Abstract
Factor XI (FXI) is a homodimeric blood coagulation protein. Each monomer comprises four tandem apple-domain repeats (A1–A4) and a serine protease domain. We report here the NMR solution structure of the A4 domain (residues 272–361), which mediates formation of the disulfide-linked FXI dimer. A4 exhibits characteristic features of the plasminogen apple nematode domain family, including a five-stranded β-sheet flanked by an α-helix on one side and a two-stranded β-sheet on the other. In addition, the solution structure reveals a second α-helix at the C terminus. Comparison with a recent crystal structure of full-length FXI, combined with molecular modeling, suggests that the C-terminal helix is formed only upon proteolytic activation. The newly formed helix disrupts interdomain contacts and reorients the catalytic domains, bringing the active sites into close proximity. This hypothesis is supported by small-angle x-ray scattering and electron microscopy data, which indicate that FXI activation is accompanied by a major change in shape. The results are consistent with biochemical evidence that activated FXI cleaves its substrate at two positions without release of an intermediate.
Keywords: blood coagulation, NMR, plasminogen apple nematode domain, small-angle x-ray scattering, EM
Factor XI (FXI) is a blood plasma protein in the intrinsic pathway of blood coagulation. In response to blood vessel injury, thrombin can convert the homodimeric FXI zymogen into its proteolytically active form, FXIa, which in turn cleaves its substrate, factor IX (FIX), resulting in a cascade of events leading to fibrin formation (1, 2). FXI may also play a role in protection of clots from fibrinolysis (3). The enzymatic activators of FXI (thrombin, FXIIa, or FXIa) cleave the Arg-369–Ile-370 bond in each monomer of FXI, yielding FXIa, which consists of a 369-residue heavy chain and a disulfide-linked serine protease domain of 238 residues. A substantial body of evidence indicates that binding interactions outside the protease domain (exosites) are important determinants of ligand and receptor interactions and of substrate affinity and specificity in coagulation reactions involving FXI/XIa (2).
The FXI heavy chain comprises four apple-domain repeats, A1–A4, each of which contains six cysteine residues involved in intradomain disulfide bonds (1, 4). An additional cysteine in A4 (Cys-321) forms a physiologically important disulfide bridge with the corresponding Cys in the other subunit of the FXI dimer. The sequence identity among the four FXI apple domains ranges from 23% to 34%. They are members of the plasminogen apple nematode (PAN) domain family of proteins (5). Various domains of FXI have been modeled by using PAN domain structures as templates (6). However, the model could not predict the quaternary structure of the A4 domain, which mediates dimerization of FXI through both noncovalent interactions as well as an intersubunit disulfide bond (Cys-321–Cys-321′). A recent crystal structure of the FXI zymogen (7) showed that all of the contacts between the two subunits of the dimer are localized to the A4 domain. A single base change in A4 resulting in an F283L mutation is commonly found in FXI-deficient patients (8), who may experience extensive hemorrhage after major trauma (2). The secreted F283L variant has normal enzymatic activity, suggesting that the FXI deficiency associated with this mutation may not be due to a malfunctioning enzyme present in the plasma, but a deficiency in secretion due to misfolding or a defect in dimer formation (9, 10). This conclusion is supported by our recent finding that the F283L mutation alters the packing of aromatic core residues and stabilizes a monomeric form of the isolated A4 domain at the expense of the native dimer (11). On the other hand, high FXI levels may comprise a significant risk factor for thrombosis, making FXI a promising target for antithrombotic therapy (2).
The time course of FXIa-catalyzed activation of FIX in solution, analyzed by SDS/PAGE, showed that cleavage of the two scissile bonds in FIX proceeds without visible accumulation of intermediates, suggesting participation of two active sites (12). The dimeric nature of FXI also has been hypothesized to be critical for normal function of FXIa on the platelet surface (13). This conclusion was based on comparative studies between wild-type FXIa and a monomeric chimera of FXIa with the A4 domain replaced by that of prekallikrein, a homologous monomeric blood protein. Although the solution phase kinetics is similar for both forms, the monomeric chimera was found to be deficient in both clotting and FIX activation assays in the presence of platelets. Thus, structural and functional studies of FXI A4 should further help in understanding the function of FXI.
We report here the NMR solution structure of the covalently linked dimer of the FXI A4 domain (residues 272–361). The structural core of each monomer consists of an α-helix and two antiparallel β-sheets resembling other members of the PAN domain family. In addition, the solution structure of the isolated A4 domain exhibits a well-defined second α-helix at the C terminus, which is not observed in the crystal structure of intact FXI (7). We postulate that formation of this C-terminal helix upon activation of FXI (via cleavage of the Arg-369–Ile-370 peptide bond), along with interactions involving the newly created C terminus at Arg-369, trigger a major rearrangement of the FXI domain structure, including disruption of some of the contacts between apple domains and repositioning of the catalytic domains. Small angle x-ray scattering (SAXS) and EM data confirmed that FXI activation is accompanied by a major transformation from an elongated to a more compact arrangement of the domains. The resulting decrease in the distance between the two catalytic domains of the FXI dimer helps to explain the observation that FXI simultaneously cleaves its physiological substrate, FIX at two different sites without the release of an intermediate (12).
