Abstract
The fact that adenoviral vectors activate innate immunity and induce type I IFNs has not been fully appreciated in the context of cancer gene therapy. Type I IFNs influence different aspects of human immune response and are believed to be crucial for efficient tumor rejection. We performed transcriptional profiling to characterize the response of cutaneous lymphomas to intralesional adenovirus-mediated IFN-γ (Ad-IFN-γ) gene transfer. Gene expression profiles of skin lesions obtained from 19 cutaneous lymphoma patients before and after treatment with Ad-IFN-γ revealed a distinct gene signature consisting of IFN-γ– and numerous IFN-α–inducible genes (type II– and type I–inducible genes, respectively). The type I IFN response appears to have been induced by the vector itself, and its complexity, in terms of immune activation, was potentiated by the IFN-γ gene insert. Intralesional IFN-γ expression together with the induction of a combined type I/II IFN response to Ad-IFN-γ gene transfer seem to underlie the objective (measurable) clinical response of the treated lesions. Biological effects of type I IFNs seem to enhance those set in motion by the transgene, in our case IFN-γ. This combination may prove to be of therapeutic importance in cytokine gene transfer using Ads.
Introduction
Primary cutaneous lymphomas (CLs) represent the second most common extranodal group of non-Hodgkin lymphomas and are characterized by an accumulation of clonal T or B lymphocytes preferentially homing to the skin (1). Most CLs are indolent diseases with good prognosis that display a chronic course, sometimes progressing over decades. This in turn creates an increasing need for treatment modalities that would effectively control the disease for a longer time period together with showing good tolerability. Immunotherapy is one such solution that currently belongs to the standard therapeutic armamentarium. The use of recombinant cytokines has found its rationale in the immunobiology of this disease group and aims to enhance antitumor immunity (1, 2). As an alternative to recombinant cytokines that are often associated with significant systemic side effects, we recently described adenovirus-mediated (Ad-mediated) delivery of human IFN-γ gene directly into CL lesions, showing good tolerability together with promising efficacy (3). Adenoviral vectors belong to the most frequently used gene delivery systems due to their ability to transduce a broad spectrum of cells and package large sequences and their low production costs (4). The fact that they do not integrate into the host genome has made them attractive in cancer immunotherapy, where only transient expression of the delivered gene is required. Even though the use of adenoviral vectors is associated with the activation of cellular immunity, their potential to activate innate immunity per se, thereby inducing type I IFNs such as IFN-α and -β (reviewed in refs. 4–6), has not been fully appreciated in the context of cancer gene therapy in humans. In addition to IFN-γ, type I IFNs are considered to have a crucial role in tumor rejection by the host immune system (7). The beneficial effect of IFN-α on antitumor immunity is easily appreciated in its frequent use in the treatment of different solid and hematologic malignancies, including CL (1, 2, 7).
Our current study sought to investigate the mechanisms involved in the regression of CL lesions treated with adenoviral IFN-γ gene transfer using high-throughput gene expression profiling. We show that induction of combined type I (IFN-α) and type II (IFN-γ) IFN response in treated lesions is predictive of objective (measurable) clinical response later in the course of the treatment.
Results
Type I and type II IFN signature in lesions treated with Ad-IFN-γ.
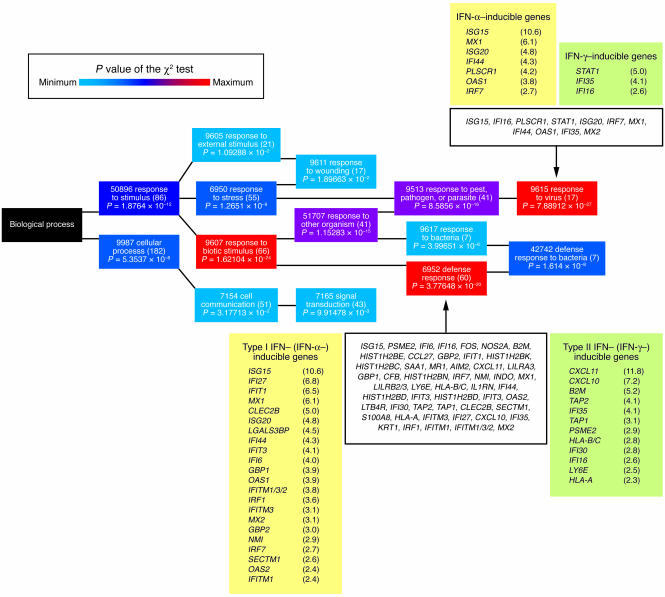
To investigate any changes induced by intralesional Ad-mediated IFN-γ (Ad-IFN-γ) gene transfer on the transcriptional profile of CL skin lesions, we performed gene expression profiling on specimens of lesional skin obtained from 19 patients before the first (baseline) and 24 hours after the third injection of Ad-IFN-γ (“posttreatment 3”). Using our filtering criteria (see Methods), we found 495 genes that were differentially expressed in response to intralesional treatment with Ad-IFN-γ. Of these, the expression of 251 (51%) genes was increased, and expression of 244 (49%) genes was decreased. To facilitate the analysis of genes by accounting for differences in overall gene expression, each gene was assigned to a defined functional category according to available annotations (gene ontology mining using biological processes at NetAffx). In the group of genes with increased expression, 213 genes of 251 uploaded probe sets could be successfully annotated using the NetAffx databank. Roughly one-third of these genes (60; 28%) was involved in the defense response, representing, at the same time, 1 of 2 gene groups with high grouping probability (χ2 test, P = 3.78 × 10–20) (Figure 1). The other gene group with the highest grouping probability included 17 genes characterizing the response to virus (χ2 test, P = 7.89 × 10–27). Of 244 genes with decreased expression, 196 had annotated functions, with preferential involvement of genes regulating various metabolic and physiological processes (χ2 test, P > 0.05). We then focused on genes whose expression could be associated with the intralesional IFN-γ gene delivery. In this context, we observed upregulation of several genes that are known to be directly induced by IFN-γ, such as CXCL10/11, IFI16, IFI30/35, B2M, HLA-A/B/C, TAP1/2, as well as different (immuno)proteasome subunits (PSMA1/3, PSMC1/2/6, PSME2). In addition to IFN-γ–inducible genes, there was also a very prominent upregulation of numerous IFN-α–stimulated genes (also known as ISGs), such as ISG15/20, myxovirus resistance 1/2 (MX1/2), GBP1/2, OAS1/2, IFIT1/3, IFN-induced transmembrane protein 1/2/3 [IFITM1/2/3], IFI6/27/44, and PLSCR1, that are known to function in the innate immune recognition of viruses (Figure 1). Treatment with Ad-IFN-γ stimulated the expression of these type I and II IFN-inducible genes regardless of the lymphoma type and the disease stage. We also did not observe differences in overall gene expression in treated lesions with respect to the dose of Ad-IFN-γ applied.
Figure 1. Functional annotation of genes upregulated after treatment with Ad-IFN-γ using the GO mining tool (NetAffx).
The GO mining tool maps 213 queried genes and groups them hierarchically according to the GO biological processes in which they participate (GO term), represented by colored rectangles (nodes). The node color represents the value of the c2 test used by the GO mining tool to measure the grouping probability of the queried genes (highest values shown in red and lower values in blue). Node text contains the GO term ID, the GO term title, and the number of queried genes belonging to that GO term shown in parentheses; the value of the c2 test for the respective GO term is shown below. Genes of interest are additionally listed in detail. Genes are identified by their gene symbols and fold change in expression (posttreatment 3 versus baseline), shown in parentheses.
Adenoviral vector accounts for the type I IFN signature.
