Abstract
Members of the KNOX gene family have important roles in plant meristems by regulating cell division and differentiation. BREVIPEDICELLUS (BP), one of seven KNOX genes in Arabidopsis, has a primary role in internode patterning. We carried out a comparison of RNA expression profiles between wild-type seedlings and bp mutants at a developmental stage prior to a visible phenotypic difference. Transcript differences were found for a number of genes in cell wall biosynthesis, especially genes in the lignin pathway. The regulation of lignin biosynthesis by BP was demonstrated by observing increased lignin deposition in bp mutants following bolting, decreased lignification in plants overexpressing BP, and aberrant lignin deposition in discrete regions of the bp stem. Furthermore, we showed that BP binds promoters of some genes in the lignin pathway. Our results provide a metabolic fingerprint for BP and identify the lignin pathway as one of the coordinate processes that BP regulates.
Keywords: Homeobox, Brevipedicellus, Knat1, lignin, microarray, meristem
In species with simple leaves, class I members of the KNOX gene family are expressed in shoot meristems and stems, but not leaves or roots. Loss-of-function phenotypes have been described for a few KNOX genes, and demonstrate a role in meristem maintenance (Long et al. 1996; Byrne et al. 2002; Vollbrecht et al. 2000) and stem elongation (Sato et al. 1999; Douglas et al. 2002; Venglat et al. 2002). Mutations in the KNOX gene BREVIPEDICELLUS (BP), result in plants with shorter internodes, downward pointing siliques, and an epidermal stripe of disorganized cells along the stem (Douglas et al. 2002; Venglat et al. 2002). BP overexpression results in changes in leaf shape and increased cytokinin levels in several species (Lincoln et al. 1994; Chuck et al. 1996; Ori et al. 2000; Frugis et al. 2001).
Results and Discussion
We compared gene expression profiles between the bp-9 loss-of-function mutant (in the Columbia ecotype, Col) and Col wild-type plants using a high-density Affymetrix oligonucleotide array representing ∼8,200 Arabidopsis genes. Two-week-old seedlings grown in short-day conditions were used as starting material. Under these conditions, bp and Col plants had no apparent phenotypic differences. We reasoned that using this approach may uncover early differences in gene expression stemming directly from loss of BP function rather than down-stream secondary effects. Two independent RNA extractions and in vitro transcriptions were conducted for each genotype. Each of the two repetitions for the mutant was compared against both repetitions of the wild type to obtain a matrix of four comparisons. A set of filtering parameters (see Supplemental Material) was used to identify significant expression changes and eliminate genes exhibiting erratic patterns. Our analysis revealed a difference in expression of 64 genes (see Supplementry Table 1).
Genes identified from the oligo array analysis were divided into functional groups (Supplementary Table 1) according to the NCBI predicted function (see Supplementary material). The predominant group of genes (37%) encodes biosynthetic enzymes. Examination of this group revealed that 54% of these enzymes are involved in cell wall biosynthesis, 61% of which are predicted to be in the lignin biosynthesis pathway (Table 1).
Table 1.