Results and Discussion
NMR Solution Structure of the FXI A4 Domain.
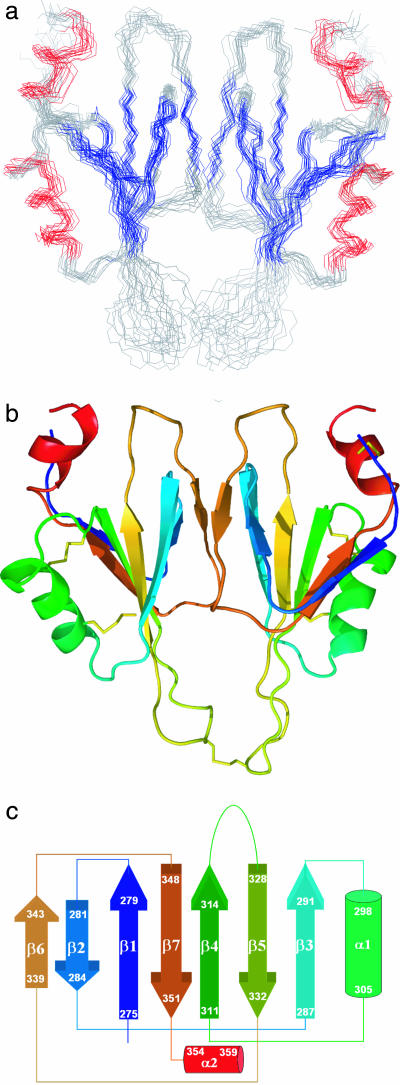
The solution structure of the covalently linked A4 dimer of FXI (residues Phe-272 to Glu-361) was determined by using NMR techniques (Fig. 1). Backbone resonance assignments for all but four residues (N-terminal F272, S320, and C321 and C-terminal Q361) and side chain assignments (93% complete) were obtained by using standard triple-resonance NMR techniques (14–16). A set of unusually weak cross-peaks assigned to residues 316–325 show evidence for exchange broadening and were ultimately found to be part of a mobile loop. Detailed analysis of these cross-peaks revealed two mutually exclusive sets of nuclear Overhauser effects (NOEs) consistent with a predominant structure in which the 316–325 loop is in an extended conformation (Fig. 1) and an alternate more compact conformation [supporting information (SI) Fig. 6]. Structure calculations were carried out by using 1,339 NOE distance constraints and 112 dihedral angle restraints (SI Table 1) derived from the E-COSY (exclusive correlation spectroscopy) experiment or TALOS (torsion angle likelihood obtained from shift) database predictions (17).
Fig. 1.
Solution structure of the A4 dimer. (a) The backbone overlay of 14 low-energy structures of FXI A4. Colored segments indicate β-sheet (blue) and helices (red). (b) Ribbon representation of the two domains of the dimer rendered in colors ranging from blue (N-terminal) to red (C-terminal). (c) Topology diagram of the NMR structure of the FXI A4 domain. The color scheme follows that in b.
The structure was further refined on the basis of 70 residual dipolar coupling (RDC) restraints per monomer (excluding mobile loop residues) measured on a weakly aligned sample in a compressed polyacrylamide gel (18, 19). The refined structure (Fig. 1) is fully consistent with the observed RDC values; a correlation plot of predicted versus observed RDC values yielded a correlation coefficient of 0.9951 and rmsd of 0.48 Hz. Fig. 1a shows backbone traces of 14 low-energy structures from 100 structures calculated on the basis of the experimental restraints (SI Table 1). Fig. 1b shows a ribbon diagram of a representative structure (the one closest to the mean structure). The quality of the structure was assessed by using PROCHECK. Ramachandran plot analysis showed that 58%, 32%, 6%, and 4% of the nonglycine residues are in the most favorable, additionally allowed, generously allowed, and disallowed regions, respectively. The intersubunit NOE constraints were authenticated by using a cross-validation procedure. Several structure calculations were carried out by randomly removing one-third of the interdomain NOE constraints. The results showed a negligible decrease in the precision (rmsd increased from 1.3 to 1.5 for alignment of an individual monomer and from 1.8 to 2.1 for the unaligned subunit of the dimer). Even with a minimal number of interfacial constraints (using only one-third of the NOEs), the overall disposition of the disulfide-bond-linked dimer remains essentially unchanged. Note that in our structure of the isolated A4 domain (Fig. 1), the terminal residues (F272 and E361) are both exposed to the solvent and are far from each other (≈18 Å), ruling out the possibility of interaction between the newly generated N- and C-terminal charges.