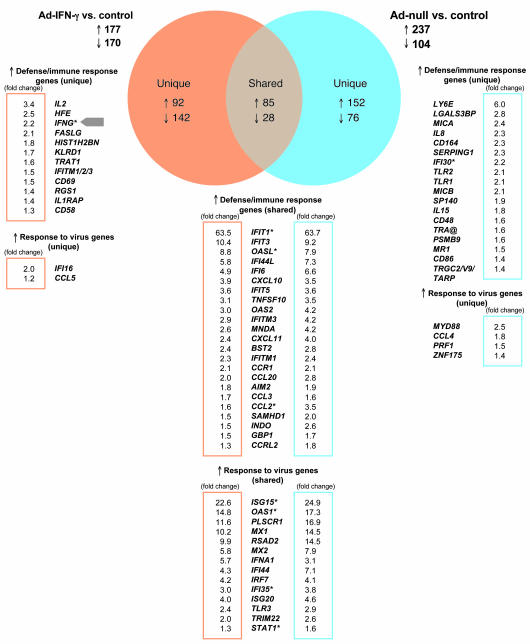
To further dissect the gene expression profile induced by the adenoviral vector itself, we performed transcriptional profiling of human PBMCs exposed to Ad-IFN-γ or to Ad-null (adenoviral vector without IFN-γ insert). A 24-hour exposure to either of the Ad vectors resulted in differential expression of almost the same number of genes: 347 genes (177 upregulated and 170 downregulated) in the case of Ad-IFN-γ versus control and 341 genes (237 upregulated and 104 downregulated) in the case of Ad-null versus control (Figure 2). There were only 113 genes (85 upregulated and 28 downregulated) whose differentially changed expression was shared by Ad-IFN-γ and Ad-null (Figure 2). By looking at the shared genes, we observed that approximately one-half of the shared upregulated genes were involved in defense and immune response to virus (e.g., various IFN-induced protein [IFI] and 2ι-5ι-oligoadenylate synthetase [OAS] isoforms, ISG15/20, MX1/2, RSAD2, IFNA1, IFN regulatory factor 7 [IRF7], TLR3) (Figure 2). Additionally, Ad-IFN-γ uniquely induced genes related to lymphocytes and cellular immunity (e.g., IL2, FASLG, TRAT1, CD69, CCL5), whereas Ad-null seemed to uniquely activate the expression of genes expressed by the innate immune effectors (e.g., IL8, MYD88, MICA/B, CD164, TLR1/2, IL15) (Figure 2). Increases in expression of IFNG gene as well as IFN-γ–inducible IFI16 gene could only be detected upon infection with Ad-IFN-γ.
Figure 2. Genes shared and uniquely expressed by Ad-IFN-γ and/or Ad-null.
Venn diagram represents genes that are shared by or specific for data sets obtained from Ad-IFN-γ–infected PBMCs (orange) and Ad-null infected PBMCs (blue). Upregulated genes related to defense/immune response and response to virus are listed, and fold change values (postinfection versus control) are indicated. Fold change values are displayed in frames (orange for Ad-IFN-γ–induced genes and blue for Ad-null–induced genes). Asterisks indicate genes whose expression levels were obtained using more than 1 probe set on Affymetrix arrays.
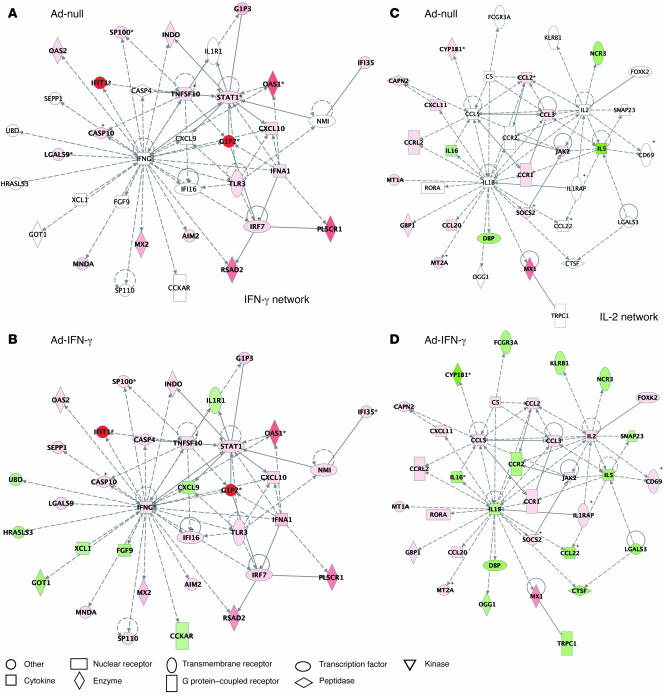
These findings were further confirmed using Ingenuity pathway analysis, which graphically depicts functional interaction of the proteins encoded by the investigated genes. In contrast to Ad-null, Ad-IFN-γ induced not only IFNG and IL2, but also other genes that enabled formation of the complex functional networks linking innate and adaptive immunity (Figure 3). We conclude that the vector itself is responsible for the induction of numerous ISGs observed in the gene profiles in lymphoma lesions treated with Ad-IFN-γ and that IFN-γ insert contributes to the activation of genes related to adaptive cellular immunity.
Figure 3. Ingenuity pathway analysis (IPA) of functional associations between IFN-γ– and IL-2–associated gene networks after Ad-null (A and C) and Ad-IFN-γ infection (B and D).
Each network is graphically displayed with genes/gene products as nodes (different shapes represent the functional classes of the gene products; see Node legend) and the biological relationships between the nodes as edges (lines). The length of an edge reflects the evidence in the literature supporting that node-to-node relationship. The intensity of the node color indicates the degree of up- (red) or downregulation (green) of the respective gene. IPA networks were generated as follows: Upon completed uploading of genes and corresponding fold-change expression values (done separately for Ad-IFN-γ and Ad-null differentially expressed genes), each gene identifier was mapped to its corresponding gene object in the IPA Knowledge Base (part of the IPA algorithm). Fold-change expression values were then used to identify genes whose expression was differentially regulated; these “focus genes” were overlaid onto a global molecular network contained in the IPA Knowledge Base. Networks of these focus genes were then algorithmically generated based on their connectivity and scored according to the number of focus genes within the network as well as according to the strength of their associations. We focused on IFNG- and IL-2–associated networks obtained after Ad-IFN-γ infection. To show the difference in genes expressed after infection with Ad-null, these networks were overlaid with Ad-null data. It is of note that the networks obtained after Ad-null infection lacked expression of several genes (white nodes) differentially regulated by Ad-IFN-γ.
Induction of mixed type I and II IFN signature predicts clinical objective response.
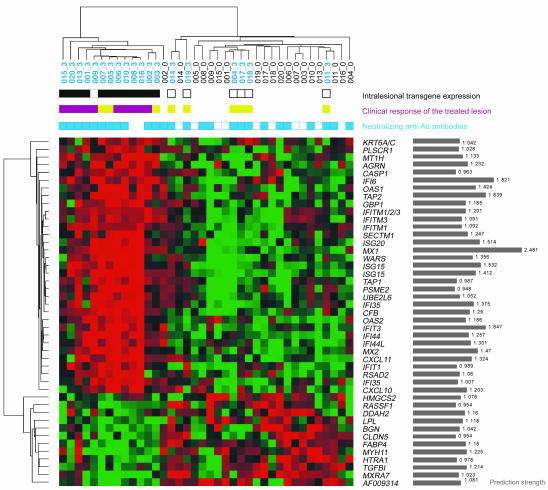
After having elucidated the effects of adenoviral vector and the transgene on overall gene expression, we next sought to identify the genes that could predict clinical objective response (OR) later in the course of treatment. For this purpose we used transcriptional profiles obtained at baseline or 24 hours after the third Ad-IFN-γ injection (day 16 of the treatment cycle), referred to as posttreatment 3. The earliest efficacy evaluation was performed 2 weeks after the posttreatment 3 biopsy for the patients whose tumors completely cleared after 1 treatment cycle; the latest efficacy assessment was performed after 9 months of treatment that followed the posttreatment 3 biopsy (average number of injections per patient, 3; 95% confidence interval, 5–11 injections; n = 19). Baseline samples proved to be uninformative in predicting OR at later points during the treatment. By using posttreatment 3 samples in the training set, on the other hand, we identified a group of 45 genes that could be predictive of an OR of treated lesions with the lowest error rate. To our surprise, the majority of predictor genes was represented by type I and type II IFN–inducible genes (“IFN-signature”), with MX1 having the highest prediction score (Figure 4).
Figure 4. Classification of the skin lesions using OR predictor genes.
The heatmap was obtained by unsupervised hierarchical clustering of the baseline and posttreatment 3 data using 45 predictor genes. The similarity of gene expression profiles among experimental samples is summarized in a dendrogram above the heatmap, in which the pattern and the length of the branches reflect the relatedness of the samples. Black dendrogram branches represent baseline samples; blue branches represent posttreatment 3 samples. Red in the heatmap stands for high relative level of expression; green for low relative level of expression. Genes are identified by their gene symbols (NetAffx). To the right of each gene symbol, the prediction strength of each gene is represented by gray-shaded bars. Positive intralesional transgene expression is shown by black squares, whereas white squares represent no transgene expression. Objective clinical response of the treated lesion is represented by violet squares, whereas lack of OR is represented by yellow squares.