Microarray results for cell wall biosynthetic genes
| Gene # | Name | Enzyme | Function | EC # | E-value | Average fold change |
|---|---|---|---|---|---|---|
| At1g01120 | KCS1 | Fatty acid elongase 3-ketoacyl-CoA synthase 1 | Wax biosynthesis | 2.3 | ||
| At1g12780 | UDGE | Uridine diphosphate glucose epimerase | Cell wall biosynthesis | 5.1.3.2 | Biochemically tested | -1.4 |
| At1g16410 | CYP710A2 | Cytochrome P450 | Lignin biosynthesis | 1.14.13.41 | 1E-115 | 1.6 |
| At1g51680 | At4CL1 | 4-Coumarate-CoA ligase 1 | Lignin biosynthesis | 6.2.1.12 | Biochemically tested | 1.5 |
| At2g30490 | AtC4H | Cinnamate 4-monooxygenase | Lignin biosynthesis | 1.14.13.11 | Biochemically tested | 1.5 |
| At2g32990 | CEL | Cellulase | Cell wall biosynthesis | 3.2.1.4 | Biochemically tested | 2.1 |
| At2g37040 | PAL1 | Phenylalanine ammonia-lyase 1 | Lignin biosynthesis | 4.3.1.5 | Biochemically tested | 1.9 |
| At3g14310 | PME3 | Pectin methyl esterase | Cell wall biosynthesis | 3.1.1.11 | Biochemically tested | -1.6 |
| At3g21770 | AtPRXR9GE | Peroxidase | Lignin biosynthesis | 1.11.1.7 | 1E-118 | -2.0 |
| At4g23600 | TRYT | Tyrosine transaminase | Lignin biosynthesis | 2.6.1.5 | 9E-57 | -1.8 |
| At4g23990 | AtCSLG1 | Cellulose synthetase | Cellulose biosynthesis | 1.6 | ||
| At4g30270 | SEN4 | Endo-xyloglucan transferase | Cell wall biosynthesis | -1.7 | ||
| At4g39330 | CAD1 | Cinnamyl-alcohol dehydrogenase 1 | Lignin biosynthesis | 1.1.1.195 | Biochemically tested | 1.9 |
EC number and E-value are derived from the mips server. E-value is a mips prediction when the enzyme is not biochemically tested. Positive fold change is an increase of expression in bp-9 compared to wild type, negative fold change is a decrease.
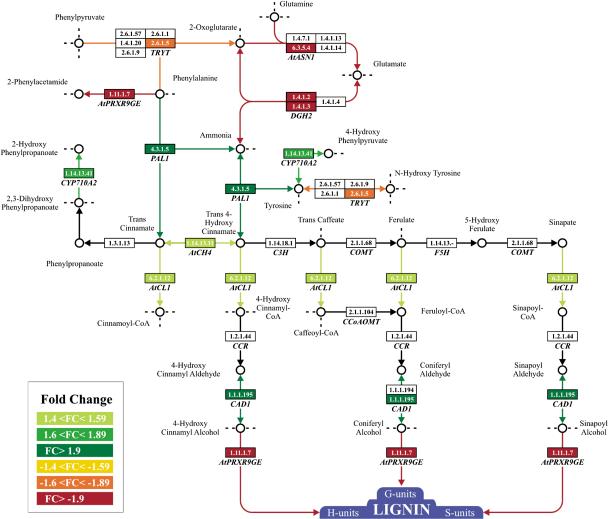
EC numbers (see Supplemental Material) were used to identify and locate the enzymes in biosynthetic pathways. A set of enzymes catalyzing the synthesis of different lignin subunits was increased at the transcriptional level in bp-9 seedlings, with the exception of the peroxidase encoded by AtPRXR9GE, which was decreased (Fig. 1). We compared lignin deposition in bp-9 and Col seedlings, grown under the conditions used in the chip experiment. Staining with phloroglucinol-HCl did not reveal any difference in lignin deposition between bp-9 and Col at this developmental stage (Supplementary Fig. 1A–D). This observation is in agreement with the low levels of RNA encoding AtPRXR9GE, which according to catalytic properties predicted by protein analysis, is one of the peroxidases that polymerizes the different subunits into lignin in the final step of the pathway.
Figure 1.
Biosynthetic pathways affected in bp-9 seedlings. Genes up-regulated in bp-9 are boxed in shades of green; genes down-regulated in bp-9 are boxed in red. Genes not affected or absent in the oligonucleotide array are boxed in white. The arrows indicate the direction of the enzymatic reaction identified by EC number. The genes At-ASN1, GDH2, CYP710A2, and TRYT belong to other pathways but share common substrates with the lignin pathway. Complete names for the genes differentially expressed are reported in Table 1 and Supplementary Table 1.
