteins (27, 28).
Is High-Affinity Binding of Agonists a Typical Feature of Cytosolic Sensor Kinases?
Ulrich et al. (1) analyzed the presence of one- and two-component regulatory systems in complete bacterial genomes and concluded that entirely cytosolic one-component systems are older, in evolutionary terms, than TCSs. The physiological reason leading to the emergence of TCSs, which frequently have an HPK with a periplasmic sensor domain, was their capacity to regulate cellular processes in response to extracytosolic signals. This capacity is of particular importance for soil bacteria such as Pseudomonas, which live in a rapidly changing environment. A subfamily of HPK has been identified that is entirely located in the cytosol and recognizes effectors (8). The physiological role of these cytosolic HPKs however, is unknown. The absence of transmembrane regions (13) and high solubility in the absence of detergents (14) are evidence that TodS belongs to this subfamily. Available data on the interaction between agonists and HPKs were obtained primarily with kinases containing a periplasmic sensor domain. The affinity of HPKs for their cognate agonists was found to be in the micromolar range, as exemplified by CitA, which has a KD of 5.5 μM for citrate (29), and NarX and PhoQ, which have apparent affinities of ≈35 μM for nitrate and ≈300 μM for Mg2+ ions, respectively (30, 31). Here we show that TodS binds agonists with much higher affinities than those just mentioned. It remains to be established whether high-affinity ligand binding is a typical feature of cytosolic HPK. This information might help to solve the enigma surrounding the factors that led to the evolution of cytosolic HPKs.
Ortho-Substitutions of Toluene: Converting an Agonist Molecule into an Antagonist.
In vivo gene-expression studies showed toluene to be one of the most efficient agonists. The substitution of toluene at the ortho position by a methyl, chloro, or amino group either abolished or reduced the inducing capacity without producing a significant impact on affinity (Table 1). Thus, we have identified a structural modification that converts a powerful agonist into an antagonist. These antagonists behave as competitive inhibitors of the regulator activity mediated by agonists. This finding is relevant to the development of inhibitors of TCS such as PhoP/Q of S. typhimurium (32, 33), which were shown to be important virulence factors (6). A large number of inhibitors of different TCSs have been developed in vitro (34). However, when tested for their antimicrobial potential in vivo, most of these compounds were not selective for signal transduction pathways, but exerted their effects through multiple mechanisms of action (35), making them unsuitable for any clinical application. Most of these inhibitors were developed by random screening, and most of them were shown to bind to the kinase domain, which might explain their restricted selectivity (36). In this study, we demonstrate that an HPK can be inhibited by structural analogues that behave as antagonists. Targeting the effector-binding domain of an HPK by specifically exploring the inhibitory activity of antagonists could thus be a productive line of research for the more rational development of TCS inhibitors.
In summary, we demonstrate that agonists and antagonists bind to the same domain of TodS with similar affinities. The binding of agonist molecules stimulates the autophosphorylation activity of TodS, whereas antagonists do not modulate this activity. The binding of antagonists inhibits agonist-mediated transcriptional activation, which is relevant to the development of more specific inhibitors of TCS. X-ray crystallographic studies of TodS in the presence of different ligands are currently in progress to establish the molecular basis for the differential action of agonists and antagonists.
Materials and Methods
Generation of a 3D Model of the N-Terminal PAS Domain of TodS.
A homology model of the N-terminal PAS domain of TodS (amino acids 31–168) was generated with the aid of 3D-JIGSAW software (37) by using the structure of FixL of Rhizobium meliloti (38) (residues 122–251; Protein Data Bank ID code 1D06) as a template. The model was validated by calculating the solvation profile with SolvX software (38), which gave a satisfactory score of −17.5. The DALI algorithm (22) was used to align this model with the structure of the sensor domain of CitA (23).
Site-Directed Mutagenesis.
Plasmids encoding TodS mutants F46A, I74A, F79A, and I114A were prepared with the QuikChange site-directed mutagenesis kit according to the manufacturer's instructions (Stratagene, La Jolla, CA), with pTodS or pMIR66 (14, 17) as a template. For each mutant, two 30-mer complementary oligonucleotides were designed that contained the desired mismatch in the center. The pTodS derivatives were used for protein expression. The entire todS coding region and the flanking regions of the resulting plasmids were verified by DNA sequencing.
ITC.