Two-way clustering analysis using these predictor genes revealed the presence of IFN-signature in patients with OR and the absence of IFN-signature in patients without OR, except in patients 003, 005, and 007 (Figure 4). The posttreatment 3 samples from patients whose treated lesions showed no OR (nos. 004, 011, 014, 017, 018, 019) clustered with baseline samples, implying that their gene profile did not change significantly during treatment. Interestingly, 3 of these patients (nos. 004, 011, 014) demonstrated global progressive disease (PD) despite the treatment. None of the samples from patients 004, 011, or 014 had detectable transgene-derived IFN-γ message as determined by real-time PCR (Figure 4). On the other hand, posttreatment 3 samples from 3 patients that lacked OR (nos. 003, 005, 007), and clustered with other posttreatment 3 samples, demonstrated the presence of transgene-derived IFN-γ message by real-time PCR (Figure 4). Only 1 posttreatment sample, from patient 009 who showed an OR, did not reveal intralesional expression of transgene-derived IFN-γ message. This is likely due to the absence of tumor cells from the lesion (infiltrate consisting mainly of inflammatory cells). These results imply that the vector-induced immune stimulation is not enough to induce regression of the lymphoma lesions. Only the combination with intralesional transgene expression appears to induce an efficient immune response reflected in clinical OR. Caution is warranted, however, when interpreting the therapeutic effects in patients with Sézary syndrome (patients 005, 007). Sézary syndrome is an aggressive generalized form of cutaneous T cell lymphoma (CTCL) characterized by whole skin, blood, and lymph node involvement. Immune stimulation via intralesional, local IFN-γ gene delivery in these 2 patients with systemic CTCL might have been insufficient to induce OR, despite the presence of the IFN-signature. Furthermore, the status of neutralizing anti-Ad antibodies either at baseline or after treatment did not have an influence on either transgene IFN-γ expression or clinical response to Ad-IFN-γ.
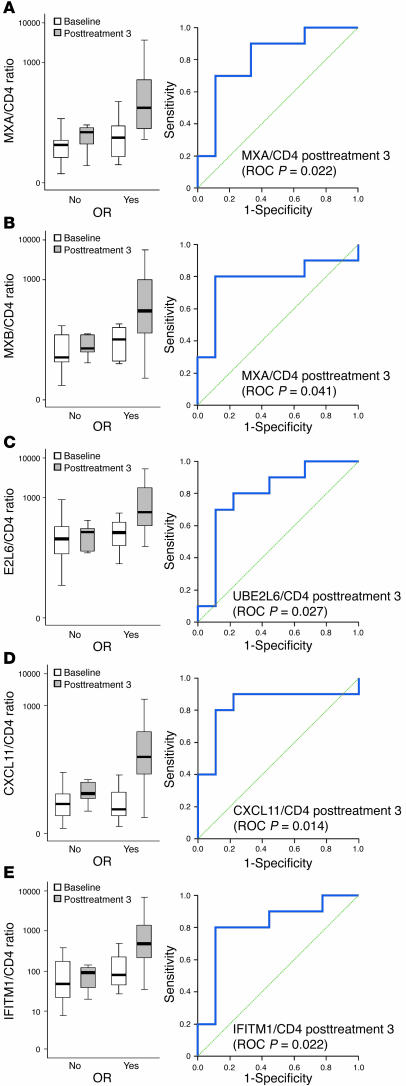
We then focused on verifying the strength of predictor genes to discriminate between patients who will and will not show a clinical OR in the course of treatment by using receiver operating characteristic (ROC) curves. We assessed the expression of several predictor ISGs in biopsies obtained before and after 3 intralesional Ad-IFN-γ injections by real-time PCR (MX1/2, ISG15, UBE2L6, IFITM1, and CXCL11) and by immunohistochemistry (MX1). Type I IFN–induced MX1 and MX2 gene products (MxA and MxB protein, respectively) are large GTPases that are induced in response to viruses (8, 9). Type I IFN–induced ISG15 encodes for an ubiquitin-like protein most highly expressed following viral infection (10, 11). ISG15 is conjugated to cellular proteins by the ubiquitin-conjugating enzyme UBE2L6, which is also inducible by type I IFNs (10, 11). CXCL11 chemokine and IFITM1 transmembrane protein with antiproliferative activity are 2 ISGs that are inducible both by type I IFNs and by IFN-γ (12, 13). Since the normalization to GAPDH lacked predictive value with ROC curve statistics (data not shown), we chose to normalize the mRNA expression levels of the validated predictor genes to CD4 gene expression, since intralesional CD4 count did not appear to change after treatment (3). The GAPDH gene is, on the other hand, known to fluctuate in activated immune cells, which explains its unsatisfactory performance in defining markers that predict OR (14). Expression of the MX1 gene normalized to CD4 gene expression (MX1/CD4 ratio) as well as MxA protein expression in the infiltrate in posttreatment 3 biopsies could easily distinguish between patients with or without OR (P = 0.022 and P = 0.008, respectively) (Figure 5A). CXCL11/CD4, MX2/CD4, IFITM1/CD4, and UBE2L6/CD4 ratio in posttreatment 3 samples performed similarly (P = 0.014, P = 0.041, P = 0.022, and P = 0.027, respectively) (Figure 5, B–E). ISG15 expression was shown to be nondiscriminative in this setting (P = 0.165).
Figure 5. Expression of MX1 (MXA), MX2 (MXB), UBE2L6, CXCL11, and IFITM1 genes that have value in predicting OR.
(A–E) The left panel shows a box plot that represents the summarized expression of the respective gene in baseline and posttreatment 3 samples (y axis) normalized to CD4 and are grouped with respect to OR (x axis). Each box shows the median (thick black line), quartile range (the box itself), and minimum/maximum (whiskers). Outliers (values between 1.5 and 3 box lengths from either end of the box) are denoted by white circles. Extreme values (values that are more than 3 box lengths from either end of the box) are marked by asterisks. The right panel represents the ROC curve generated for the expression of the respective gene at posttreatment 3. For each gene, the P value of area under the curve is shown in parentheses.
Type I IFN resistance in nonresponders.
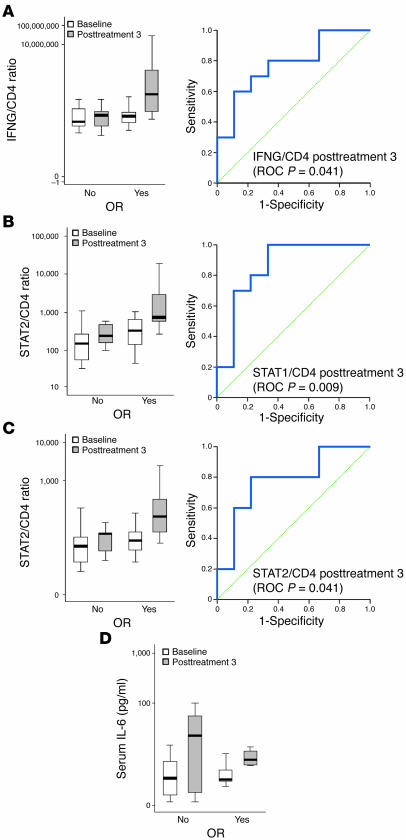
Tumor cells in CTCL have been described to have defects at different levels in type I/II IFN signaling (including altered ISG expression), a phenomenon termed IFN resistance (15–18). Given that all of the patients who did not mount an OR had CTCL, we investigated the expression of an additional panel of genes involved in type I/II IFN signaling (IFN-α/γ, IRF7, STAT1/2, JAK1/2, IFNAR1/2, IFNGR1/2) in baseline and posttreatment 3 skin samples by real-time PCR. Furthermore, we assessed whether the expression level of these genes can predict OR by using ROC curve statistics. Type I IFNs bind a receptor that consists of the subunits IFNAR1 and IFNAR2, which are constitutively associated with JAKs TYK2 and JAK1, respectively (19). Type II IFN (IFN-γ), on the other hand, binds to a receptor that is composed of 2 subunits, IFNGR1 and IFNGR2, which are associated with JAK1 and JAK2, respectively. Upon activation, JAKs phosphorylate STAT1 and STAT2, both of which then associate with IRF9 to form a trimeric complex, ISGF3, that translocates to the nucleus and binds to IFN-stimulated response elements in DNA to initiate transcription of various ISGs. Both type I and II IFNs also induce the formation of STAT1-STAT1 homodimers that translocate to the nucleus and bind to the IFN-γ–activated site element present in the promoter of certain ISGs, thereby initiating their transcription. IRF7 is a key transcription factor for the induction of type I IFNs upon viral challenge, and its expression is in turn inducible by type I IFNs, creating a positive feedback mechanism (20, 21). We first assessed the expression of IFN-α and IFN-γ mRNA in the treated CL lesions at baseline and posttreatment 3. Expression of IFN-α and IFN-γ messages was detectable and increased in posttreatment 3 samples as compared with baseline, even though the difference was statistically insignificant (paired Student’s t test; IFNA/CD4, P = 0.318; IFNG/CD4, P = 0.255; Figure 6A). In terms of predicting OR in the course of the treatment, IFNG/CD4, STAT1/CD4, and STAT2/CD4 expression ratios in posttreatment 3 samples discriminated between patients with and without OR upon Ad-IFN-γ gene delivery (P = 0.041, P = 0.009, and P = 0.041, respectively) (Figure 6, A–C). Moreover, the expression of transgene IFN-γ/CD4 in posttreatment 3 samples was also predictive for OR (P = 0.003). IFNA, IFNAR1/2, IFNGR1/2, JAK1/2, and IRF7 gene expression was not able to discriminate between responding and nonresponding patients (P > 0.05). Together with the findings described above, demonstrating a discriminative expression of several ISGs (MX1, MX2, IFITM1, UBE2L6, CXCL11), these results suggest that tumor cells forming skin lesions in patients with CTCL truly display gene expression alterations consistent with IFN resistance. This phenomenon may therefore underlie the lack of OR observed in these CTCL patients.