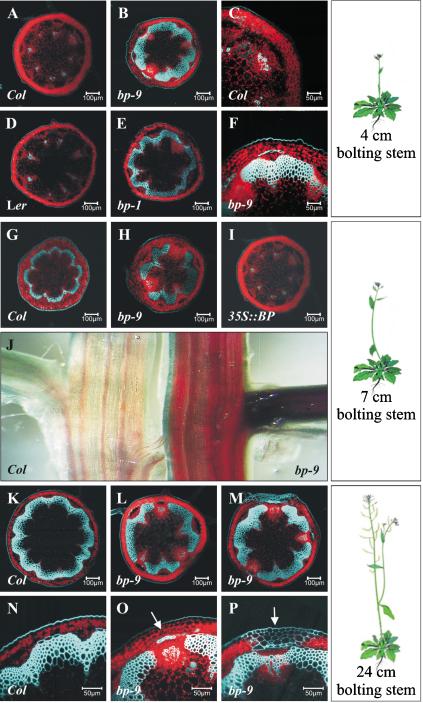
Given the prediction from the oligonucleotide array for differences in lignin, we examined elongated stems, where lignin is normally deposited. Sections at the base of 4-cm bp-9 stems showed interfascicular (between the vascular bundles) lignin deposition (Fig. 2B), whereas the wild-type stem still showed lignin deposition only in vasculature (Fig. 2A). This interfascicular lignin deposition in bp-9 is characteristic of more mature stem development in wild-type plants (Fig. 2G). In addition, bp-9 xylem elements appeared larger than equivalent bundles in wild type (Fig. 2C,F). The premature lignin deposition and bundle differentiation was verified with another allele, bp-1, in the Landsberg erecta (Ler) background. Sections of 4-cm Ler stems showed the same pattern as Col with lignin deposition only in vascular tissue (Fig. 2D). Similar to bp-9, bp-1 showed interfascicular lignin deposition between the bundles (Fig. 2E). Thus, bp seedlings prior to bolting do not accumulate more lignin than wild type, whereas bp plants that have bolted show a premature increase in lignin deposition.
Figure 2.
Lignin deposition during plant development. (A–I,K–P) Confocal microscopy observations taken at the base of plant stems just above the rosette leaves. The red auto-florescence represents chlorophyll localization; the light-blue auto-florescence represents lignin deposition. (A) Four-centimeter Col stem. (B) Four-centimeter bp-9 stem. (C) Magnification of 4-cm Col bundle. (D) Four-centimeter Ler stem. (E) Four-centimeter bp-1 stem. (F) Magnification of highly lignified 4-cm bp-9 bundle. (G) Seven-centimeter Col stem. (H) Seven-centimeter bp-9 stem. (I) Seven-centimeter 35S::BP stem. (J) Whole mounts of 7-cm Col and bp-9 with phloroglucinol-HCl staining (red color represents lignin deposition). (K) Twenty-four-centimeter Col stem showing a continuous lignified ring. (L) Twenty-four-centimeter bp-9 stem showing gaps in the lignin ring. (M) bp-9 lignin mislocalization in the epidermis and cortex associated with the unlignified bundle at the base of 24-cm stem. (N) Magnification of wild-type bundle at the base of 24-cm stem. (O) Magnification of bp-9 unlignified bundle with lignin mislocalization in the cortical cells (arrow) at the base of 24-cm stem. (P) Magnification of bp-9 lignin mislocalization in the epidermis and cortex (arrow) at the base of 24-cm stem.
We analyzed slightly older plants (7 cm) to determine whether lignin deposition continued to differ from wild type. Sections at the base of 7-cm stems revealed a higher level of lignin deposition in bp-9 compared with Col (Fig. 2G,H). Whole mounts of Col and bp-9 stems also showed a difference in lignin accumulation; Col stems showed staining in the bundles and weak staining localized to the interfascicular cells, whereas bp-9 stems showed intense lignin staining of both bundles and interfascicular cells (Fig. 2J).
The increased lignin in bp stems suggested that overexpression of BP might delay lignification. Cross-sections at the base of 7-cm 35S::BP stems showed lignin deposition only in the bundles (Fig. 2I), whereas wild-type plants at the same stage also showed lignin deposition in the interfascicular cells (Fig. 2G). Lignin deposition is considered a signature for irreversible cell differentiation (Groover and Jones 1999); thus it is not surprising that BP, which promotes indeterminate cell fates (Chuck et al. 1996), represses lignin deposition and premature cell differentiation.