TodS and its mutant variants, NTodS and CTodS, were purified as described (14). ITC assays were performed with freshly purified protein by using a VP-microcalorimeter (Microcal, Amherst, MA) (39). Protein was dialyzed into ITC buffer [50 mM Tris·HCl/200 mM KCl/2 mM MgCl2/2 mM DTT/0.1 mM EDTA (pH 7.5)]. Typically, the TodS concentration was in the range of 12–15 μM. Ligand solutions were prepared as described (14). Typically, TodS was titrated with 1.6-μl aliquots of ligand solution. If no binding heats were detected, the experiment was repeated with larger injection volumes (≤12 μl). The mean enthalpies measured from the injection of agonists/antagonists into the buffer were subtracted from raw titration data before data analysis with ORIGIN software.
Expression of PtodX in vivo.
The β-gal measurements reported in Table 1 were obtained as described previously (14). The measurements reported in Fig. 3 were obtained with P. putida DOT-T1E bearing pMIR66 (17) (containing todST) or its mutant derivatives and pMIR77 (containing a PtodX::lacZ fusion). Cultures were grown on LB supplemented with 100 μg/ml gentamycin and 10 μg/ml tetracycline in the absence and presence of agonists and antagonists. The β-gal activity was determined in permeabilized whole cells as described (25).
in vitro Autophosphorylation Assay.
Assays were carried out as described (14), except that 300 pmol of TodS and 4 μCi of [32P]ATP were used.
Supplementary Material
Acknowledgments
We thank C. Lorente and M. Fandila for secretarial assistance, A. Hurtado for DNA sequencing, A. J. Molina-Henares for statistical analysis, G. Rivas and C. Alfonso for ultracentrifugation analyses, C. Daniels for critically reading the manuscript, and K. Shashok for improving the English in the manuscript. This work was supported by Ministry of Science and Education (Spain) Grant BIO-2006-05668, European Union Grant Sysmo GEN-2006-27750-CS-5B, and Junta de Andalucía Grant CVI-344.
Abbreviations
- HPK
histidine protein kinase
- ITC
isothermal titration calorimetry
- PAS
periodic circadian-Ah receptor single-minded protein
- RR
response regulator
- TCS
two-component system
- TMB
trimethylbenzene
- TOD
toluene dioxygenase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701547104/DC1.
References
- 1.Ulrich E, Koonin EV, Zhulin IB. Trends Microbiol. 2005;13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashby MK. FEMS Microbiol Lett. 2004;231:277–281. doi: 10.1016/S0378-1097(04)00004-7. [DOI] [PubMed] [Google Scholar]
- 3.Stock AM, Robinson VL, Goudreau PN. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 4.Koretke KK, Lupas A, Warren PV, Rosenberg M, Brown JR. Mol Biol Evol. 2000;17:1956–1970. doi: 10.1093/oxfordjournals.molbev.a026297. [DOI] [PubMed] [Google Scholar]
- 5.Galperin MY. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calva E, Oropeza R. Microbial Ecol. 2006;51:166–176. doi: 10.1007/s00248-005-0087-1. [DOI] [PubMed] [Google Scholar]
- 7.Zhou L, Lei X-H, Bocher BR, Wanner BL. J Bacteriol. 2003;185:4956–4972. doi: 10.1128/JB.185.16.4956-4972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mascher T, Helmann JD, Unden G. Microbiol Mol Biol Rev. 2006;70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.García Véscovi E, Soncini F, Groisman EA. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 10.Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, Fang FC, Miller SI. Mol Microbiol. 2003;50:219–230. doi: 10.1046/j.1365-2958.2003.03675.x. [DOI] [PubMed] [Google Scholar]
- 11.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 12.Cho US, Bader MW, Amaya MF, Daley ME, Klevit RE, Miller SI, Xu W. J Mol Biol. 2006;356:1193–1206. doi: 10.1016/j.jmb.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 13.Lau PCK, Wang Y, Patel A, Labbé D, Bergeron H, Brousseau R, Konishi Y, Rawlings M. Proc Natl Acad Sci USA. 1997;94:1453–1458. doi: 10.1073/pnas.94.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacal J, Busch A, Guazzaroni M, Krell T, Ramos JL. Proc Natl Acad Sci USA. 2006;103:8191–8196. doi: 10.1073/pnas.0602902103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zylstra GJ, Gibson DT. J Biol Chem. 1989;264:14940–14941. [PubMed] [Google Scholar]