Figure 6. Expression of the genes involved in innate immune response to adenoviral vectors.
(A–C) Expression of the genes involved in type I and II IFN signaling that have value in prediction of OR. The left panel shows a box plot that represents the summarized expression of the respective gene in baseline and posttreatment 3 samples (y axis) normalized to CD4 and are grouped with respect to OR (x axis). Right panel represents the ROC curve generated for the expression of the respective gene at posttreatment 3. For each gene, the P value of area under the curve statistic is shown in parentheses. (D) IL-6 concentration in patients’ sera. Values are summarized and shown as box plots that represent serum IL-6 values as pg/ml (y axis) and are grouped according to status of the OR (x axis). For a description of the box plot parameters, see Figure 4.
As production of type I IFNs is triggered by an innate immune response to adenoviral structures, we also assessed the early response to the adenoviral vector by measuring serum IL-6 levels. Increases in circulating proinflammatory cytokines, such as IL-6, have been observed within 6 hours of systemic adenoviral challenge (4–6). There was an increase in serum IL-6 levels in all patients 6 hours following the third Ad-IFN-γ injection as compared with baseline, irrespective of their clinical responsiveness (Figure 6D). Furthermore, all patients revealed peaking of serum IL-6 that was assessed 6 hours after every intralesional Ad-IFN-γ injection, implying the existence of unimpaired innate recognition mechanism(s) (data not shown). As all patients were “recognizing” the vector, we next sought to measure circulating IFN-α, which has also been reported to increase in patients undergoing gene therapy with adenoviral vectors (22). In our patient set, we could not detect any changes in serum IFN-α during the treatment with Ad-IFN-γ (data not shown), suggesting minimal systemic activation induced by Ad-IFN-γ. This is consistent with the lack of systemic toxicity observed after intralesional Ad-IFN-γ administration and supports the safety of this local immunotherapy approach.
Discussion
First-generation adenoviral vectors are considered to be very immunogenic, as they induce potent humoral and cellular responses against various components of the vector (reviewed in refs. 4–6). Such immune responses are associated with the elimination of the vector by neutralizing antibodies and vector-infected cells by cytotoxic cells. One main consequence of the anti-vector immune reaction is the loss of transgene expression and therapeutic efficacy. Anti-vector immunity is therefore considered to hamper systemic gene therapy approaches (reviewed in refs. 4–6). In addition to adaptive immunity, adenoviral vectors also activate innate immunity, leading to the production of type I IFNs. This effect is dependent on the viral vector dose, does not require adenoviral vector internalization/replication/transcription, and is partly dependent on TLR9 (23–26), suggesting the existence of another DNA recognition pathway for Ads (possibly a member of the RIG family; ref. 27). Adenoviral vectors activate DCs, principally plasmacytoid DCs, to produce type I IFNs, which in turn drives their maturation as well as the maturation of conventional, myeloid DCs (22, 28), resulting in enhanced recognition of tumor antigens by tumor-specific lymphocytes (29). This is the reason why this bystander adenoviral activation of the innate immune system is increasingly looked upon as favorable in the context of cancer immunotherapy (5, 6, 30, 31). In this study we demonstrate in vivo induction of type I (vector-dependent) and type II IFN (transgene-dependent) gene signature in skin CL lesions after adenoviral IFN-γ gene transfer using microarray technology and provide evidence supporting the beneficial role of vector-induced innate response in tumor regression.
Almost 2 decades ago, Reich et al. described induction of 3 ISGs, IFIT1, IFIT2, and ISG15, by Ad type 5–derived particles in HeLa cell cultures (32). Since then, high-throughput microarray technology has been employed on several occasions to elucidate transcriptional changes upon infection with adenoviral vectors. It is of note that for this purpose, nonimmune human (e.g., lung cell carcinoma cell line [A549], ref. 33; mammary epithelial cells [HB2], ref. 34; normal lung cells [WI-38], ref. 35; lung fibroblasts [IMR-90], ref. 36) and mouse (e.g., embryo fibroblasts, ref. 33; liver, ref. 25) cells have been used, making the interpretation of the results difficult with respect to the activation of innate immunity. Similar to our data obtained with Ad-IFN-γ in vivo and Ad-IFN-γ and Ad-null in vitro, different type 5 replication–deficient adenoviral vectors induced numerous genes, such as GBP1/2, IFIT1/2/3, IRF7, MYD88, OAS1, LGAL3BP, FOS, NMI, B2M, and TAP1 (among which were many ISGs), reflecting the activation of innate immunity and induction of antiviral program (33–36). Moreover, gene expression profiling of PBMCs from individuals with febrile respiratory illness caused by wild-type replicating Ad revealed upregulation of the same ISGs, such as IFI27/35/44, IFIT1, OAS1/2, GBP1, RSAD2, ISG15, and MX1 (37). Adenovirus-induced changes in the transcriptome appear to primarily depend on the complexity of the innate recognition machinery of a particular cell rather than on the cell type itself. Adenovirus-induced gene expression profiles, described herein and by others, show a striking resemblance to the profiles we obtained after stimulation with TLR7 ligands (38, 39). This further supports the concept of TLR triggering upon Ad recognition (TLR7 is another intracellular virus sentinel and uses the same signaling pathway as TLR9 to activate production of proinflammatory cytokines) (26). Furthermore, TLR triggering in humans, as observed in the current and our previous studies, induces TLR agonist–specific transcriptional profiles that are independent of the underlying tumor biology (38, 39). Furthermore, the dose of injected Ad-IFN-γ particles did not appear to have an influence on the intensity/complexity of the transcriptional response. Apart from defining the profiles induced by the vector backbone, our study shows that the presence of IFN-γ transgene via selective induction of IFNG, IL2, and other proinflammatory genes may additionally enhance and polarize the immune response toward the Th1 arm, which is the key event in the initiation of antitumor immunity.
Our report is the first to our knowledge to show that concomitant immune modulation by the transgene (type II IFN) and by the vector (type I IFNs) appears to underlie the ORs in CL patients treated with cytokine gene transfer. Bystander TLR signaling with subsequent IFN-α production could, in this context, provide locally absent danger signals that would ultimately contribute to abrogation of local immunosuppression, a phenomenon observed in different cancer types, including CL (1, 7, 40, 41). In addition to its antiproliferative and antiangiogenic activity, IFN-α promotes efficient cross-presentation through DC activation, increases cytolytic activity of macrophages and NK cells, primes T cells, and increases their survival (7). CpG oligonucleotides act similarly by mimicking bacterial DNA recognized by TLR9 and are currently used as potent adjuvants in cancer immunotherapy (42). IFN-γ, on the other hand, is a proinflammatory cytokine intimately involved in innate and acquired immune responses. It activates a broad spectrum of cell types and induces a shift toward Th1 phenotype by simultaneously inhibiting the production of immunosuppressive Th2 cytokines (7, 43). Recent reports in the literature seem to corroborate our findings on the favorable effects of TLR triggering and cytokine delivery. Wysocka et al. demonstrate that the use of TLR agonists (inducing type I IFNs) combined with recombinant cytokines (e.g., IFN-γ) leads to potent and broad enhancement of the cellular immune response in CTCL patients (44, 45). Their data imply that there could be a benefit in using a combination of several immunostimulators to achieve clinically significant activation of an antitumor immune response. Furthermore, Sun and Ding describe that MyD88 adaptor protein, in addition to its essential role in TLR signaling, is also active in IFN-γ signaling (46). MyD88 associates with IFNGR1 upon IFN-γ stimulation and stabilizes IFN-γ–induced mRNA transcripts encoding TNF and IFN-γ–inducible protein CXCL10/IP-10 (46). It is conceivable that in this fashion, IFN-γ could enhance the expression of genes encoding for proinflammatory cytokines induced via TLR/NF-κB signaling. IFN-γ is also capable of priming macrophages (main sentinels for adenoviral vectors) for more rapid and heightened responses to TLR ligands as well as increasing cellular sensitivity to type I IFNs (reviewed in ref. 47).