The influence of BP on cell wall properties was also observed as lignin mislocalization and inappropriate cell differentiation in discrete regions of bp stems. Near the base of mature Col stems, lignin was deposited in fully developed bundles and interfascicular cells, forming a continuous ring of lignin (Fig. 2K,N). In contrast, at the base of bp-9 stems, the vascular ring was interrupted by three gaps with non-lignified, chlorophyll-containing cells (Fig. 2L,O). Previous studies described differentiation-defective cell stripes along the bp epidermis (Douglas et al. 2002; Venglat et al. 2002). We observed that these stripes were always positioned over the gaps in the vascular ring. The cells in these epidermal stripes were lignified, as were the adjacent cortical cells (Fig. 2M,P), and had vasculature features (Douglas et al. 2002; Venglat et al. 2002). It appears that the absence of BP led to a switch in cell fates in discrete regions; cells that should have differentiated as epidermis and cortex were lignified, whereas cells in the gaps, which should have lignified, retained chlorophyll.
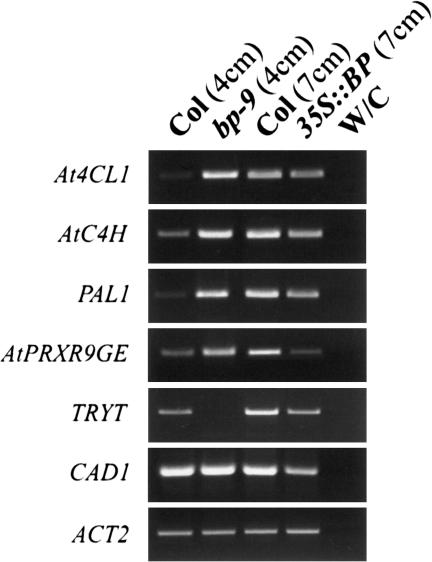
To determine how BP regulates lignin accumulation, we assayed transcript levels of the genes previously identified by oligonucleotide array analysis in bp-9 and 35S::BP bolting plants. Consistent with premature lignin deposition in 4-cm bp stems, the expression level of four genes—At4CL1 (4-coumarate:CoA ligase 1), AtC4H (cinnamate 4-monooxygenase), PAL1 (phenylalanine ammonia-lyase 1), and AtPRXR9GE (peroxidase)—was higher in bp-9. The expression of TRYT (tyrosine transaminase) was less in 4-cm bp-9 bolting stems and the level of CAD1 (cinnamyl-alcohol dehydrogenase) RNA was stable (Fig. 3). We also examined RNA from 7-cm stems of 35S::BP plants. Consistent with the delay in lignin deposition, reverse transcriptase PCR (RT–PCR) analysis revealed that transcripts for all the genes examined were decreased in 35S::BP stems compared with wild type (Fig. 3). The comparison of transcripts at seedling (Fig. 1) and adult stages (Fig. 3) reveals the important role of the AtPRXR9GE peroxidase. AtPRXR9GE RNA levels were lower in bp-9 at the seedling stage when there was no lignin difference, and increased in plants with bolted stems. Furthermore, AtPRXR9GE transcripts were also regulated in a BP-dependent manner. Taken together, these data suggest that AtPRXR9GE may be a key protein in lignin polymerization in Arabidopsis.
Figure 3.
RT–PCR analysis of genes in the lignin pathway. Expression levels were tested in the following samples: 4-cm Col stem, 4-cm bp-9 stem, 7-cm Col stem, 7-cm 35S::BP stem, and amplification with no cDNA. Control amplification with ACT2 indicates that equal amounts of cDNA were amplified.
