Abstract
The recent rapid growth of protein sequence databases is outpacing the capacity of researchers to biochemically and structurally characterize new proteins. Accordingly, new methods for recognition of motifs and homologies in protein primary sequences may be useful in determining how these proteins might function. We have applied such a method, an iterative learning algorithm, to analyze possible coiled coil domains in histidine kinase receptors. The potential coiled coils have not yet been structurally characterized in any histidine kinase, and they appear outside previously noted kinase homology regions. The learning algorithm uses a combination of established sequence patterns in known coiled coil proteins and histidine kinase sequence data to learn to recognize efficiently this coiled coil-like motif in the histidine kinases. The common appearance of the structural motif in a functionally important part of the receptors suggests hypotheses for kinase regulation and signal transduction.
Bacteria are remarkably adept at sensing and adjusting to the conditions in their immediate environments. Frequently, such processes are mediated by two-component regulatory systems, consisting of a histidine kinase sensor and an aspartic acid receiver (1). Most often, the histidine kinase is a transmembrane receptor, with a periplasmic sensory domain and a cytoplasmic kinase domain, and the receiver is a transcription factor. Sensory inputs are coupled to gene expression by regulation of kinase activity. Processes mediated by more elaborate two-component systems include bacterial chemotaxis (2), in which chemoeffectors are sensed by a complex of transmembrane chemoreceptor, CheW, and the cytoplasmic histidine kinase CheA. The receiver protein CheY carries the signal to the flagellar switch. Although most two-component systems so far discovered are bacterial, they occur in archea and eukaryotes as well (3, 4).
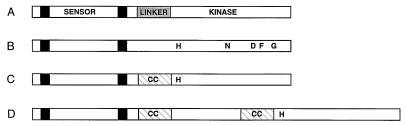
In those cases examined, histidine kinases have been found to be dimers with autophosphorylation occurring in trans between monomers (5). However, sensory regulation of this process is not yet well understood. No structures have been reported for any of the kinases, although a crystal structure of the C-terminal phosphotransfer domain of ArcB (6) and NMR structures of the phosphotransfer (7, 8) and CheY-binding (9, 10) domains of CheA have been determined. In a number of systems, genetic studies identify a segment of the cytoplasmic domain as critical for signal transmission (11–14). This region, termed the “linker” domain, links the final transmembrane domain to the kinase domain (Fig. 1A).
Figure 1.
Common domain arrangements in histidine kinases. (A) Simple transmembrane histidine kinases with two membrane-spanning segments (black boxes). The sensor component is periplasmic, and a cytoplasmic “linker” domain precedes the receptor kinase domain. (B) Approximate spacing of conserved, short sequence motifs characteristic of histidine kinases (5). The H block motif includes the histidine autophosphorylation site. (C) Location of many high likelihood coiled coils in the cytoplasmic linker domain before the H block motif. (D) Common domain pattern in longer kinases. Two coiled coils are predicted, occurring before the H block motif and shortly after the final transmembrane segment.
Histidine kinases are a highly diverse group of proteins, ranging from ≈350 to well over 2,000 amino acids in length. However, they can be recognized by several short blocks of sequence similarity within an ≈250-residue core kinase domain (5). These have been designated the H , N , D , F , and G blocks (5) (or H , N , G1 , F , and G2 blocks) (15) (Fig. 1B). The sequences within the five blocks are somewhat variable, as is their spacing within the protein. Occasionally, a block is missing in a particular kinase sequence. Other than these consensus motifs, no general sequence features have been reported for this family of receptors. However, an intriguing similarity recently has been noted between histidine kinase consensus motifs and dimeric ATP-binding domains of topoisomerases (16).
We find that the cytoplasmic linker domains of many histidine kinases appear to contain coiled coils, when evaluated with algorithms developed to identify these motifs within primary sequences.** It is intriguing that many of the predicted coiled coils would precede directly the histidine autophosphorylation sites, terminating at the conserved proline within the 16-residue H block consensus motif (Fig. 1C). Coils in this position are predicted in only ≈30% of the sequences examined by newcoils (17) or paircoil (18). Nonetheless, it is possible to identify heptad patterns in many of the remaining kinase sequences by visual inspection. The coiled coil databases used by newcoils and paircoil include primarily dimeric, parallel coils from α-fibrous proteins such as myosin. Thus, these algorithms may not be optimal for identifying and evaluating other types of coiled coil proteins (19).