Type I IFNs lead to transcriptional activation/repression of hundreds of ISGs, whose concerted action leads to the generation of the antiviral state (8, 9, 48). ISGs are principally induced through IFN-αR1/2-Jak1/Tyk2-Stat1/2 pathways, even though some ISGs can be induced independently of IFNs. The Mx-resistance genes MX1 and MX2 are among the best characterized ISGs. They encode for Mx proteins, specifically type I IFN–induced large GTPases that inhibit viral replication (8, 9). MxA protein expression has often been used as surrogate marker for the local type I IFN activity in the skin (38, 49, 50). Our current results reveal induction of MX1 and MX2 genes as well as MxA protein expression in CL patients presenting with OR, underlining the role of type I IFNs in the regression of treated lesions. ISG15 is another preferentially type I IFN–induced ISG encoding for IFN-stimulated protein of 15 kDa (10, 11). ISG15 is an ubiquitin-like protein that becomes conjugated to many cellular proteins, a process termed ISGylation. ISGylation of a wide array of cellular proteins, among which several are encoded by ISGs (MxA, GBP1, IFIT1/2/3/5, STAT1), greatly extends the repertoire of cellular functions that are affected by type I IFNs (51). UBE2L6, also known as UbcH8, is a hydrolase responsible for the conjugation of ISG15 to cellular proteins (10, 11). Based on its specific induction by type I IFNs, UBE2L6 expression was able to discriminate between patients with and without OR. ISG15, on the other hand, did not fulfill this criteria, likely due to the nature of its induction through different pathways (JAK/STAT- or IRF3-dependent) (10, 11). CXCL11 was originally identified as an IFN-γ–inducible chemokine that stimulates a chemotactic response in activated T cells (12). To a lesser extent, type I IFNs are also capable of inducing CXCL11 through NF-κB signaling (52). Similar to other IFN-γ–inducible chemokines, CXCL9/MIG and CXCL10/IP-10, CXCL11 displays angiostatic properties. These chemokines, of which we show induction of CXCL10 and CXCL11 upon IFN-γ gene delivery, represent the key players in “immuno-angiostasis,” a connection between Th1 antitumor immunity and inhibition of tumor-induced angiogenesis (53). Furthermore, our data reveal that patients with OR of the treated lesion upregulate CXCL11 gene expression in contrast to nonresponders, implying the importance of this IFN-γ–inducible chemokine in the regression of CL lesions. IFITM1, also known as 9-27 and Leu13, is a member of the IFITM protein family that is responsive to both IFN-α and IFN-γ (13). In lymphoid tissues, IFITM1 is a part of the membrane complex involved in the transduction of antiproliferative and homotypic adhesions signals (54, 55). Yang et al. have recently shown that IFITM1 mediates direct antiproliferative activity of IFN-γ by activating tumor-suppressor gene p53 (56). Similar to previous predictor genes we assessed, IFITM1 expression was preferentially induced in patients that developed OR to local cytokine gene transfer.
Taken together, our results show that the response to this type of immunomodulatory treatment requires a coordinated action of type I and type II IFN–inducible genes, exerting as proteins immunomodulatory, antiangiogenic, and antiproliferative activity and leading to regression of skin CL lesions.
Even though the majority of the CTCL patients and all of the patients with cutaneous B cell lymphoma (CBCL) responded to the treatment with intralesional IFN-γ, some patients with CTCL did not. Tumor cells in CTCL have been shown to have various defects in type I/II IFN signaling and subsequent induction of ISGs (15–18). Downregulation of STAT1 expression is one of the most frequently described defects accounting for resistance to IFN-α and IFN-γ stimulation (15, 16). Our current data are in line with these observations and show lack of STAT1 and STAT2 induction in nonresponder CTCL patients. Despite STAT1 activation by both IFN signaling pathways (19), type II IFN (IFN-γ) is considered to primarily activate STAT1, whereas STAT2 is regarded as the essential component of type I IFN signaling (57). STAT2 associates with STAT1 to form heterodimers that govern the biological responsiveness to type I IFNs (57, 58). STAT1 is currently appreciated as one of the major regulators of innate and adaptive immune responses (7, 58). Moreover, STAT1 has an antioncogenic feature that is partially dependent on p53 (59). Induction of the IFITM1 protein was shown to be dependent on STAT1 and may therefore explain the connection between STAT1 and p53 activated by IFITM1 (56). Through altered expression of these key regulatory factors, tumor cells in patients with CTCL are rendered unresponsive to complex immunomodulatory and antiproliferative effects of type I and II IFNs, which may explain the lack of clinical response by those patients observed in our study.
The use of first-generation adenoviral vectors in systemic gene therapy approaches has been hampered by their inherent immunogenicity (4–6). Despite its presumably negative impact on transgene expression, adenoviral immunogenicity has led to their development as vaccine vehicles for infectious diseases (HIV/AIDS) and cancer, where such immunostimulating properties may be of advantage. Recognition of adenoviral structures leads to systemic production of IL-6, which is largely attributed to the activation of target cells, such as macrophages. IL-6 is therefore often monitored as a marker of systemic toxicity for adenoviral vectors. As any other viral (pathogen) structure, adenoviral vectors also induce production of type I IFNs, which, according to Zhu et al., also contributes to impaired transgene expression (26). In contrast to this finding observed in a mouse system and corroborated by similar reports in the literature (24, 60), Huarte et al. showed recently that type I IFNs do not interfere with transgene expression in both humans and mice either in vitro or in vivo. Furthermore, Huarte et al. provided evidence that vector-induced serum IFN-α activity increases within 24 hours (maximum peak at 6 hours; ref. 61) and is undetectable 24 hours later without affecting intratumoral transgene expression in patients receiving gene therapy (22). Failure to detect IFN-α in patients’ sera following injection in our study may be also attributed to the administration route (minimal spill after intralesional application; ref. 3), short half-life of the IFN-α protein in the circulation (62), and its fast uptake by all nucleated cells expressing the cognate receptor (63, 64). This would leave a very small time window for IFN-α to affect transgene expression significantly and is thus unlikely to play an important role. Despite existing neutralizing anti-Ad antibodies, we show in the current, as well as in our previous study (3), sustained transgene expression in the treated lesion. Furthermore, the majority of our patients revealed intralesional type I IFN activity together with transgene expression, which is in line with observations by Huarte et al. (22). It is of note that the majority of the previous studies investigated the effects of host immunity on transgenes not involved in the immune response, such as different coagulation factors (24, 60) or β-galactosidase (24) in controlled animal systems. We show in our current study that intratumoral IFN-γ gene transfer using an adenoviral vector results in systemic recognition of the vector (as witnessed by serum IL-6 levels) together with local (MxA protein expression) type I IFN response but does not involve systemic type I IFNs affecting transgene expression.
In conclusion, we demonstrate that both the expression of the transgene and the adenoviral vector itself contribute to the regression of skin lesions in CL patients treated with adenoviral IFN-γ gene transfer. IFN-γ seems to primarily affect an IFN-γ–responsive tumor by promoting its immunogenicity. Type I IFNs, on the other hand, exhibit profound antitumor effects by acting on host immune cells (a concept developed by Dunn et al.; ref. 7). Thus, biological effects of type I IFNs induced by the adenoviral vector seem to enhance and complement those set in motion by the IFN-γ transgene. Future studies will provide more information on whether this immunomodulatory combination may have therapeutic advantage in the treatment of CL.
Methods
Study design.
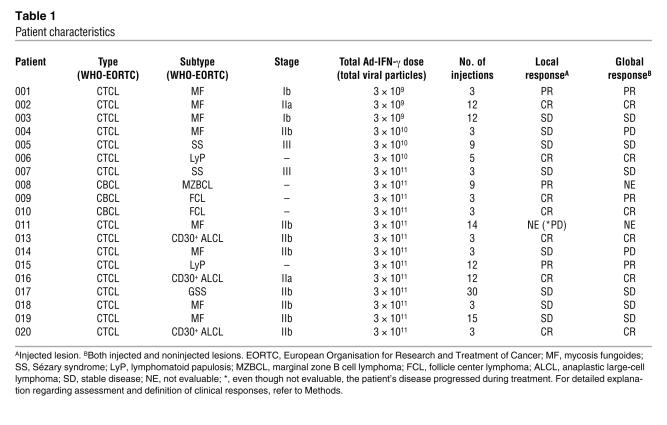
Material for gene expression profiling was available from the first 20 patients, excluding patient 012, enrolled in a phase I/II, open-label trial of repeated intratumoral administration of TG1042 (Ad-IFN-γ) in patients with advanced CTCL–mycosis fungoides and other CTCL- and multilesional CBCLs, supported by Transgene S.A. (3). The study was approved by the institutional ethical committee of the University Hospital Zurich, the Swiss Agency for Therapeutic Products (Swissmedic), and the Swiss Expert Committee for Biosafety (EFBS; formerly SKBS)/Federal Office for the Environment (FOEN). Patient characteristics are presented in Table 1. In order to be eligible for the study, patients had to fulfill various criteria, including histologically proven CTCL or CBCL; performance status of 0, 1, or 2 on the Eastern Cooperative Oncology Group (ECOG) scale; stage Ib or higher; failure of local tumor control by at least 2 first-line treatments; no therapy (systemic or topical) against CTCL and CBCL within 3 weeks preceding the first administration of TG1042; adequate hematologic, hepatic, and renal function; minimum life expectancy longer than 3 months. Prior to entering the study, all patients provided written informed consent.