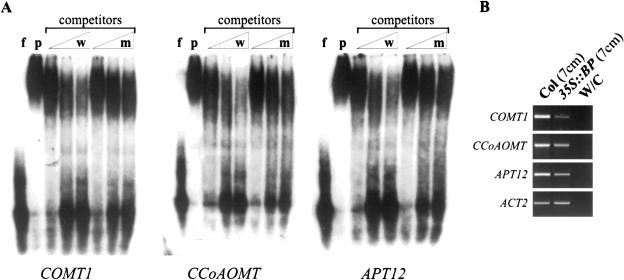
As a further test for the regulation of lignin biosynthesis by BP, we examined promoters of genes in the lignin pathway for the presence of KN1-binding sites (Smith et al. 2002), which are also bound by BP (H. Smith and S. Hake, unpubl.). Three promoters that contained binding sites (Supplementary Table 2) were tested by electrophoretic mobility shift assays (EMSA). Promoter fragments containing the binding site were competitively bound by recombinant BP protein (Fig. 4A). These three genes, not previously identified by our oligonucleotide expression analysis, show decreased expression in 35S::BP plants at bolting (Fig. 4B). Three other genes involved in the lignin pathway with KNOX-binding sites in the promoter region were also identified (Supplementary Table 2).
Figure 4.
EMSA and expression analyses of COMT1, CCoAOMT, and APT12. (A) EMSA analysis. (f) free probe; (p) BP incubated with COMT1, CCoAOMT, and APT12 probes. DNA-binding specificity was determined with unlabeled wild-type DNA (w) or mutant DNA (m) at 10-, 50-, and 100-fold molar excess over labeled probe. (B) RNA levels of COMT1, CcoAOMT, and APT12 were tested in the following samples: 7-cm Col stem, 7-cm 35S::BP stem, and amplification with no cDNA. Control amplification with ACT2 indicates that equal amounts of cDNA were amplified.
Lignin metabolism has been intensely investigated in numerous species to identify the participating enzymes and regulatory elements that control the pathway (Ehlting et al. 1999; Grima-Pettenati and Goffner 1999; Zhong et al. 2000; Anterola and Lewis 2002). Although a number of genes encoding enzymes in the phenylpropanoid and lignin pathways are regulated by MYB transcription factors (Sablowski et al. 1994, 1995; Borevitz et al. 2000), the regulation has been specific for certain enzymes, rather than global (Tamagnone et al. 1998). We show that BP affects the expression of a large number of genes, suggesting that it may act early in the lignin regulatory cascade. BP may also directly regulate the quality and quantity of lignin. Caffeic acid O-methyltransferases and caffeoyl-CoA O-methyltransferases, two proteins whose promoters are bound by BP, are thought to mediate the flux of intermediate products between the branches that form the three different lignin subunits regulating lignin composition. Laccases and peroxidases, also containing BP-binding sites in their promoters, polymerize the lignin subunits and are considered the final steps of lignin biosynthesis (Grima-Pettenati and Goffner 1999). Given the cross-talk between the lignin branches (Meyer et al. 1998; Ralph et al. 1998) and the fact that the target genes are members of gene families, some of which do not contain the KNOX-binding site, the regulation of lignin biosynthesis is likely to be very complex.
The KNOX family of transcription factors is thought to maintain cells in an indeterminate state based on expression and mutant analysis (Kerstetter et al. 1997; Tsiantis 2001). Preventing the premature accumulation of lignin is an appropriate task for the regulation of determinacy. A link between KNOX genes and secondary cell wall metabolism is also supported by a microarray study that examined a trans-section of a poplar tree trunk (Hertzberg et al. 2001). The expression of KNAP2, a BP homolog, was decreased in the region of the stem where secondary cell walls are synthesized, suggesting that its presence might prohibit the induction of secondary cell wall synthesis. Our data suggest that the modification of cell walls, and in particular the regulation of lignin deposition and quality, is one of the coordinated processes that BP regulates to achieve proper cell differentiation. Although it is likely that other BP functions, such as internode patterning, occur outside of the context of lignin regulation, it remains possible that they also occur through the regulation of additional metabolic pathways and cell wall components.
In conclusion, our comparison of transcript levels prior to a visible morphological difference allowed us to predict changes in lignin deposition. This prediction led to the discovery that BP regulates the lignin pathway, thereby providing a glimpse into the cross-talk required for proper cell differentiation during plant development. This research opens new fields of investigation aimed at understanding how BP affects other components of the cell wall and the relationship of other KNOX genes to metabolic pathways.