The A4 homodimer, covalently linked via the naturally occurring Cys-321–Cys-321′ disulfide bond, exhibits twofold molecular symmetry. Each subunit is stabilized by three intrachain disulfide bonds (273–356, 299–328, and 303–309). The structural core consists of an α-helix and two layers of antiparallel β-sheet containing five and two strands, respectively, consistent with the PAN domain fold (5). However, in contrast to other members of the PAN domain family, the solution structure of A4 contains an extended loop (residues 316–326) connecting strands 4 and 5 of the β-sheet and a second α-helix at the C terminus (residues 354–360). Fig. 1c shows a topology diagram indicating the connectivity of the seven β-strands and two α-helices. Since the secondary structural elements of A4 are stacked in layers, a number of hydrophobic clusters are observed, rather than a single hydrophobic core. The hydrophobic contacts observed between aromatic residues (Phe-312 and Tyr-314) and aliphatic residues (Ala-298, Leu-302, and Leu-330) mediate stacking of the disulfide bonded α-helical segment, α1 (298–305), over the five-strand β-sheet. Similarly, the two-strand β-sheet comprising β2 (281–284) and β6 (339–343) interacts with β1 (275–279) and β7 (348–351) of the five-strand β-sheet, forming a second hydrophobic core. The dimer interface of A4 involves hydrophobic interactions between residues Leu-284, Leu-288, Ile-290, and Leu-342 (SI Fig. 7). Other interactions stabilizing the dimer include a hydrogen bond between the OH groups of Tyr-329 from each monomer and a charge–charge interaction at the periphery of the interface between Lys-340 of one monomer and Glu-286 of the other.
Residues 316–325 of the A4 dimer form a pair of adjacent loops linked at their apex via the Cys-321–Cys-321′ disulfide bond. Loop residues show unusually weak cross-peaks, suggesting that the loop samples alternative conformations on a relatively slow (millisecond) time scale, giving rise to exchange broadening. The presence of conformational heterogeneity was confirmed by NMR measurements as a function of temperatures (SI Fig. 8). The fact that our NMR data recorded at 37°C show only one set of peaks for residues of the mobile loop is consistent with relatively rapid conformational exchange. However, we observed a number of NOEs (SI Table 2) that are inconsistent with a fully extended loop conformation as depicted in Fig. 1. An example of such a loop-in conformation is shown in SI Fig. 6). The mobile loop appears to be a characteristic feature of the A4 domain only. Comparison of the amino acid sequence of this loop region with other apple domains (SI Fig. 7a) reveals that only A4 has a proline residue at the end of β4 and possesses a characteristic turn sequence (–GXGX–) at the beginning of β5. Other apple domains have proline residues involved in reverse turns near the tip of the loop. It is not clear at this time whether these motions have any functional significance other than entropic stabilization of the native dimer.
The C-terminal Helix.
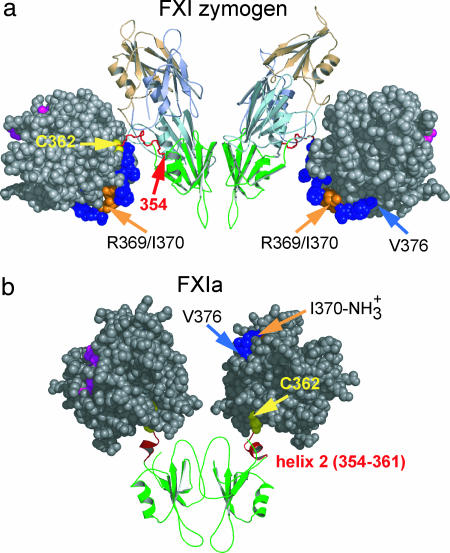
The scissile bond in FXI, Arg-369–Ile-370, which is cleaved to form active enzyme (FXIa) from the inactive zymogen (1, 2), is located in the linker region connecting the A4 and catalytic domains (7). Once activated, the only covalent linkage between the heavy chain of FXIa (residues 1–369) and the catalytic domain (residues 370–607) is through a disulfide bond between Cys-362 and Cys-482 (4). The newly generated NH3+ group of the N-terminal Ile-370 moves over a distance of ≈20 Å into close proximity to the catalytic site and interacts electrostatically with active-site residues, including Asp-194 (20, 21). Although this phenomenon has long been known to play a key role in the activation of serine proteases (22), the fate of the newly created C-terminal end of the heavy chain and its influence on the regulatory domains remain to be elucidated.
