Abstract
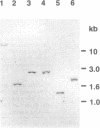
Rhodothermus marinus, a gram-negative heterotrophic marine thermophile, has been the subject of several recent studies. Isolation, sequencing, and analyses of a 16S rRNA gene have shown that R. marinus diverges sharply from major bacterial phyla and is most closely allied to the Flexibacter-Cytophaga-Bacteroides group. Further analyses revealed that the R. marinus chromosome contains a single rRNA operon with a 16S-23S intergenic region coding for tRNA(Ile) and tRNA(Ala).
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achenbach-Richter L., Gupta R., Stetter K. O., Woese C. R. Were the original eubacteria thermophiles? Syst Appl Microbiol. 1987;9:34–39. doi: 10.1016/s0723-2020(87)80053-x. [DOI] [PubMed] [Google Scholar]
- Achenbach-Richter L., Woese C. R. The ribosomal gene spacer region in archaebacteria. Syst Appl Microbiol. 1988;10:211–214. doi: 10.1016/s0723-2020(88)80002-x. [DOI] [PubMed] [Google Scholar]
- Burggraf S., Olsen G. J., Stetter K. O., Woese C. R. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 1992 Aug;15(3):352–356. doi: 10.1016/S0723-2020(11)80207-9. [DOI] [PubMed] [Google Scholar]
- Feingold J., Bellofatto V., Shapiro L., Amemiya K. Organization and nucleotide sequence analysis of an rRNA and tRNA gene cluster from Caulobacter crescentus. J Bacteriol. 1985 Jul;163(1):155–166. doi: 10.1128/jb.163.1.155-166.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GANDELMAN B., ZAMENHOF S., CHARGAFF E. The desoxypentose nucleic acids of three strains of Escherichia coli. Biochim Biophys Acta. 1952 Oct;9(4):399–401. doi: 10.1016/0006-3002(52)90183-2. [DOI] [PubMed] [Google Scholar]
- Garnier T., Canard B., Cole S. T. Cloning, mapping, and molecular characterization of the rRNA operons of Clostridium perfringens. J Bacteriol. 1991 Sep;173(17):5431–5438. doi: 10.1128/jb.173.17.5431-5438.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen M. A., Webster J. A., Straus N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol. 1993 Apr;59(4):945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A. A rate-independent technique for analysis of nucleic acid sequences: evolutionary parsimony. Mol Biol Evol. 1987 Mar;4(2):167–191. doi: 10.1093/oxfordjournals.molbev.a040433. [DOI] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Olsen G. J., Maidak B. L., McCaughey M. J., Overbeek R., Macke T. J., Marsh T. L., Woese C. R. The ribosomal database project. Nucleic Acids Res. 1993 Jul 1;21(13):3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan E. A., Ikemura T., Nomura M. Identification of spacer tRNA genes in individual ribosomal RNA transcription units of Escherichia coli. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2710–2714. doi: 10.1073/pnas.74.7.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefs J. M., Van de Peer Y., De Rijk P., Goris A., De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):1987–2015. doi: 10.1093/nar/19.suppl.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Matsuda H., Hagstrom R., Overbeek R. fastDNAmL: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994 Feb;10(1):41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaiwa F., Sugiura M. Nucleotide sequence of the 16S - 23S spacer region in an rRNA gene cluster from tobacco chloroplast DNA. Nucleic Acids Res. 1982 Apr 24;10(8):2665–2676. doi: 10.1093/nar/10.8.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada A., Ohta H., Kulthanan K., Hiramatsu K. Molecular cloning and mapping of 16S-23S rRNA gene complexes of Staphylococcus aureus. J Bacteriol. 1993 Nov;175(22):7483–7487. doi: 10.1128/jb.175.22.7483-7487.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W. G., Oyaizu Y., Oyaizu H., Woese C. R. Natural relationship between bacteroides and flavobacteria. J Bacteriol. 1985 Oct;164(1):230–236. doi: 10.1128/jb.164.1.230-236.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Gutell R., Gupta R., Noller H. F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983 Dec;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]