We decided to apply an iterative learning algorithm (learncoil)‡‡ (24) to detect potential coiled coils in the histidine kinases. The assumption was made that, despite limited sequence homology in the coiled coil region, there is likely to be some structural and mechanistic similarity among the kinases and, therefore, patterns that may not be readily apparent through multiple sequence alignment. In the learning algorithm, an initial evaluation of the kinase sequences is made by using an “off-the-shelf” table of coiled coil probabilities. Those sequences with likelihoods above 0 are selected by a randomized procedure to update the probability table. This process is repeated until the table converges. We find that, after use of learncoil, 76% of kinases are predicted (likelihood ≥0.5) as coiled coils, or coiled coil-like helical structures, in the region preceding the H block. Evaluation of the sequences of proteins in the Protein Data Bank (release February 1994) yielded no likelihoods >0.31 for proteins known not to contain coiled coils. We therefore propose addition of this “coiled coil block” (CC block) to the list of histidine kinase consensus motifs and further suggest how conformational changes in this region might mediate signal transduction.
METHODS
A collection of 189 histidine kinase sequences was assembled from online sequence databases by using the Entrez browser.†† (CheA sequences were excluded.) Extremely close homologs (those with 30 or more consecutive identical residues) were allowed only once into the iteration test set, leaving 168 learncoil test sequences. Before using learncoil, sequences were evaluated for coiled coil motifs by using the paircoil (18) and newcoils (17) algorithms.
The learncoil program uses the pairwise correlation scoring method of paircoil as a subroutine. This scoring method computes scores for sliding sequence windows (generally 28 residues in length) by using estimates of pairwise and singles probabilities (i.e., probabilities of finding each pair of residues a given distance apart in a coiled coil and of finding each residue at a particular heptad position in a coiled coil). Once learncoil scores a window, the score is converted into a likelihood. The initial database of two-stranded coiled coils and the procedures for computing probabilities from frequency tables, scoring, and computing likelihoods were as described (18).
The learncoil program iteratively scans the test sequences to build a new database of potential coiled coil regions. At the start of each iteration, this new database contains no residues. Once scores and likelihoods are computed, sequences are selected for the new database with probabilities proportional to their likelihoods. That is, a number is drawn uniformly at random from the interval {0,1}, and if the number drawn is less than or equal to the likelihood of a sequence (taken from the highest scoring window), then the sequence is selected for the new database. If a sequence is selected, only regions of the sequence where the residues have likelihoods greater than or equal to either the sequence likelihood (if <0.5) or 0.5 are included in the new database. Residues at the ends of scoring windows are not included in the new database if consecutive residue likelihoods drop by >0.1.
In the first iteration, probabilities are estimated from a two-stranded coiled coil database (18). Then, at the end of an iteration, estimates of the probabilities are updated by using the new database. The probabilities are a weighted average of those computed from the original database (weight 0.1) and those computed from the current database (weight 0.9); these updated probabilities affect the scoring of sequences in the next iteration. In each iteration after the first, any sequence included in the database from the previous iteration is removed from the database (and the probabilities are adjusted) before that sequence is scored.
This procedure repeats until the number of residues in the database differs from the previous iteration by <3%. In the final iteration, regions that have likelihoods >0.5 are selected for the final database. The algorithm was run five times on the histidine kinase sequences, giving five final databases. Five to seven iterations were required for convergence. Each residue was then scored by taking an average of five probability scores (each computed as described above), and final likelihoods were derived. Scores from the five runs agreed closely in most cases, but considerable variability was observed for some sequences, particularly those of moderate likelihood (near 0.5). Finally, to check the robustness of the learncoil results, a subset of the test sequences (142 sequences chosen at random) was used to generate a second probability table; this second table was used to evaluate the 26 excluded sequences.