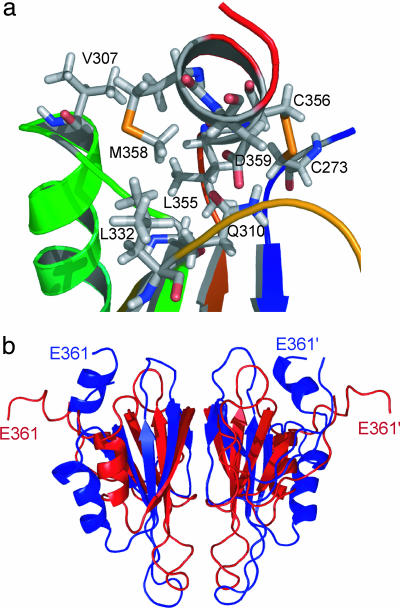
Our solution structure of A4 includes part of the linker region up to Glu-361 preceding the Cys-362–Cys-482 disulfide bond. The C-terminal segment (354–360) forms a well defined α-helix (α2) involved in intimate interactions with the rest of the structure, including several hydrophobic and polar contacts (Fig. 2a). Helix α2 lies across a groove flanked by two loops connecting α1 to β4 and β5 to β6, respectively. This structural feature is in marked contrast to the crystal structure of FXI (7), where the segment connecting the A4 and the catalytic domains (354–362) assumes an extended conformation with some 310-helical character (Fig. 2b, red structure). Superposition of the NMR structure onto the A4 domain of the x-ray structure (using the five-stranded β-sheet core as a reference) shows that the C-terminal Glu-361 has moved by ≈25 Å toward the twofold symmetry axis of the dimer (Fig. 2b). Moreover, helix α1 has moved away from the five-stranded β-sheet to accommodate the C-terminal helix, whereas the β-sheet core and the dimer interface remain intact. Many of the residues that contact the C-terminal helix in the NMR structure of the isolated A4 domain (V307, R308, Q310, and N332–N335) are engaged in interactions with the A1 domain in the crystal structure of whole FXI (7). Thus, formation of a second α-helix at the C terminus of A4 is incompatible with the native domain structure of the zymogen. Given the large number of contacts and intricate network of interactions that anchor the C-terminal helix to the body of the A4 domain (Fig. 2a), this is likely to be an evolved structural feature rather than a consequence of isolating the A4 dimer from the context of full-length FXI. Thus, we propose that our solution structure reflects the conformation of the A4 domain within the context of the activated enzyme, FXIa, which differs from that of the same segment within the zymogen.
Fig. 2.
C-terminal helix. (a) Local environment of the C-terminal helix (α2) in the NMR structure of A4. Residues involved in interactions between the C-terminal helix and other parts of the protein are shown. (b) Overlay of a representative NMR solution structure (blue) and crystal structure (red) of the A4 dimer. The C-terminal segment (354–361), which is part of an extended connecting loop in the crystal structure, assumes an α-helical conformation in the NMR structure, resulting in an ≈20 Å displacement of the C-terminal Glu-361.
Domain Structure of FXI and FXIa Observed by SAXS and EM.
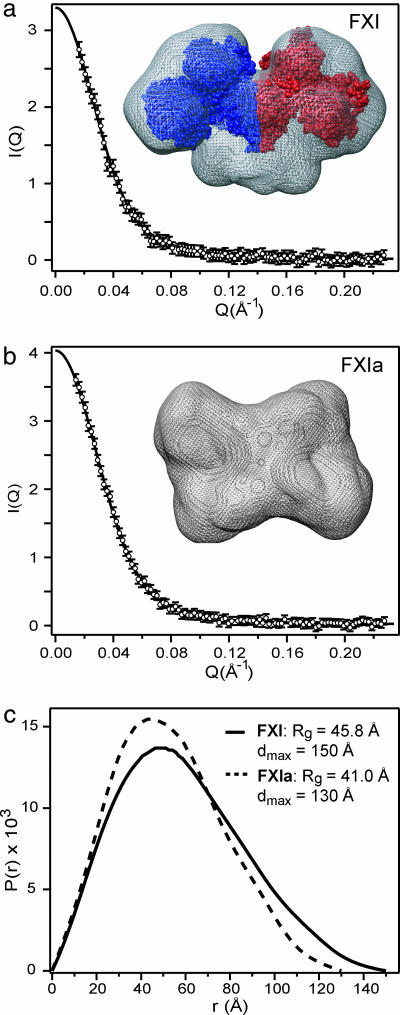
To detect any changes in the shape of the FXI dimer associated with zymogen activation, we used SAXS, which provides information on the overall size and shape of macromolecules in solution (23). To avoid contributions to x-ray scattering from the structurally poorly defined N-linked carbohydrate chains, we used the enzyme N-glycanase (PNGase F) to deglycosylate FXI and FXIa (SI Fig. 9). Functional assays before and after deglycosylation showed that removal of carbohydrate does not have a significant effect on FXIa activity (see the legend of SI Fig. 9). Dynamic light scattering measurements yielded essentially single-exponential autocorrelation decays for both FXI and FXIa samples, indicating that the solutions are monodisperse with negligible amounts of aggregated material. The x-ray scattering profiles for FXI and FXIa are shown in Fig. 3 a and b, respectively. Guinier analysis of the SAXS data in the low-angle region (see Materials and Methods) indicates that conversion of the FXI zymogen into active enzyme is accompanied by a decrease in the radius of gyration, Rg, from 45.9 ± 0.8 Å to 42.2 ± 0.9 Å, consistent with formation of a more compact structure. Fig. 3c shows the corresponding pair distance distribution function, P(r), which represents the probability of finding two scattering points at a given distance r from each other in the macromolecule (24). The Rg values computed from P(r), 45.8 ± 0.3 Å and 41.0 ± 0.5 Å for FXI and FXIa, respectively, are in good agreement with those obtained by using