- 16.Mosqueda G, Ramos-González MI, Ramos JL. Gene. 1999;232:69–76. doi: 10.1016/s0378-1119(99)00113-4. [DOI] [PubMed] [Google Scholar]
- 17.Ramos-González MI, Olson M, Gatenby AA, Mosqueda G, Manzanera M, Campos MJ, Víchez S, Ramos JL. J Bacteriol. 2002;184:7062–7067. doi: 10.1128/JB.184.24.7062-7067.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coschigano PW, Young LY. Appl Environ Microbiol. 1997;63:652–660. doi: 10.1128/aem.63.2.652-660.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velasco A, Alonso S, García JL, Perera J, Díaz E. J Bacteriol. 1998;180:1063–1071. doi: 10.1128/jb.180.5.1063-1071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vreede J, van der Horst MA, Hellingwerf KJ, Crielaard W, van Aalten DM. J Biol Chem. 2003;278:18434–18439. doi: 10.1074/jbc.M301701200. [DOI] [PubMed] [Google Scholar]
- 21.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dietmann S, Park J, Notredame C, Heger A, Lappe M, Holm L. Nucleic Acids Res. 2001;29:55–57. doi: 10.1093/nar/29.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reinelt S, Hofmann E, Gerharz T, Bott M, Madden DR. J Biol Chem. 2003;278:39189–39196. doi: 10.1074/jbc.M305864200. [DOI] [PubMed] [Google Scholar]
- 24.Rojas A, Segura A, Guazzaroni M-E, Terán W, Hurtado A, Gallegos MT, Ramos JL. J Bacteriol. 2003;185:4755–4763. doi: 10.1128/JB.185.16.4755-4763.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guazzaroni ME, Krell T, Felipe A, Ruiz R, Meng C, Zhang X, Gallegos MT, Ramos JL. J Biol Chem. 2005;280:20887–20893. doi: 10.1074/jbc.M500783200. [DOI] [PubMed] [Google Scholar]
- 26.Guazzaroni M-E, Gallegos M-T, Ramos JL, Krell T. J Biol Chem. 2007;282:16308–16316. doi: 10.1074/jbc.M610032200. [DOI] [PubMed] [Google Scholar]
- 27.Boehr DD, Farley AR, Wright GD, Cox JR. Chem Biol. 2002;9:1209–1217. doi: 10.1016/s1074-5521(02)00245-4. [DOI] [PubMed] [Google Scholar]
- 28.Calero G, Wilson KF, Ly T, Rios-Steiner JL, Clardy JC, Cerione RA. Nat Struct Biol. 2002;9:912–917. doi: 10.1038/nsb874. [DOI] [PubMed] [Google Scholar]
- 29.Gerharz T, Reinelt S, Kaspar S, Scapozza L, Bott M. Biochemistry. 2003;42:5917–5924. doi: 10.1021/bi0340595. [DOI] [PubMed] [Google Scholar]
- 30.Lee AI, Delgado A, Gunsalus RP. J Bacteriol. 1999;181:5309–5316. doi: 10.1128/jb.181.17.5309-5316.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lesley JA, Waldburger CD. J Biol Chem. 2001;276:30827–30833. doi: 10.1074/jbc.M104262200. [DOI] [PubMed] [Google Scholar]
- 32.Foussard M, Cabantous S, Pedelacq J, Guillet V, Tranier S, Mourey L, Birck C, Samama J. Microbes Infect. 2001;3:417–424. doi: 10.1016/s1286-4579(01)01390-9. [DOI] [PubMed] [Google Scholar]
- 33.Groisman EA, Mouslim C. Nat Rev Microbiol. 2006;4:705–709. doi: 10.1038/nrmicro1478. [DOI] [PubMed] [Google Scholar]
- 34.Matsushita M, Janda KD. Biorg Med Chem. 2002;10:855–867. doi: 10.1016/s0968-0896(01)00355-8. [DOI] [PubMed] [Google Scholar]
- 35.Stephensen K, Hoch JA. Curr Med Chem. 2004;11:765–773. doi: 10.2174/0929867043455765. [DOI] [PubMed] [Google Scholar]
- 36.Stephensen K, Yamaguchi Y, Hoch JA. J Biol Chem. 2000;275:38900–38904. doi: 10.1074/jbc.M006633200. [DOI] [PubMed] [Google Scholar]
- 37.Bates PA, Kelley LA, MacCallum RM, Sternberg MJE. Proteins S. 2001;5:39–46. doi: 10.1002/prot.1168. [DOI] [PubMed] [Google Scholar]
- 38.Miyatake H, Mukai M, Park S-Y, Adachi S, Tamura K, Nakamura H, Nakamura K, Tsuchiya T, Iizuka T, Shiro Y. J Mol Biol. 2000;301:415–431. doi: 10.1006/jmbi.2000.3954. [DOI] [PubMed] [Google Scholar]
- 39.Holm L, Sander C. J Mol Biol. 1992;225:93–105. doi: 10.1016/0022-2836(92)91028-n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.