RESULTS
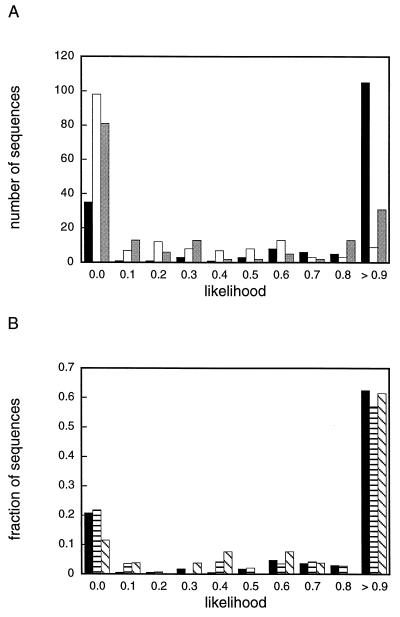
The distribution of likelihoods for the kinase region preceding the H block is shown in Fig. 2A. paircoil (18) and newcoils (17) fail to detect coiled coil motifs in the majority of cases. Likelihoods of <0.5 are calculated for 79% and 68% of the kinases (paircoil and newcoils, respectively), with ≈70% of these negative sequences having a likelihood of 0. Much higher likelihoods are seen in the learncoil distribution. Only 24% of kinase sequences have calculated likelihoods <0.5, with nearly all of these falling at likelihood 0. Of those sequences scoring 0, 9 of 35 have a proline within the region (not tolerated in coiled coils by the prediction algorithms). As a negative control, noncoiled coil sequences in the Protein Data Bank (PDB-minus) (18) were evaluated with the final histidine kinase table. No likelihoods >0.31 were obtained. However, the model coiled coil peptide from yeast GCN4 is detected with likelihood 0.7 (paircoil likelihood = 1). This suggests that the histidine kinase motif shares features with the coiled coils in the original database. It is important to note that exclusion of a subset of sequences from the iteration test set does not significantly change the distribution of likelihoods. A comparison of the original distribution to the distribution for the smaller iteration test set and the distribution for the excluded sequences is shown in Fig. 2B.
Figure 2.
Distribution of coiled coil likelihoods for 168 histidine kinase sequences. The likelihoods are for the kinase region directly preceding the H block. (A) Three prediction methods are compared: learncoil (24) using the final probabilities obtained as described in Methods (black), paircoil (18) (white), and newcoils (17) (gray). (B) Comparison of normalized learncoil likelihood distributions for the entire histidine kinase test set (168 sequences, black), a subset run separately as described in Methods (142 sequences, horizontal stripes), and the sequences excluded from the subset (26 sequences, diagonal stripes).
Additional regions in many of the kinases are predicted to be coiled coils by newcoils or by paircoil, often with likelihoods that are comparable to, or higher than, those of the target regions. An especially common pattern is seen in longer kinases: the H block occurs some distance into the cytoplasmic domain, yet a coiled coil motif also appears shortly after the last predicted transmembrane segment (Fig. 1D). In some cases, there are more than two coiled coil motifs preceding the H block or an additional motif following it. These sequences can contribute to the final database in the same way as do sequences from the target regions. After running learncoil, some of these additional high likelihood regions persist, and others do not (data not shown). Of all sequences with learncoil likelihoods ≥0.1 in any region, the highest likelihood region occurs most often (93%) before the H block, suggesting that this CC block is selectively recognized.
High likelihood CC block motifs occur broadly in the histidine kinases, appearing in all reported eukaryotic proteins, as well as in most of those from Escherichia coli and Salmonella typhimurium. In addition, learncoil predicts this coiled coil-like motif for the Caulobacter crescentus kinases PleC and FlbE implicated in asymmetric cell division (25–27), Bacillus subtilis sporulation phosphorelay sensors KinA and KinC (27, 28), the chromatic adaptation sensor RcaE (29), the activator of Bordetella virulence BvgS (30), the heme-containing oxygen sensor FixL (31), and the vancomycin resistance sensors VanS and VanSB (32, 33) (data not shown).
An especially interesting and long studied group of bacterial kinases includes the osmosensor EnvZ (34), the chemotaxis protein CheA (2), and the phosphate and nitrogen regulatory proteins PhoR and NtrB (35, 36). In general, these sequences are not recognized as coiled coil proteins by using standard methods, perhaps explaining why this motif has not been noted previously. However, by using learncoil, the PhoR and EnvZ likelihoods increase substantially, indicating that these sequences share features with the more obvious coiled coil motifs of other kinases.
The phospho-accepting domain of the chemotaxis kinase CheA is not recognized when evaluated with kinase-derived probability tables (data not shown). CheA is a cytoplasmic kinase, with a nonstandard H block motif located at the amino terminus of the protein, well away from the other kinase consensus motifs. Furthermore, NMR studies of the CheA phosphotransfer domain reveal a 5-helix bundle rather than the more extended structure of a coiled coil (7, 8). (For these reasons, CheA sequences were excluded from the learncoil iteration test set.)
learncoil does recognize NtrB sequences, although not consistently: four sequences (plus two homologs not included in the iteration test set) have calculated likelihoods >0.6, but three have likelihood 0. One reason for this may be evolutionary distance; the three highly homologous, zero-likelihood sequences (GenBank accession nos. Z37984, X71436, and M14227) are from Proteobacteria, α subdivision organisms, whereas the others are from γ subdivision organisms (e.g., E. coli). Alternatively, the explanation may be mechanistic. NtrB is a soluble cytoplasmic kinase, whereas almost all members of the test set are transmembrane kinases. Some structural details may differ between the two classes, resulting in a somewhat weaker prediction for NtrB.