Table 1 .
Patient characteristics
TG1042 is a nonreplicating (E1 and E3 regions deleted) Ad type 5 (group C) vector containing the human IFN-γ cDNA insert under the control of a CMV promoter. Patients received intratumoral TG1042 injections into the designated lesion(s) once weekly for 3 weeks with no injection in the fourth week (i.e., 1 treatment cycle). The treatment cycle was repeated thereafter if there was no evidence of PD. Information regarding the TG1042 concentrations that patients received is shown in Table 1.
Assessment of the clinical response to TG1042 (Ad-IFN-γ).
Initial tumor assessment of the target lesions (treated) as well as other existing lymphoma lesions (nontreated) was performed at a baseline visit that took place 2 weeks before treatment initiation. The lesions were documented using a transparency and by recording lesion size (i.e., longest diameter of the lesion × second-longest lesion diameter perpendicular to the first). The patients were seen for the following clinical tumor assessment 2 weeks after the third injection of the respective treatment cycle (in 4-week intervals). At that point the lesions were measured to record changes in the size of the injected lesion(s) as well as nontreated lesions. In the case of PD (defined as appearance of new lesions; a 25% increase in the size of previously existing lesions; a change from macular to plaque-like or nodular appearance in more than 25% of previously existing lesions; or any evidence for blood/visceral involvement), the treatment was stopped. The patients were treated with Ad-IFN-γ until complete disappearance of the injected lesions or until they received the maximal number of injections allowed by the protocol. Final assessment and definition of the local and global clinical response were made after the treatment with Ad-IFN-γ had been completed (Table 1). Complete response (CR) was defined as a clinical disappearance of the evaluated lesion. Partial response (PR) was defined as a greater than 50% decrease in the size of the evaluated lesion; reversal of more than 50% nodular or plaque-like lesions to macular lesions. Stable disease (SD) was defined as any response that did not meet the criteria for CR, PR, or PD.
Gene expression profiling.
Tissue samples used in this study were obtained before (baseline) and 24 hours after the third Ad-IFN-γ injection (posttreatment 3) from 19 patients, by excisional biopsy, and were immediately snap-frozen in liquid nitrogen. Total RNA was isolated from tissues using TRI zol reagent (Invitrogen). Double-stranded cDNA was generated with a cDNA synthesis kit (MessageAmp; Ambion) and a (dT)24 primer containing a T7 RNA polymerase promoter at the 3ι end. Labeled complementary RNA (cRNA) was prepared from double-stranded cDNA by in vitro transcription with a T7 polymerase in a reaction mixture containing biotin-11-CTP and biotin-16-UTP (Enzo Diagnostics). cRNA was purified with RNA clean-up columns according to the manufacturer’s instructions (MessageAmp kit; Ambion). Biotinylated cRNA (15 μg) was fragmented and hybridized to HG-U95Av2 GeneChip arrays (Affymetrix) containing probe sets representing approximately 12,000 genes. Chips were hybridized, washed, and stained according to protocols provided by Affymetrix. The overall fluorescence intensity was scaled to a global intensity of 500 to enable the comparison between chips.
Data analysis.
Raw data were imported into GeneSpring GX Expression Analysis software (version 7.3.1; Silicon Genetics). Normalization of the raw data was performed as follows: each signal measurement was divided by the 50th percentile of all measurements in that sample; the signal for each gene was divided by the median of its measurements in all samples; if the median of the raw signal values was below 10, then each measurement for that gene was divided by 10, if the numerator was above 10, otherwise the measurement was not taken into account; values below 0.001 were set to 0.001; posttreatment 3 samples were normalized to respective baseline samples; each measurement for each gene in post treatment 3 samples was divided by the median of that gene’s measurements in the corresponding baseline samples. Median polishing was done, wherein each chip was normalized to its median and each gene was normalized to its median. This was repeated until the medians converged. Prior to median polishing, raw values below 10 were replaced with 10.
The following criteria were used to select differentially regulated genes in posttreatment 3 versus baseline specimens: (a) a nonparametric test with Benjamini-Hochberg multiple-testing correction and a statistical significance level at P < 0.05; (b) a detection “present” call that coincided with the regulation in question — e.g., if a candidate gene was called “upregulated” in posttreatment 3 samples, at least 1 of the samples in the posttreatment 3 group had to be called present; and, vice versa, if a gene was called “downregulated,” at least 1 of the samples in the baseline group had to be called present. The selected differentially expressed genes were normalized to a mean of 0 and a standard deviation of 1, log2-transformed, and subjected to average-linkage hierarchical clustering by use of the uncentered Pearson similarity matrix. Clustering analysis was performed with the CLUSTER program, and the display of the rearranged data was generated with the TreeView program (both programs are available through http://rana.lbl.gov). The selected genes were annotated and classified using NetAffx Internet analysis system provided by Affymetrix (65). Functional associations between differentially expressed genes were analyzed using Ingenuity Pathways Analysis (IPA), version 4.0 (Ingenuity Systems). Complete microarray data sets are available online (see Supplemental Data 1 and 2; available online with this article; doi:10.1172/JCI32077DS1).
To identify genes that may predict objective (measurable) response beyond the first treatment cycle, support vector machine (SVM) algorithm (66) integrated in GeneSpring software was used. The training set comprised either baseline or posttreatment 3 data from patients 004, 011, 014, 019 (no OR) and from patients 006, 008, 010, 016 (OR). The authors decided to assess the objective (measurable) versus lack of OR situation, given the purely quantitative but not biological nature of differences between PR and CR. Patients with PR or CR represented the group with OR, whereas patients with SD or PD represented the group with no OR. From 495 differentially expressed genes that were included initially in the class prediction analysis, 1 group of genes was first selected for the subsequent analysis because it had large between-group variability and small within-group variability. Second and finally, a set of predictor genes was defined based on the lowest prediction error rate obtained with both training and test sets (including both baseline and posttreatment 3 samples from 19 patients) using either only baseline or posttreatment 3 samples.
Real-time quantitative PCR.
Approximately 1 μg of total RNA was reverse transcribed with the First Strand cDNA Synthesis Kit for real-time PCR (Roche Diagnostics) at 42°C for 90 minutes. PCR amplifications were carried out with the HotStart system (LightCycler FastStart DNA Master SYBR Green I; Roche Diagnostics) in the LightCycler thermocycler (Roche Diagnostics). Primers and standards for MX1 and IRF7 (38), IFN signaling (67), and transgene-derived IFN-γ (3) were described previously; IFN-α, IFN-γ, MX2, CXCL11, ISG15, UBE2L6, IFITM1, and CD4 ready-to-use primer sets were purchased from Search LC.
Immunohistochemistry.
Paraffin-embedded tissue sections were stained with anti-MxA protein antibody (clone CL143), a kind gift from Jovan Pavlovic, University of Zurich (68). After antigen retrieval, immunohistochemistry was performed using the alkaline phosphatase–anti-alkaline phosphatase technique, as previously described (3).
In vitro exposure of PBMCs to adenoviral vectors.
Human PBMCs from 3 donors were cultured at 37°C with 5% CO2 in flat-bottomed 6-well culture plates (Nalge Nunc International) at a concentration of 3 × 106 cells per well in 1 ml of DMEM (Invitrogen) supplemented with 10% inactivated FBS (Sigma-Aldrich). After 16 hours of culture, nonadherent cells were infected with Ad-IFN-γ or with the same construct without the IFN-γ insert (Ad-null). All experiments were performed at an MOI of 100 by incubating cells with adenoviral vectors in 100 ml of PBS for 30 minutes (Sigma-Aldrich) supplemented with 2% inactivated FBS and 1% cations. Cells were then plated in complete fresh medium supplemented with 3% inactivated FBS for 24 hours at 37°C with 5% CO2. Control PBMC samples were treated similarly but without exposure to either of the vectors. Cells were then harvested, washed in PBS, and recovered in RNA NOW (Ozyme) at a final concentration of 1 × 106 cells/ml. Total RNA obtained from these samples (Ad-IFN-γ, Ad-null, control) was used to generate cDNA and was hybridized to HG-U95Av2 GeneChip arrays using the methodology described above (see Gene expression profiling). Raw data were normalized using GeneSpring and analyzed for differentially expressed genes between control and Ad-IFN-γ, control and Ad-null, and Ad-null and Ad-IFN-γ using Student’s t test at statistical significance level of P < 0.05. Generated lists of differentially expressed genes were then imported into a significance analysis of microarrays (SAM) application (http://www-stat.stanford.edu/~tibs/SAM) (69) and controlled for the false discovery rate using the default parameters (δ value was adjusted to obtain a maximum number of genes, while maintaining a conservative false discovery rate of 0).