Materials and methods
Genetic material
SLAT filters, containing inverse PCR products from pools of 50 each of the Jonathan Jones Sainsbury Lab Arabidopsis Transposant lines (Tissier) were kindly provided by Drs. Steven Rutherford and Ian Moore at Oxford University. The filters were screened with a full-length BP cDNA probe, and one positive pool was detected. PCR amplification with dSpm-specific primers and BP 5′ and 3′ primers confirmed the presence of a dSpm insertion in the positive pool. Sequencing of the PCR fragment revealed an insertion in the first intron of BP. To identify the mutant line within the pool, 180 individuals were screened by PCR. A single plant was identified that contained the dSpm insertion. Following allelism tests with bp-1, the dSpm insertion in BP was named bp-9. The 35S::BP plants used in these experiments are in a Columbia background.
Plant material and growth conditions
The Affymetrix chip experiment was carried out on brevipedicellus (bp-9) in a Columbia background and wild-type Columbia seeds, grown in a growth chamber under short-day condition (8 h light and 16 h dark) for 2 wk. Entire plants were harvested and frozen in liquid nitrogen. For the lignin staining of seedlings, the seeds were planted in a short-day chamber under the same conditions used for the chip experiment. For the lignin staining on bolted stems, plants were grown in the greenhouse under long-day conditions, 16 h light and 8 h dark. Observations were made on plants at the same age, with the same number of internodes and same height. Day temperature was 20°C and night temperature was 18°C in both short- and long-day conditions for all experiments.
Tissue preparation, histochemical stains, light and confocal microscopy
Fifty micrometers of fresh tissue sections were cut by a vibratome in the presence of 0.4 M Mannitol and 50 mM HEPES. For lignin visualization, the tissues were stained with phloroglucinol-HCl [1% (wt/vol) phloroglucinol in 6N HCl] for 5 min and then observed under a light microscope. The lignin auto-florescence was detected by Zeiss 510 UV Confocal Laser Scanning Microscope.
Semi-quantitative RT–PCR
One microgram of total RNA that had been extracted with RNeasy (Qiagen) and treated with DNase was used for cDNA synthesis. After retrotranscription reaction with SuperScript RT (Invitrogen) the samples were purified by PCR purification Kit (Qiagen) and diluted eight times. AtC4H, AtPRXR9GE, At4CL1, CAD1, PAL1, TRYT, COMT1, CCoAOMT, ATP12A, and ACTIN2 (ACT2, At3g18780) products were amplified with 25–30 PCR cycles and with an annealing temperature between 60°C and 63°C. Experiments were conducted at least three times with equivalent results. For the PCR primer sequences see Supplemental Material.
EMSA
Recombinant expressed and purified BP protein was mixed with 50,000 cpm of wild-type or mutant DNA probes, 2 μg poly [d(I-C)] in 1× EMSA buffer (10 mM NaCl, 0.5 mM DTT, 0.5 mM EDTA, 1 mM MgCl2, 5% glycerol and 0.15% NP-40) for 2 h at 4°C in 20 μL reaction volume. For wild-type (WT) and mutant (MT) probe sequences see Supplemental Material. The binding complexes were resolved by EMSA in 5% polyacrylamide gel (70:1 ratio of acrylamide to bis-acrylamide) in 0.5× TBE buffer. After electrophoresis, the gels were dried, exposed O/N, and analyzed.
Acknowledgments
We thank Jonathan Jones for making the SLAT lines available; Hans Holtan and Steve Ruzin for help with confocal microscopy; Maurice Ahyow for assistance in lab protocols; Jim Tepperman for help with Affymetrix analysis; and Bassem Al-Sady, Elena Monte, Carla Ticconi and Hake laboratory members for helpful discussions and critical reading of the manuscript. Special thanks to Harley Smith for his help with the biochemistry. This work was supported by a BSF grant (2000109) to N.O., a National Science Foundation grant (IBN031431) to S.H., and the U.S. Department of Agriculture-Agricultural Research Service.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www. genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1120003.