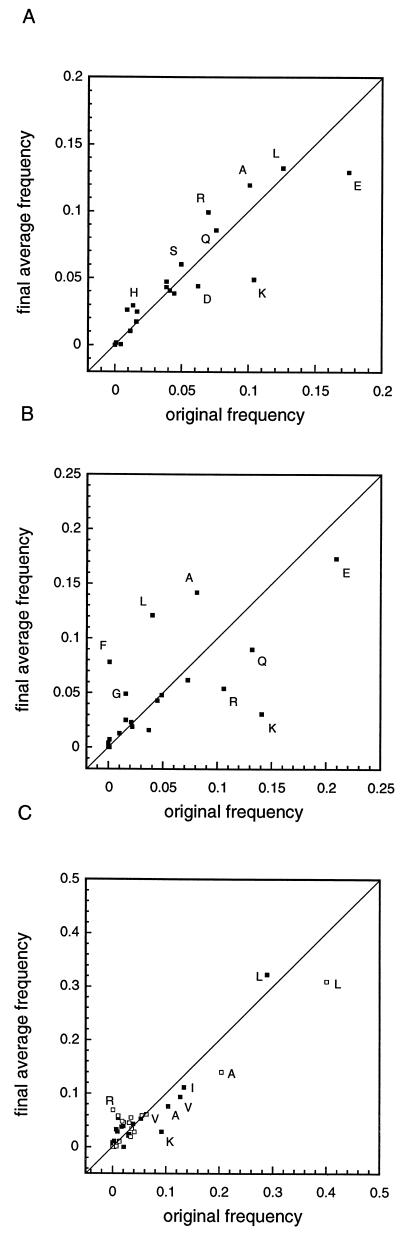
Comparison of the learncoil-derived singles and pairwise probabilities (18) to the initial coiled coil values suggests some ways in which the new motif differs from typical two-stranded coiled coils. The relative frequencies of the 20 amino acids in the initial coiled coil database (18) and the iterated database are shown in Fig. 3A. The most significant change is an overall decrease in the abundance of lysine and glutamic acid in the new database. Histidine, a relatively rare amino acid in coiled coils (1.4%), occurs at higher frequency in the new motif (2.9%) because of its absolute conservation in the H block sequence. Comparison of the singles probabilities at the individual heptad positions a–g reveals that residue distributions change more at exterior heptad positions (see, e.g., Fig. 3B) than at the interior hydrophobic positions a and d (Fig. 3C). An especially interesting trend is seen for position g (Fig. 3B), which is more hydrophobic in the new database. Comparison of pairwise probabilities (data not shown) reveals that many positive correlations for the histidine kinases can be traced to homology in the H block region. In particular, pairs including histidine at position f increase in probability, as do other pairs within the single most frequent H block sequence (FLANMSHELRT). Other positively correlated pairs have no connection to the H block sequence but appear to be related to trends in the singles probabilities. Examples include arginine at sequential c positions and glutamine at sequential f positions; these pairs occur at significantly higher frequencies in the histidine kinase motif.
Figure 3.
Comparison of singles frequencies (see Methods) for the 20 amino acids before and after learncoil iteration (original frequency and final average frequency, respectively). A diagonal line is indicated for clarity. (A) Amino acid content at all heptad positions. Points for histidine and for the more abundant amino acids (frequency >5%) are labeled. (B) Amino acid composition of position g. Points are labeled for amino acids whose frequency changes most noticeably. (C) Amino acid composition of the hydrophobic interior positions a (filled squares) and d (open squares). Points for the more abundant amino acids are labeled.
DISCUSSION
The learncoil program is a general iterative method that extends the two-stranded coiled coil prediction program paircoil to the identification of other types of coiled coils. Previously, the learncoil program successfully identified three-stranded coiled coils (24).