the Guinier approximation. The point on the x axis where P(r) reaches zero represents the maximum diameter, Dmax, averaged over all orientations. The decrease in Dmax from 150 to 130 Å observed upon zymogen activation represents a significant contraction indicative of a change from an elongated (dumbbell-shaped) structure to a more compact domain arrangement. Also shown in Fig. 3 are molecular envelopes of FXI (Fig. 3a) and FXIa (Fig. 3b) reconstructed on the basis of the SAXS data by using the ab initio method of Svergun (25). The SAXS-based structure of the FXI zymogen (mesh surface in Fig. 3a) is in reasonable agreement with the overall dimensions and shape of the crystal structure (7) shown in space-filling representation. In contrast to the elongated shape of the zymogen, the SAXS-derived surface of FXIa (Fig. 3b) has a box-like appearance consistent with a more compact domain arrangement.
Fig. 3.
Conformational changes associated with activation of FXI monitored by SAXS. (a and b) The scattering intensity, I(Q), is plotted vs. scattering vector, Q, for FXI (a) and FXIa (b). (c) The lines represent the predicted scattering profiles corresponding to the P(r) functions. Low-resolution structures derived from the SAXS profiles by ab initio simulated annealing calculations (25) for FXI and FXIa are shown in a and b, respectively, using mesh surface rendering. For comparison, a space-filling representation of FXI based on the crystal structure (7) is included in a.
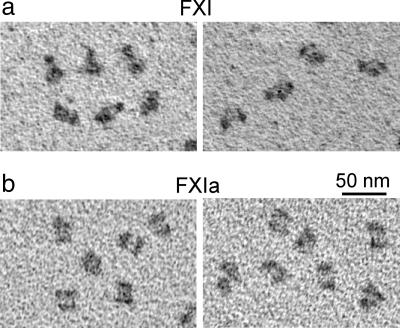
To visualize the molecules more directly, we examined FXI and FXIa by EM after rotary shadowing for contrast enhancement (26). Examination of large areas with high protein density revealed particles with sizes consistent with a dimeric structure. A detailed comparison between FXI and FXIa particles revealed striking differences in the shape and fine structure. As illustrated by the examples in Fig. 4, many of the FXI particles consist of three nodules in a shallow triangular arrangement whereas the FXIa particles have a more box-like appearance with two elongated nodules arranged side-by-side. The dimensions were measured for a total of 400 randomly selected particles (five blinded evaluators). Measured dimensions of FXI particles are 18.1 ± 0.8 nm × 10.8 ± 0.9 nm (n = 200), corresponding to a length-to-width ratio of 1.68 ± 0.16. After accounting for the tungsten coating (≈3 nm), these measurements agree quite well with dimensions of the FXI crystal structure (≈15 × 8 nm). In contrast, FXIa particles shows dimensions of 16.2 ± 1.3 nm × 12.7 ± 1.2 nm (n = 200), corresponding to a significantly smaller aspect ratio of 1.28 ± 0.15 (P < 0.001). The characteristic box-like shape of the FXIa dimer bears a remarkable resemblance to the molecular envelope determined on the basis of the SAXS data (Fig. 3b).
Fig. 4.
Electron micrographs showing domain rearrangements associated with FXI zymogen activation. Representative electron micrographs are shown for samples of FXI (a) and FXIa (b) prepared by using rotary shadowing with tungsten.
Model of FXI Activation.
To understand the impact of the C-terminal helix on the overall structure of FXIa, we used molecular modeling to explore possible arrangements of the catalytic domains relative to the A4 dimer that are sterically compatible with the NMR structure (SI Materials and Methods). The coordinates of the isolated catalytic domain of FXIa determined by x-ray crystallography (20) (PDB ID code 1ZJD) were joined to each subunit of the A4 dimer through a disulfide bond between Cys-362 and Cys-482. To link the catalytic domains to the A4 dimer as observed by NMR while avoiding steric clashes, they have to be reoriented relative to their position in the crystal structure (Fig. 5a) and moved toward the twofold symmetry axis (Fig. 5b). Although the exact orientation of the catalytic domains cannot be determined, the model predicts a minimum distance between the two active sites of 40 Å, and a maximum separation of ≈75 Å is allowed while maintaining the 362–482 disulfide linkage between A4 and the catalytic domain.