Serum cytokine levels and neutralizing anti-Ad antibodies.
IL-6 levels in patients’ sera were measured by an enzyme immunoassay, as previously described (3). Patients’ sera were collected for IL-6 measurement at baseline/prior to injection and 6 hours after the injection.
Serum IFN-α concentration was assayed by ELISA (Bender MedSystems GmbH). To detect neutralizing anti-Ad antibodies, we assessed dilutions of patients’ sera for their ability to block in vitro cellular infection by infectious Ad particles, as previously described (70). Serum samples at baseline and at day 29 (tumor assessment) were available for IFN-α and neutralizing anti-Ad antibody titer measurement.
Statistics.
Statistical analysis was performed with the SPSS software package, version 12.0. The strength of a defined gene predictor to differentiate between patients with and without OR was evaluated using ROC curves. Statistical differences between baseline and posttreatment 3 samples were assessed using paired 2-tailed Student’s t test. P values of less than 0.05 were considered statistically significant. Box plots and ROC curve graphs were generated using SPSS.
Supplementary Material
Acknowledgments
The authors would like to thank Monika Lusky for critical review of the manuscript.
Footnotes
Nonstandard abbreviations used: Ad, adenovirus; CBCL, cutaneous B cell lymphoma; CL, cutaneous lymphoma; CR, complete response; CTCL, cutaneous T cell lymphoma; IFI, IFN-induced protein; IFITM1, IFN-induced transmembrane protein 1; IRF7, IFN regulatory factor 7; ISG, IFN-stimulated gene; Mx, myxovirus resistance; OAS, 2ι-5ι-oligoadenylate synthetase; OR, objective response; PD, progressive disease; PR, partial response; ROC, receiver operating characteristic.
Conflict of interest: This work was supported by Transgene SA. B. Calmels and B. Acres are employees of Transgene SA.
Citation for this article: J. Clin. Invest. 117:2834–2846 (2007). doi:10.1172/JCI32077
Kazuyasu Fujii’s present address is: Department of Dermatology, Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama, Japan.
References
- 1.Dummer R., Cozzio A., Urosevic M. Pathogenesis and therapy of cutaneous lymphomas-progress or impasse? Exp. Dermatol. 2006;15:392–400. doi: 10.1111/j.0906-6705.2006.00419.x. [DOI] [PubMed] [Google Scholar]
- 2.Dummer R., et al. Standard and experimental therapy in cutaneous T-cell lymphomas. J. Cutan. Pathol. 2006;33(Suppl. 1):52–57. doi: 10.1111/j.0303-6987.2006.00497.x. [DOI] [PubMed] [Google Scholar]
- 3.Dummer R., et al. Adenovirus-mediated intralesional interferon-gamma gene transfer induces tumor regressions in cutaneous lymphomas. Blood. 2004;104:1631–1638. doi: 10.1182/blood-2004-01-0360. [DOI] [PubMed] [Google Scholar]
- 4.McConnell M.J., Imperiale M.J. Biology of adenovirus and its use as a vector for gene therapy. Hum. Gene Ther. 2004;15:1022–1033. doi: 10.1089/hum.2004.15.1022. [DOI] [PubMed] [Google Scholar]
- 5.Bessis N., GarciaCozar F.J., Boissier M.C. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther. 2004;11(Suppl. 1):S10–S17. doi: 10.1038/sj.gt.3302364. [DOI] [PubMed] [Google Scholar]
- 6.Muruve D.A. The innate immune response to adenovirus vectors. Hum. Gene Ther. 2004;15:1157–1166. doi: 10.1089/hum.2004.15.1157. [DOI] [PubMed] [Google Scholar]
- 7.Dunn G.P., Koebel C.M., Schreiber R.D. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 8.Katze M.G., He Y., Gale M., Jr. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 9.Kunzi M.S., Pitha P.M. Interferon targeted genes in host defense. Autoimmunity. 2003;36:457–461. doi: 10.1080/08916930310001605855. [DOI] [PubMed] [Google Scholar]
- 10.Ritchie K.J., Zhang D.E. ISG15: the immunological kin of ubiquitin. Semin. Cell Dev. Biol. 2004;15:237–246. doi: 10.1016/j.semcdb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Martensen P.M., Justesen J. Small ISGs coming forward. J. Interferon Cytokine Res. 2004;24:1–19. doi: 10.1089/107999004772719864. [DOI] [PubMed] [Google Scholar]
- 12.Cole K.E., et al. Interferon-inducible T cell alpha chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J. Exp. Med. 1998;187:2009–2021. doi: 10.1084/jem.187.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaffe E.A., et al. IFN-gamma and IFN-alpha induce the expression and synthesis of Leu 13 antigen by cultured human endothelial cells. J. Immunol. 1989;143:3961–3966. [PubMed] [Google Scholar]
- 14.Bas A., Forsberg G., Hammarstrom S., Hammarstrom M.L. Utility of the housekeeping genes 18S rRNA, beta-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand. J. Immunol. 2004;59:566–573. doi: 10.1111/j.0300-9475.2004.01440.x. [DOI] [PubMed] [Google Scholar]
- 15.Dummer R., et al. Interferon resistance of cutaneous T-cell lymphoma-derived clonal T-helper 2 cells allows selective viral replication. Blood. 2001;97:523–527. doi: 10.1182/blood.v97.2.523. [DOI] [PubMed] [Google Scholar]
- 16.Sun W.H., et al. Interferon-alpha resistance in a cutaneous T-cell lymphoma cell line is associated with lack of STAT1 expression. Blood. 1998;91:570–576. [PubMed] [Google Scholar]
- 17.Tracey L., et al. Transcriptional response of T cells to IFN-alpha: changes induced in IFN-alpha-sensitive and resistant cutaneous T cell lymphoma. J. Interferon Cytokine Res. 2004;24:185–195. doi: 10.1089/107999004322917034. [DOI] [PubMed] [Google Scholar]
- 18.Tracey L., et al. Identification of genes involved in resistance to interferon-alpha in cutaneous T-cell lymphoma. Am. J. Pathol. 2002;161:1825–1837. doi: 10.1016/s0002-9440(10)64459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 20.Honda K., et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 21.Kawai T., et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 2004;5:1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- 22.Huarte E., et al. Recombinant adenoviral vectors turn on the type I interferon system without inhibition of transgene expression and viral replication. Mol. Ther. 2006;14:129–138. doi: 10.1016/j.ymthe.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 23.Li S., et al. Induction of IFN-regulated factors and antitumoral surveillance by transfected placebo plasmid DNA. Mol. Ther. 2005;11:112–119. doi: 10.1016/j.ymthe.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 24.Cerullo V., et al. Toll-like receptor 9 triggers an innate immune response to helper-dependent adenoviral vectors. Mol. Ther. 2007;15:378–385. doi: 10.1038/sj.mt.6300031. [DOI] [PubMed] [Google Scholar]
- 25.Hartman Z.C., et al. Adenovirus infection triggers a rapid, MyD88 regulated, transcriptome response critical to acute phase and adaptive immune responses in vivo. J. Virol. 2007;81:1796–1812. doi: 10.1128/JVI.01936-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu J., Huang X., Yang Y. Innate immune response to adenoviral vectors is mediated by both TLR-dependent and -independent pathways. J. Virol. 2007;81:3170–3180. doi: 10.1128/JVI.02192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner H., Bauer S. All is not Toll: new pathways in DNA recognition. J. Exp. Med. 2006;203:265–268. doi: 10.1084/jem.20052191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hensley S.E., Giles-Davis W., McCoy K.C., Weninger W., Ertl H.C. Dendritic cell maturation, but not CD8+ T cell induction, is dependent on type I IFN signaling during vaccination with adenovirus vectors. J. Immunol. 2005;175:6032–6041. doi: 10.4049/jimmunol.175.9.6032. [DOI] [PubMed] [Google Scholar]
- 29.Geutskens S.B., et al. Recombinant adenoviral vectors have adjuvant activity and stimulate T cell responses against tumor cells. Gene Ther. 2000;7:1410–1416. doi: 10.1038/sj.gt.3301251. [DOI] [PubMed] [Google Scholar]
- 30.Molinier-Frenkel V., et al. Adenovirus hexon protein is a potent adjuvant for activation of a cellular immune response. J. Virol. 2002;76:127–135. doi: 10.1128/JVI.76.1.127-135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruzek M.C., Kavanagh B.F., Scaria A., Richards S.M., Garman R.D. Adenoviral vectors stimulate murine natural killer cell responses and demonstrate antitumor activities in the absence of transgene expression. Mol. Ther. 2002;5:115–124. doi: 10.1006/mthe.2002.0529. [DOI] [PubMed] [Google Scholar]