Iterative approaches similar to learncoil have been applied to sequence alignment and motif recognition (16, 37–41). Each method repeats two steps. First, a scoring algorithm is used to scan a database of sequences for regions thought to represent the motif of interest. Second, selected residues are used to update the parameters of the scoring algorithm (e.g., a weight matrix, profile, or probability table). The updated parameters change the regions identified in the next iteration, and usually these two steps are repeated until convergence occurs.
learncoil combines several strategies to achieve good performance without identifying false-positive sequences (24). First, instead of choosing all sequences that score above a cutoff, learncoil adds a randomized selection step to determine which sequences are used to update the scoring parameters; empirically, this has proven more effective. Second, learncoil calculates the new scoring parameters by taking a weighted average of the original parameters and the parameters estimated from the sequences selected in the latest iteration. In this manner, the initial scoring parameters have a greater effect than in traditional Bayesian approaches. None of the previously reported approaches uses weighting, and only one uses randomness (38). Finally, learncoil uses pairwise frequency data, whereas all of the other approaches are based on single frequency data. Pairwise interactions between heptad repeat positions are important for distinguishing coiled coils from false-positive sequences (18).
Using learncoil, we have identified a common but previously undescribed coiled coil-like motif (CC block) in the histidine kinase family. The frequency of this motif is striking, given the very high diversity of histidine kinase primary sequences and domain arrangements. The CC block directly precedes the conserved H block motif of the kinase domain and often coincides with a cytoplasmic region termed the “linker” domain (Fig. 1). In some cases, these regions are separated by intervening sequences, and there are two predicted motifs (Fig. 1D). Several lines of evidence indicate that these regions are of functional importance in regulating signal transduction (15).
Genetic studies in a number of systems have yielded mutant histidine kinases with extreme, poorly regulated, or constitutive signaling phenotypes. Many of these mutations cause single amino acid substitutions in the linker regions of the receptors. For example, glnL (NtrB) mutations that suppress a defect in uridylylation of the NtrB ligand PII (12) map largely (9 of 16) to the predicted coiled coil-like motif. Another two mutations map to the 12 amino acids following the motif. Single amino acid substitutions in the linker region of Bordetella pertussis BvgS cause constitutive expression of the virulence regulon (13). These mutations map near the ends of the 161-amino acid linker region; BvgS has two prominent, high likelihood scoring regions, one at each end of its linker (Fig. 1D). Finally, a mutation in the linker region of the yeast osmosensor Sln1p activates expression of Mcm1-dependent genes while conferring an osmosensitive phenotype (14); this mutation occurs within the predicted coiled coil-like motif. It is possible that these mutations alter the stability or structure of an extended helical domain.
The importance of kinase linker regions is indicated further in a genetic screen for inhibitory receptor subdomains. Random carboxyl-terminal truncations of the vancomycin-resistance sensor VanS yield a fragment spanning residues 95–174 that can inhibit VanS phosphorylation of PhoB in vivo, possibly by disrupting receptor dimerization (42). It is intriguing that the H block ends at residue 174, and residues 120–160 are predicted to form a coiled coil (by all three methods used here).
Comparisons between histidine kinases and bacterial chemotaxis receptors have suggested strongly a common signaling mechanism, despite the lack of a kinase domain in the latter. Alignment of the linker regions of the nitrate sensor NarX and the serine chemotaxis receptor Tsr revealed slight, but statistically significant, homology (11). More important, it was noted that signaling mutations in both genes mapped to these linkers, in two cases causing the same changes in identical residues (11). NarX and NarQ (a second E. coli nitrate sensor) have unusual H block sequences with domains arranged as in Fig. 1D. The NarX and NarQ linkers are predicted as coiled coils before iteration, and their likelihoods increase upon iteration, supporting the idea that the NarX/Q linkers are equivalent to the CC blocks of other kinases.
Chimeric receptor experiments have demonstrated that fusions between the chemotaxis receptors Tar and Trg and the kinase domain of the osmosensor EnvZ can transmit signals in response to chemoattractants (43, 44). Again, this suggests a shared signaling mechanism between the two classes of proteins. The point of fusion in these experiments is a conserved methionine within the linker region. The homology between EnvZ and chemoreceptor linker regions is rather limited, and thus, it has been suggested that packing of side chains across the fusion junction is unlikely (44). No coiled coils are predicted for EnvZ or chemotaxis receptor linkers by using standard methods (17, 18). However, an amphipathic heptad pattern does occur in these regions (45), and learncoil identifies this motif as coiled coil-like in EnvZ. Such a structure is consistent with the proposal that an extended secondary structural element is the critical feature shared by EnvZ and the chemotaxis receptors (44, 46).