Fig. 5.
Changes in domain structure upon zymogen activation. (a) Domain arrangement of the FXI zymogen, based on the crystal structure of Papagrigoriou et al. (7). The A4 domain (green) mediating dimerization and the three other apple domains are shown in ribbon representation. The two catalytic domains (gray, space-filling) are connected to the A4 domain via an extended loop (red, residues 354–362). (b) A model representing one of the possible conformations of the catalytic and A4 domains in FXIa triggered by formation of a second α-helix at the C terminus of the A4 domain (red). Upon cleavage of the R369-I370 bond (orange), the 362–482 disulfide bond (yellow) is the only covalent link between the pair of catalytic domains and A4 domains. Residues in the active-site cleft are colored magenta.
Molecular modeling was also used in conjunction with the program CRYSOL (27), which yields predicted x-ray scattering profiles that can be compared with the experimental SAXS data (Fig. 3). To build a complete model of the FXIa dimer, we began with our initial model of the A4 dimer linked with a pair of activated catalytic domains (Fig. 5b), which is consistent with the NMR data, and added the remaining apple domains (A1–A3) in various sterically allowed arrangements that satisfy the covalent linkage between the domains, as detailed in SI Fig. 10. The predicted scattering profile for each trial structure was compared with the corresponding experimental data (SI Fig. 10a). The final model that minimizes the deviations between the predicted and measured scattering intensities in terms of the rmsd and residuals is shown in SI Fig. 10b. Comparison with the crystal structure of FXI (SI Fig. 10c) confirms that zymogen activation is accompanied by drastic changes in domain organization and packing.
Our model is consistent with prior findings indicating that dimeric FXIa can cleave FIX at its two scissile bonds without detectable accumulation of a free intermediate, suggesting a concerted cleavage mechanism (12). This requires that the catalytic domains of FXIa are appropriately oriented to allow simultaneous cleavage of the two scissile bonds in FIX separated by 35 residues (Ala-146–Arg-180). Because no structural information is currently available for the FIX zymogen, the actual distance between these two scissile bonds is not known. It should be noted that in the crystal structure of FIXa (28), the C-terminal arginine of the light chain and the N-terminal isoleucine of the catalytic domain are located at a distance of 25 Å; however, activation of FIX is likely to be accompanied by conformational changes. In the crystal structure of the FXI zymogen (7), the two catalytic domains are at opposite ends of the dimer with the active sites pointing away from the twofold symmetry axis of the molecule (Fig. 5a). Given the large distance (≈120 Å) between the two active sites, this conformation is inconsistent with a concerted cleavage mechanism. Even if the Ala-146–Arg-180 segment of FIX were in a fully extended conformation, it would be unable to reach from one active site of FXI to the other. On the other hand, our model of FXIa (Fig. 5b) predicts a much shorter distance between the two active sites consistent with a concerted cleavage mechanism.
We propose the following scenario to describe the conformational events associated with activation of FXI. In the FXI zymogen, the covalent linkage between A4 and the catalytic domain prevents formation of a helix at the C terminus. Cleavage of the Arg-369–Ile-370 peptide bond activates the catalytic domain by inserting the newly generated N-terminal amino group into the active site region (Fig. 5b). This releases the locally strained loop connecting A4 to the catalytic domain (Fig. 5b, residues 354–362; shown in red) and allows formation of helix α2 at the C terminus of A4. Docking of α2 onto the surface of A4 disrupts inter-domain contacts with the A1 domain and causes a 25-Å shift in the position of the C terminus, along with the disulfide-linked catalytic domains, resulting in a major shortening of the distance between the active sites. In addition, cleavage of the scissile bond creates a new C terminus and liberates the segment between Cys-362 and Arg-369, which is now free to engage in alternative interactions that may compete with some of the native domain contacts, along with the docking of the C-terminal helix into its binding pocket on A4.
Our conclusion that FXI zymogen activation is accompanied by major changes in domain structure is further supported by biochemical data indicating that the binding properties of FXIa differ from those of the zymogen with respect to its substrate, FIX, and the platelet surface (29–31). These observations are consistent with domain rearrangements exposing new binding sites upon conversion of FXI into FXIa.