- 32.Reich N., Pine R., Levy D., Darnell J.E., Jr. Transcription of interferon-stimulated genes is induced by adenovirus particles but is suppressed by E1A gene products. J. Virol. 1988;62:114–119. doi: 10.1128/jvi.62.1.114-119.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartman Z.C., Black E.P., Amalfitano A. Adenoviral infection induces a multi-faceted innate cellular immune response that is mediated by the toll-like receptor pathway in A549 cells. Virology. 2007;358:357–372. doi: 10.1016/j.virol.2006.08.041. [DOI] [PubMed] [Google Scholar]
- 34.Scibetta A.G., Copier J., Barrett A., Chaplin T., Taylor-Papadimitriou J. Gene expression changes induced by a recombinant E1-/E3- adenovirus type 5 vector in human mammary epithelial cells. Intervirology. 2005;48:350–361. doi: 10.1159/000086062. [DOI] [PubMed] [Google Scholar]
- 35.Rao X.M., et al. Gene expression profiles of normal human lung cells affected by adenoviral E1B. Virology. 2006;350:418–428. doi: 10.1016/j.virol.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 36.Stilwell J.L., McCarty D.M., Negishi A., Superfine R., Samulski R.J. Development and characterization of novel empty adenovirus capsids and their impact on cellular gene expression. J. Virol. 2003;77:12881–12885. doi: 10.1128/JVI.77.23.12881-12885.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thach D.C., et al. Surveillance of transcriptomes in basic military trainees with normal, febrile respiratory illness, and convalescent phenotypes. Genes Immun. 2005;6:588–595. doi: 10.1038/sj.gene.6364244. [DOI] [PubMed] [Google Scholar]
- 38.Urosevic M., et al. Disease-independent skin recruitment and activation of plasmacytoid predendritic cells following imiquimod treatment. J. Natl. Cancer Inst. 2005;97:1143–1153. doi: 10.1093/jnci/dji207. [DOI] [PubMed] [Google Scholar]
- 39.Urosevic M., et al. Imiquimod treatment induces expression of opioid growth factor receptor: a novel tumor antigen induced by interferon-alpha? Clin. Cancer Res. 2004;10:4959–4970. doi: 10.1158/1078-0432.CCR-04-0193. [DOI] [PubMed] [Google Scholar]
- 40.Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat. Rev. Cancer. 2005;5:263–274. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- 41.Tsan M.F. Toll-like receptors, inflammation and cancer. Semin. Cancer Biol. 2006;16:32–37. doi: 10.1016/j.semcancer.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 42.Killeen S.D., Wang J.H., Andrews E.J., Redmond H.P. Exploitation of the Toll-like receptor system in cancer: a doubled-edged sword? Br. J. Cancer. 2006;95:247–252. doi: 10.1038/sj.bjc.6603275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schroder K., Hertzog P.J., Ravasi T., Hume D.A. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 44.Wysocka M., et al. Enhancement of the host immune responses in cutaneous T-cell lymphoma by CpG oligodeoxynucleotides and IL-15. Blood. 2004;104:4142–4149. doi: 10.1182/blood-2004-03-1190. [DOI] [PubMed] [Google Scholar]
- 45.Wysocka M., et al. Imidazoquinolines are toll receptor agonists which potently and broadly enhance the cellular immune response of immunodepressed cutaneous T-cell lymphoma patients: synergy with interferon gamma and IL-18 [abstract]. J. Invest. Dermatol. 2006;126:38. [Google Scholar]
- 46.Sun D., Ding A. MyD88-mediated stabilization of interferon-gamma-induced cytokine and chemokine mRNA. Nat. Immunol. 2006;7:375–381. doi: 10.1038/ni1308. [DOI] [PubMed] [Google Scholar]
- 47.Schroder K., Sweet M.J., Hume D.A. Signal integration between IFNgamma and TLR signalling pathways in macrophages. Immunobiology. 2006;211:511–524. doi: 10.1016/j.imbio.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 48.Der S.D., Zhou A., Williams B.R., Silverman R.H. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urosevic M., et al. CD4+CD56+ hematodermic neoplasms bear a plasmacytoid dendritic cell phenotype. Hum. Pathol. 2005;36:1020–1024. doi: 10.1016/j.humpath.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 50.Wenzel J., et al. Enhanced type I interferon signalling promotes Th1-biased inflammation in cutaneous lupus erythematosus. J. Pathol. 2005;205:435–442. doi: 10.1002/path.1721. [DOI] [PubMed] [Google Scholar]
- 51.Zhao C., Denison C., Huibregtse J.M., Gygi S., Krug R.M. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10200–10205. doi: 10.1073/pnas.0504754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rani M.R., Hibbert L., Sizemore N., Stark G.R., Ransohoff R.M. Requirement of phosphoinositide 3-kinase and Akt for interferon-beta-mediated induction of the beta-R1 (SCYB11) gene. J. Biol. Chem. 2002;277:38456–38461. doi: 10.1074/jbc.M203204200. [DOI] [PubMed] [Google Scholar]
- 53.Strieter R.M., et al. Cancer CXC chemokine networks and tumour angiogenesis. Eur. J. Cancer. 2006;42:768–778. doi: 10.1016/j.ejca.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 54.Deblandre G.A., et al. Expression cloning of an interferon-inducible 17-kDa membrane protein implicated in the control of cell growth. J. Biol. Chem. 1995;270:23860–23866. doi: 10.1074/jbc.270.40.23860. [DOI] [PubMed] [Google Scholar]
- 55.Evans S.S., Collea R.P., Leasure J.A., Lee D.B. IFN-alpha induces homotypic adhesion and Leu-13 expression in human B lymphoid cells. J. Immunol. 1993;150:736–747. [PubMed] [Google Scholar]
- 56.Yang G., Xu Y., Chen X., Hu G. IFITM1 plays an essential role in the antiproliferative action of interferon-gamma. Oncogene. 2006;26:594–603. doi: 10.1038/sj.onc.1209807. [DOI] [PubMed] [Google Scholar]
- 57.Brierley M.M., Fish E.N. Stats: multifaceted regulators of transcription. J. Interferon Cytokine Res. 2005;25:733–744. doi: 10.1089/jir.2005.25.733. [DOI] [PubMed] [Google Scholar]
- 58.Garcia-Sastre A., Biron C.A. Type 1 interferons and the virus-host relationship: a lesson in detente. Science. 2006;312:879–882. doi: 10.1126/science.1125676. [DOI] [PubMed] [Google Scholar]
- 59.Yoshimura A. Signal transduction of inflammatory cytokines and tumor development. Cancer Sci. 2006;97:439–447. doi: 10.1111/j.1349-7006.2006.00197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dai Y., et al. Cellular and humoral immune responses to adenoviral vectors containing factor IX gene: tolerization of factor IX and vector antigens allows for long-term expression. Proc. Natl. Acad. Sci. U. S. A. 1995;92:1401–1405. doi: 10.1073/pnas.92.5.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Y.J. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 62.Shechter Y., Preciado-Patt L., Schreiber G., Fridkin M. Prolonging the half-life of human interferon-alpha 2 in circulation: design, preparation, and analysis of (2-sulfo-9-fluorenylmethoxycarbonyl)7-interferon-alpha 2. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1212–1217. doi: 10.1073/pnas.98.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Platanias L.C., Fish E.N. Signaling pathways activated by interferons. Exp. Hematol. 1999;27:1583–1592. doi: 10.1016/s0301-472x(99)00109-5. [DOI] [PubMed] [Google Scholar]
- 64.Takaoka A., Yanai H. Interferon signalling network in innate defence. Cell. Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- 65.Liu G., et al. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res. 2003;31:82–86. doi: 10.1093/nar/gkg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brown M.P., et al. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl. Acad. Sci. U. S. A. 2000;97:262–267. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martinelli S., et al. Induction of genes mediating interferon-dependent extracellular traps formation during neutrophil differentiation. J. Biol. Chem. 2004;279:44123–44132. doi: 10.1074/jbc.M405883200. [DOI] [PubMed] [Google Scholar]
- 68.Fah J., Pavlovic J., Burg G. Expression of MxA protein in inflammatory dermatoses. J. Histochem. Cytochem. 1995;43:47–52. doi: 10.1177/43.1.7822763. [DOI] [PubMed] [Google Scholar]
- 69.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slos P., De Meyer M., Leroy P., Rousseau C., Acres B. Immunotherapy of established tumors in mice by intratumoral injection of an adenovirus vector harboring the human IL-2 cDNA: induction of CD8(+) T-cell immunity and NK activity. Cancer Gene Ther. 2001;8:321–332. doi: 10.1038/sj.cgt.7700309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.