Thus far, we have been largely unsuccessful in using kinase-derived tables to predict coiled coils in the chemotaxis receptor linker regions.§§ This suggests that there are some differences in sequence patterns between the two types of receptors, despite the indications that their linker regions are functionally similar. Coiled coils previously have been predicted for the two regulatory methylation domains of the chemotaxis receptors (17, 47), the first of which immediately follows the linker. learncoil evaluation of chemoreceptors alone (as described for the kinases) predicts extended coiled coils that include the linker region through the first methylation domain (data not shown). A similar assignment of extended helical structure has been made on the basis of multiple sequence alignment (48).
How might a coiled coil mediate signal transduction? An obvious answer is that the coiled coil (or a coiled coil-like, extended helical bundle) might form an oligomerization interface between receptor monomers. A less obvious possibility is that the coiled coil might be a structural relay. Both of these appear to play a role in signaling by the aspartate chemotaxis receptor (Tar). Soluble chimeric receptors have been made by fusing dimeric coiled coil peptides to the cytoplasmic domain of the aspartate receptor at the linker (53, 54). These receptors can assemble with CheW and CheA into complexes capable of phosphorylating CheY. When the coiled coil peptide is connected either directly (53) or by a flexible tether (54) to the Tar cytoplasmic domain, kinase activity is higher than observed in the presence of monomeric cytoplasmic domain, demonstrating that dimerization is essential for full stimulatory activity. Furthermore, when the coiled coil peptide is directly fused to the presumably helical linker domain, stimulatory activity is strongly dependent on the helical register between the two (53). Centering the hydrophobic faces of the linker and the peptide yields a receptor of moderate stimulatory activity. Insertion of three or four amino acids at the junction point yields nonactivating and highly activating receptors, respectively (53). These insertions should shift the receptor interface to either side of center at the point of fusion.
A mechanism for triggering interface shifts in coiled coils is suggested by a recent analysis of myosin heptad patterns (55). Myosins are structural proteins with extremely long coiled coils that have conserved skips. The most common discontinuities have been termed “stutters” and “stammers”; these correspond to deletions from the heptad repeat pattern of 3 or 4 residues (or insertion of 4 or 3 residues), respectively. Thus, instead of the usual alternating 4–3 hydrophobic repeat, an occasional 4–4 or 3–3 appears. These discontinuities would result in an interface shift of one sevenfold helical wheel position (e.g., position d to a, or ≈50°). Such shifting has been observed directly in the crystal structure of the low pH form of influenza hemagglutinin (TBHA2) (56). It has been proposed that such shifts could occur gradually, without disruption of the helix, by a change in supercoiling about the dimer axis; these gradual shifts would require an intermediate region of relatively weak interhelical packing (55).
Analogously, one might imagine that a relatively unstable coiled coil could be dynamically switched between alternate interfaces by a small perturbation at one end. Such an interface shift might be expected to cause significant reorientation of kinase domains at the other end of the coiled coil, capable of suppressing or stimulating phosphorylation (53). Indeed, recent mutagenesis of EnvZ suggests that the linker region is essential not for dimerization but for proper orientation of receptor monomers within the dimer (45). The location of the histidine autophosphorylation site near the end of a coiled coil-like structure suggests that an interface perturbation might control the exposure of the histidine to an ATP-binding domain. Alternatively, the conserved H block residues might form an active site whose structure and catalytic competence are mediated by shifts in a coiled coil-like intersubunit interface.
Acknowledgments
M.S. is a Center for Discrete Mathematics and Theoretical Computer Science (DIMACS) postdoctoral fellow. DIMACS is a partnership of Rutgers University, Princeton University, AT&T Labs, Bellcore, and Bell Labs. DIMACS is a National Science Foundation Science and Technology Center, funded under contract STS-91–1999 and receives support from the New Jersey Commission on Science and Technology. B.B. is supported by a National Science Foundation Career Award. J.M.B. is a Whitehead Fellow and acknowledges support from the W. M. Keck Foundation. P.S.K. is an Associate Investigator of the Howard Hughes Medical Institute.
ABBREVIATION
- CC block
coiled coil block
Footnotes
Coiled coil motifs previously have been noted in unique amino-terminal domains of the histidine kinases Nik-1 (20) and TodS (21). Predicted coiled coil domains in the nonreceptor serine/threonine kinase TOUSLED (22) and tyrosine kinase Fes (23) are important for oligomerization and autophosphorylation.
Information on the learncoil algorithm may be obtained by email to learncoil@theory.lcs.mit.edu. The histidine kinase probability table and the paircoil program are available at http://theory.lcs.mit.edu, in the learncoil and paircoil directories, respectively.
www.ncbi.nlm.nih.gov/Entrez/ A list of histidine kinase sequences used in this study may be obtained from A.G.C.