Conclusions
The NMR structure of the isolated A4 domain of FXI resembles members of the PAN domain family. However, a detailed comparison with the crystal structure of the corresponding domain within the context of the whole FXI zymogen (7) reveals important structural differences in the C-terminal region of the A4 domain. The NMR structure has a well defined α-helix at the C terminus, which packs against the loops of the A4 domain. In the crystal structure of FXI, the C-terminal segment of A4 is part of an extended linker region connecting the A4 and catalytic domains. While the covalent linkage between A4 and the catalytic domain prevents formation of a C-terminal helix in the zymogen structure, cleavage of the Arg-369–Ile-370 scissile bond removes this constraint and allows the C-terminal helix to form and pack against the A4 domain. SAXS measurements and rotary shadowing EM data confirm that the FXI dimer undergoes a change in shape from an elongated to a more compact domain structure. In conjunction with molecular modeling, our findings support the hypothesis that, upon activation, the distance between the active sites on adjacent catalytic domains is shortened sufficiently to allow simultaneous cleavage of both scissile bonds in FIX, as previously reported (12). Our biophysical evidence that FXI zymogen activation causes a large-scale rearrangement of its domains is supported by biochemical data indicating that the binding properties of FXIa differ from those of the zymogen. For example, FXIa positions FIX for efficient macromolecular substrate hydrolysis by exposing a substrate-binding site for FIX within the FXIa heavy chain (29, 31). Thus, conversion of FXI to FXIa may obscure some zymogen-specific ligand binding sites within FXI and expose a new set of enzyme-specific binding sites within FXIa.
Materials and Methods
Recombinant FXI A4 and uniformly labeled (15N and/or 13C) samples of A4 were prepared as described (11). All NMR spectra were collected at 37°C in 20 mM sodium pyrophosphate, pH 6.0/100 mM NaCl. Additional information on NMR data acquisition, processing, and structure calculation can be found in SI Materials and Methods. Atomic coordinates have been deposited in the Protein Data Bank (ID codes 2J8J and 2J8L). FXI and FXIa samples (Haematologic Technologies, Essex Junction, VT) for solution scattering measurements were purified by gel filtration and deglycosylated using ≈2,000 units of the endoglycosidase N-glycanase (PNGase F; New England Biolabs, Ipswich, MA) at 37°C for 14 h. SDS/PAGE analysis confirmed the complete deglycosylation of the samples (SI Fig. 9). Autocatalytic activation of FXI was inhibited with 1 mM benzamidine. Gel electrophoresis under reducing conditions before and after the experiments showed no evidence for activation of FXI and confirmed that FXIa was fully activated. SAXS measurements on 3.2 μM FXI and 3.5 μM FXIa (in 20 mM Hepes/100 mM NaCl, pH 7.4) were performed at room temperature with an in-house apparatus (32), as detailed in SI Materials and Methods.
Transmission EM after low-angle rotary shadowing is a simple and effective method for obtaining low-resolution structural data on protein shape and domain organization (26, 33). FXI and FXIa samples were diluted to a concentration of 0.01–0.10 mg/ml into 0.05 M ammonium formate buffer (pH 7.4) containing 30% glycerol (to prevent surface artifacts). These solvent conditions are widely used for EM and generally do not affect protein structure. The solution was sprayed onto freshly cleaved mica, and the molecules were rotary shadowed at an angle of 5–8° with tungsten in a Denton DV502 vacuum evaporator. The samples, stabilized by a thin film of carbon, were floated off the mica and picked up onto EM grids. Images of randomly chosen fields of individual molecules were recorded on a Phillips 400 electron microscope.
Supplementary Material
Acknowledgments
We thank Jonas Emsley for sharing crystal structure coordinates of FXI before publication, Roland Dunbrack for helpful discussions, and the Spectroscopy Support, Molecular Modeling, and Biochemistry and Biotechnology Facilities at the Fox Chase Cancer Center for support. This work was supported by National Institutes of Health Grants HL46213 (to P.N.W.), GM56250 and CA06927 (to H.R.), and HL30954 (to J.W.W.) and by an appropriation from the Commonwealth of Pennsylvania to the Fox Chase Cancer Center.
Abbreviations
- PAN
plasminogen apple nematode
- NOE
nuclear Overhauser effect
- SAXS
small-angle x-ray scattering.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2J8J and 2J8L).
This article contains supporting information online at www.pnas.org/cgi/content/full/0703080104/DC1.