References
- Anterola A.M. and Lewis, N.G. 2002. Trends in lignin modification: A comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry 61: 221–294. [DOI] [PubMed] [Google Scholar]
- Borevitz J.O., Xia, Y., Blount, J., Dixon, R.A., and Lamb, C. 2000. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M.E., Simorowski, J., and Martienssen, R.A. 2002. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129: 1957–1965. [DOI] [PubMed] [Google Scholar]
- Chuck G., Lincoln, C., and Hake, S. 1996. KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8: 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas S.J., Chuck, G., Dengler, R.E., Pelecanda, L., and Riggs, C.D. 2002. KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14: 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J., Buttner, D., Wang, Q., Douglas, C.J., Somssich, I.E., and Kombrink, E. 1999. Three 4-coumarate:Coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 19: 9–20. [DOI] [PubMed] [Google Scholar]
- Frugis G., Giannino, D., Mele, G., Nicolodi, C., Chiappetta, A., Bitonti, M.B., Innocenti, A.M., Dewitte, W., Van Onckelen, H., and Mariotti, D. 2001. Overexpression of KNAT1 in lettuce shifts leaf determinate growth to a shoot-like indeterminate growth associated with an accumulation of isopentenyl-type cytokinins. Plant Physiol. 126: 1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima-Pettenati J. and Goffner, D. 1999. Lignin genetic engineering revisited. Plant Science 145: 51–65. [Google Scholar]
- Groover A. and Jones, A.M. 1999. Tracheary element differentiation uses a novel mechanism coordinating programmed cell death and secondary cell wall synthesis. Plant Physiol. 119: 375–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg M., Aspeborg, H., Schrader, J., Andersson, A., Erlandsson, R., Blomqvist, K., Bhalerao, R., Uhlen, M., Teeri, T.T., Lundeberg, J., et al. 2001. A transcriptional roadmap to wood formation. Proc. Natl. Acad. Sci. 98: 14732–14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter R.A., Laudencia-Chingcuanco, D., Smith, L.G., and Hake, S. 1997. Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development 124: 3045–3054. [DOI] [PubMed] [Google Scholar]
- Lincoln C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. 1994. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.A., Moan, E.I., Medford, J.I., and Barton, M.K. 1996. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69. [DOI] [PubMed] [Google Scholar]
- Meyer K., Shirley, A.M., Cusumano, J.C., Bell-Lelong, D.A., and Chapple, C. 1998. Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc. Natl. Acad. Sci. 95: 6619–6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. 2000. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127: 5523–5532. [DOI] [PubMed] [Google Scholar]
- Ralph J., Hatfield, R.D., Piquemal, J., Yahiaoui, N., Pean, M., Lapierre, C., and Boudet, A.M. 1998. NMR characterization of altered lignins extracted from tobacco plants down-regulated for lignification enzymes cinnamylalcohol dehydrogenase and cinnamoyl-CoA reductase. Proc. Natl. Acad. Sci. 95: 12803–12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R.W., Moyano, E., Culianez-Macia, F.A., Schuch, W., Martin, C., and Bevan, M. 1994. A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 13: 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablowski R.W., Baulcombe, D.C., and Bevan, M. 1995. Expression of a flower-specific Myb protein in leaf cells using a viral vector causes ectopic activation of a target promoter. Proc. Natl. Acad. Sci. 92: 6901–6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Sentoku, N., Miura, Y., Hirochika, H., Kitano, H., and Matsuoka, M. 1999. Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J. 18: 992–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H.M., Boschke, I., and Hake, S. 2002. Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc. Natl. Acad. Sci. 99: 9579–9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnone L., Merida, A., Parr, A., Mackay, S., Culianez-Macia, F.A., Roberts, K., and Martin, C. 1998. The AmMYB308 and AmMYB330 transcription factors from antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10: 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiantis M. 2001. Control of shoot cell fate: Beyond homeoboxes. Plant Cell 13: 733–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venglat S.P., Dumonceaux, T., Rozwadowski, K., Parnell, L., Babic, V., Keller, W., Martienssen, R., Selvaraj, G., and Datla, R. 2002. The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. 99: 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht E., Reiser, L., and Hake, S. 2000. Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127: 3161–3172. [DOI] [PubMed] [Google Scholar]
- Zhong R., Ripperger, A., and Ye, Z.H. 2000. Ectopic deposition of lignin in the pith of stems of two Arabidopsis mutants. Plant Physiol. 123: 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]