The exception is halobacterial phototaxis transducers (Htr family) (49–52). newcoils and paircoil predict numerous high likelihood coiled coils throughout Htr cytoplasmic domains, whereas learncoil-derived histidine kinase tables assign the linker regions much higher likelihoods than other regions (data not shown).
References
- 1.Hoch J A, Silhavy T J, editors. Two-Component Signal Transduction. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 2.Stock J B, Surette M G. In: Escherichia coli and Salmonella typhimurium. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1103–1129. [Google Scholar]
- 3.Rudolph J, Tolliday N, Schmitt C, Schuster S C, Oesterhelt D. EMBO J. 1995;14:4249–4257. doi: 10.1002/j.1460-2075.1995.tb00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wurgler-Murphy S M, Saito H. Trends Biochem Sci. 1997;22:172–176. doi: 10.1016/s0968-0004(97)01036-0. [DOI] [PubMed] [Google Scholar]
- 5.Stock J B, Surette M G, Levit M, Park P. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 25–51. [Google Scholar]
- 6.Kato M, Mizuno T, Shimizu T, Hakoshima T. Cell. 1997;88:717–723. doi: 10.1016/s0092-8674(00)81914-5. [DOI] [PubMed] [Google Scholar]
- 7.Zhou H, Lowry D F, Swanson R V, Simon M I, Dahlquist F W. Biochemistry. 1995;34:13858–13870. doi: 10.1021/bi00042a018. [DOI] [PubMed] [Google Scholar]
- 8.Zhou H, Dahlquist F W. Biochemistry. 1997;36:699–710. doi: 10.1021/bi961663p. [DOI] [PubMed] [Google Scholar]
- 9.McEvoy M M, Zhou H, Roth A F, Lowry D F, Morrison T B, Kay L E, Dahlquist F W. Biochemistry. 1995;34:13871–13880. doi: 10.1021/bi00042a019. [DOI] [PubMed] [Google Scholar]
- 10.McEvoy M M, Muhandiram D R, Kay L E, Dahlquist F W. Biochemistry. 1996;35:5633–5640. doi: 10.1021/bi952707h. [DOI] [PubMed] [Google Scholar]
- 11.Collins L A, Egan S M, Stewart V. J Bacteriol. 1992;174:3667–3675. doi: 10.1128/jb.174.11.3667-3675.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atkinson M R, Ninfa A J. J Bacteriol. 1992;174:4538–4548. doi: 10.1128/jb.174.14.4538-4548.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller, J. F., Johnson, S. A., Black, W. J., Beattie, D. T., Mekalanos, J. J. & Falkow, S. (1992) J. Bacteriol. 970–979. [DOI] [PMC free article] [PubMed]
- 14.Fassler J S, Gray W M, Malone C L, Tao W, Lin H, Deschenes R J. J Biol Chem. 1997;272:13365–13371. doi: 10.1074/jbc.272.20.13365. [DOI] [PubMed] [Google Scholar]
- 15.Parkinson J S, Kofoid E C. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 16.Mushegian A R, Bassett D E J, Boguski M S, Bork P, Koonin E V. Proc Natl Acad Sci USA. 1997;94:5831–5836. doi: 10.1073/pnas.94.11.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 18.Berger B, Wilson D B, Wolf E, Tonchev T, Milla M, Kim P S. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolf E, Kim P S, Berger B. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alex L A, Borkovich K A, Simon M I. Proc Natl Acad Sci USA. 1996;93:3426–3421. doi: 10.1073/pnas.93.8.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau P C K, Wang Y, Patel A, Labbe D, Bergeron H, Brousseau R, Konishi Y, Rawlings M. Proc Natl Acad Sci USA. 1997;94:1453–1458. doi: 10.1073/pnas.94.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roe J L, Durfee T, Zupan J R, Repetti P P, McLean B G, Zambryski P C. J Biol Chem. 1997;272:5838–5845. doi: 10.1074/jbc.272.9.5838. [DOI] [PubMed] [Google Scholar]
- 23.Read R D, Lionberger J M, Smithgall T E. J Biol Chem. 1997;272:18498–18503. doi: 10.1074/jbc.272.29.18498. [DOI] [PubMed] [Google Scholar]
- 24.Berger B, Singh M. J Comp Biol. 1997;4:261–273. doi: 10.1089/cmb.1997.4.261. [DOI] [PubMed] [Google Scholar]
- 25.Lane T, Benson A, Hecht G B, Burton G J, Newton A. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 403–417. [Google Scholar]
- 26.Wingrove J A, Gober J W. Science. 1996;274:597–601. doi: 10.1126/science.274.5287.597. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro L, Losick R. Science. 1997;276:712–718. doi: 10.1126/science.276.5313.712. [DOI] [PubMed] [Google Scholar]