References
- 1.Fujikawa K, Chung DW, Hendrickson LE, Davie EW. Biochemistry. 1986;25:2417–2424. doi: 10.1021/bi00357a018. [DOI] [PubMed] [Google Scholar]
- 2.Walsh PN, Gailani D. In: Hemostasis and Thrombosis: Basic Principles and Clinical Practice. Colman RW, Hirsh J, Marder VJ, Clowes AW, George JN, editors. Philadelphia: Lippincott; 2006. Chap 12. [Google Scholar]
- 3.von dem Borne PA, Meijers JC, Bouma BN. Blood. 1995;86:3035–3042. [PubMed] [Google Scholar]
- 4.McMullen BA, Fujikawa K, Davie EW. Biochemistry. 1991;30:2056–2060. doi: 10.1021/bi00222a008. [DOI] [PubMed] [Google Scholar]
- 5.Brown PJ, Gill AC, Nugent PG, McVey JH, Tomley FM. FEBS Lett. 2001;497:31–38. doi: 10.1016/s0014-5793(01)02424-3. [DOI] [PubMed] [Google Scholar]
- 6.O'Connell NM, Saunders RE, Lee CA, Perry DJ, Perkins SJ. J Thromb Haemost. 2005;3:127–138. doi: 10.1111/j.1538-7836.2004.01088.x. [DOI] [PubMed] [Google Scholar]
- 7.Papagrigoriou E, McEwan PA, Walsh PN, Emsley J. Nat Struct Mol Biol. 2006;13:557–558. doi: 10.1038/nsmb1095. [DOI] [PubMed] [Google Scholar]
- 8.Asakai R, Chung DW, Ratnoff OD, Davie EW. Proc Natl Acad Sci USA. 1989;86:7667–7671. doi: 10.1073/pnas.86.20.7667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meijers JC, Davie EW, Chung DW. Blood. 1992;79:1435–1440. [PubMed] [Google Scholar]
- 10.Yang DT, Flanders MM, Rodgers GM. Blood. 2005;106:2626. [Google Scholar]
- 11.Riley PW, Cheng H, Samuel D, Roder H, Walsh PN. J Mol Biol. 2006;367:558–573. doi: 10.1016/j.jmb.2006.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolberg AS, Morris DP, Stafford DW. Biochemistry. 1997;36:4074–4079. doi: 10.1021/bi962274y. [DOI] [PubMed] [Google Scholar]
- 13.Gailani D, Ho D, Sun MF, Cheng Q, Walsh PN. Blood. 2001;97:3117–3122. doi: 10.1182/blood.v97.10.3117. [DOI] [PubMed] [Google Scholar]
- 14.Montelione GT, Wagner G. J Magn Reson. 1990;87:183–188. [Google Scholar]
- 15.Ikura M, Kay LE, Tschudin R, Bax A. J Magn Reson. 1990;86:204–209. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Bax A, Clore GM, Driscoll PC, Gronenborn AM, Mitsuhiko I, Kay LE. J Magn Reson. 1991;87:620–627. [Google Scholar]
- 17.Cornilescu G, Delaglio F, Bax A. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 18.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Proc Natl Acad Sci USA. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tjandra N, Omichinski JG, Gronenborn AM, Clore GM, Bax A. Nat Struct Biol. 1997;4:732–738. doi: 10.1038/nsb0997-732. [DOI] [PubMed] [Google Scholar]
- 20.Navaneetham D, Jin L, Pandey P, Strickler JE, Babine RE, Abdel-Meguid SS, Walsh PN. J Biol Chem. 2005;280:36165–36175. doi: 10.1074/jbc.M504990200. [DOI] [PubMed] [Google Scholar]
- 21.Jin L, Pandey P, Babine RE, Gorga JC, Seidl KJ, Gelfand E, Weaver DT, Abdel-Meguid SS, Strickler JE. J Biol Chem. 2005;280:4704–4712. doi: 10.1074/jbc.M411309200. [DOI] [PubMed] [Google Scholar]
- 22.Stroud RM, Kossiakoff AA, Chambers JL. Annu Rev Biophys Bioeng. 1977;6:177–193. doi: 10.1146/annurev.bb.06.060177.001141. [DOI] [PubMed] [Google Scholar]
- 23.Wall ME, Gallagher SC, Trewhella J. Annu Rev Phys Chem. 2000;51:355–380. doi: 10.1146/annurev.physchem.51.1.355. [DOI] [PubMed] [Google Scholar]
- 24.Semenyuk AV, Svergun DI. J Appl Crystallogr. 1991;24:537–540. [Google Scholar]
- 25.Svergun DI. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weisel JW, Nagaswami C, Woodhead JL, DeLa Cadena RA, Page JD, Colman RW. J Biol Chem. 1994;269:10100–10106. [PubMed] [Google Scholar]
- 27.Svergun D, Barberato C, Koch MHJ. J Appl Crystallogr. 1995;1995:768–773. [Google Scholar]
- 28.Bode W, Brandstetter H, Mather T, Stubbs MT. Thromb Haemost. 1997;78:501–511. [PubMed] [Google Scholar]
- 29.Sinha D, Koshy A, Seaman FS, Walsh PN. J Biol Chem. 1985;260:10714–10719. [PubMed] [Google Scholar]
- 30.Sinha D, Seaman FS, Walsh PN. Biochemistry. 1987;26:3768–3775. doi: 10.1021/bi00387a005. [DOI] [PubMed] [Google Scholar]
- 31.Sun MF, Zhao M, Gailani D. J Biol Chem. 1999;274:36373–36378. doi: 10.1074/jbc.274.51.36373. [DOI] [PubMed] [Google Scholar]
- 32.Bu Z, Koide S, Engelman DM. Protein Sci. 1998;7:2681–2683. doi: 10.1002/pro.5560071223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veklich YI, Gorkun OV, Medved LV, Nieuwenhuizen W, Weisel JW. J Biol Chem. 1993;268:13577–13585. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.