- 28.Hoch J A. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 129–144. [Google Scholar]
- 29.Kehoe D M, Grossman A R. Science. 1996;273:1409–1412. doi: 10.1126/science.273.5280.1409. [DOI] [PubMed] [Google Scholar]
- 30.Uhl M A, Miller J F. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 333–349. [Google Scholar]
- 31.Agron P G, Helsinki D R. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 275–287. [Google Scholar]
- 32.Arthur M, Depardieu F, Holman T, Wright G, Walsh C T, Courvalin P. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 387–391. [Google Scholar]
- 33.Evers S, Courvalin P. J Bacteriol. 1996;178:1302–1309. doi: 10.1128/jb.178.5.1302-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pratt L A, Silhavy T J. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 105–127. [Google Scholar]
- 35.Wanner B L. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 203–221. [Google Scholar]
- 36.Ninfa A J, Atkinson M R, Kamberov E S, Feng J, Ninfa E G. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 67–88. [Google Scholar]
- 37.Tatsuov R, Altschul S, Koonin E. Proc Natl Acad Sci USA. 1994;91:12091–12095. doi: 10.1073/pnas.91.25.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawrence C E, Altschul S, Boguski M, Liu J, Neuwald A, Wooten J. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- 39.Attwood T K, Findlay J B C. Protein Eng. 1992;6:167–176. doi: 10.1093/protein/6.2.167. [DOI] [PubMed] [Google Scholar]
- 40.Gribskov M. Gene. 1992;119:107–111. doi: 10.1016/0378-1119(92)90073-x. [DOI] [PubMed] [Google Scholar]
- 41.Dodd I, Egan J B. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fisher S L, Jiang W, Wanner B L, Walsh C T. J Biol Chem. 1995;270:23143–23149. doi: 10.1074/jbc.270.39.23143. [DOI] [PubMed] [Google Scholar]
- 43.Utsumi R, Brissette R E, Rampersaud A, Forst S A, Oosawa K, Inouye M. Science. 1989;245:1246–1249. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- 44.Baumgartner J W, Kim C, Brissette R E, Inouye M, Park C, Hazelbauer G L. J Bacteriol. 1994;176:1157–1163. doi: 10.1128/jb.176.4.1157-1163.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park H, Inouye M. J Bacteriol. 1997;179:4382–4390. doi: 10.1128/jb.179.13.4382-4390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin T, Inouye M. J Mol Biol. 1994;244:477–481. doi: 10.1006/jmbi.1994.1746. [DOI] [PubMed] [Google Scholar]
- 47.Stock J B, Lukat G S, Stock A M. Annu Rev Biophys Biophys Chem. 1991;20:109–136. doi: 10.1146/annurev.bb.20.060191.000545. [DOI] [PubMed] [Google Scholar]
- 48.Le Moual H, Koshland D E., Jr J Mol Biol. 1996;261:568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- 49.Yao V J, Spudich J L. Proc Natl Acad Sci USA. 1992;89:11915–11919. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seidel R, Scharf B, Gautel M, Kleine K, Oesterhelt D, Engelhard M. Proc Natl Acad Sci USA. 1995;92:3036–3040. doi: 10.1073/pnas.92.7.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang W, Brooun A, Mueller M M, Alam M. Proc Natl Acad Sci USA. 1996;93:8230–8235. doi: 10.1073/pnas.93.16.8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, W., Brooun, A., McCandless, J., Banda, P. & Alam, M. (1996) Proc. Natl. Acad. Sci. USA 4649–4654. [DOI] [PMC free article] [PubMed]
- 53.Cochran A G, Kim P S. Science. 1996;271:1113–1116. doi: 10.1126/science.271.5252.1113. [DOI] [PubMed] [Google Scholar]
- 54.Surette M G, Stock J B. J Biol Chem. 1996;271:17966–17973. doi: 10.1074/jbc.271.30.17966. [DOI] [PubMed] [Google Scholar]
- 55.Brown J H, Cohen C, Parry D A D. Proteins. 1996;26:134–145. doi: 10.1002/(SICI)1097-0134(199610)26:2<134::AID-PROT3>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 56.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Nature (London) 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]