Abstract
Telomerase is the ribonucleoprotein enzyme that elongates telomeres to counteract telomere shortening. The core enzyme consists of a reverse transcriptase protein subunit and an RNA subunit. The RNA subunit contains a short region that is used as a template by the reverse transcriptase to add short, tandem, G-rich repeats to the 3′ ends of telomeres. By coexpressing two RNA subunits that differ in the telomeric repeat sequence specified and examining the telomere extensions after one cell cycle, we determined that Saccharomyces cerevisiae telomerase can dissociate and reassociate from a given telomere during one cell cycle. We also confirmed that telomerase is nonprocessive in terms of telomeric repeat addition. However, repeat addition processivity is significantly increased at extremely short telomeres, a process that is dependent on the ATM-ortholog Tel1. We propose that this enhancement of telomerase processivity at short telomeres serves to rapidly elongate critically short telomeres.
Keywords: Telomerase, processivity, Saccharomyces cerevisiae, TLC1, Tel1
Telomeres, the physical ends of eukaryotic chromosomes, protect chromosome ends from end fusions and degradation (Ferreira et al. 2004). Telomere length is maintained by a dynamic process of lengthening and shortening (Hug and Lingner 2006). Shortening can occur as a result of nucleolytic degradation and incomplete DNA replication. Lengthening is primarily accomplished by the action of a specialized reverse transcriptase called telomerase, whose core consists of a catalytic protein subunit and an RNA subunit (Greider and Blackburn 1985, 1987, 1989; Lingner et al. 1997). Telomerase extends telomeres by repeated reverse transcription of a short sequence, using the RNA subunit as a template, to the 3′ ends of telomeres (Greider and Blackburn 1989; Yu et al. 1990; Singer and Gottschling 1994). Lagging strand synthesis then presumably fills in the complementary 5′ strand. Cells lacking telomerase have telomeres that shorten with each cell division, which can ultimately lead to replicative senescence (Lundblad and Szostak 1989; Harley et al. 1990; Yu et al. 1990).
Telomerase complexes isolated from ciliated protozoa and humans are processive in vitro, capable of elongating a given telomeric primer by hundreds of nucleotides through multiple rounds of telomeric repeat addition (Morin 1989; Greider 1991). In contrast, in other organisms such as mouse, Chinese hamster, and various fungi, telomerase appears to be less or nonprocessive in vitro (Prowse et al. 1993; Cohn and Blackburn 1995; Lingner et al. 1997; Lue and Peng 1997, 1998; Fulton and Blackburn 1998). The budding yeast Saccharomyces cerevisiae telomerase enzyme remains stably bound to the telomere substrate after a single round of telomere elongation in vitro (Cohn and Blackburn 1995; Prescott and Blackburn 1997a). However, in vivo analysis suggests that yeast telomerase can add >100 nucleotides (nt) in a single cell cycle (Teixeira et al. 2004). Thus, in vivo, the enzyme is either processive or it turns over very rapidly.
In vivo analysis in S. cerevisiae has also shown that telomerase does not act on every telomere in each cell cycle but instead exhibits an increasing preference for telomeres as their lengths decline, suggesting that telomeres switch between nonextendible and extendible states (Teixeira et al. 2004). Although the number of nucleotides added per extension event does not significantly correlate with telomere length, telomere extension length is strongly increased at very short telomeres (Teixeira et al. 2004). However, it was not determined whether or not these long extension events were due to processive telomerase action.
To analyze telomerase repeat addition processivity in vivo, and to determine whether telomere length affects processivity, we coexpressed a mutant tlc1 that specifies altered telomeric sequences together with wild-type TLC1 and analyzed telomere elongation events after one cell cycle. This analysis indicated that telomerase can use both RNA templates to extend a telomere during one cell cycle, and that the telomerase complex can dissociate and reassociate with telomeres during one round of telomere elongation. We found that telomerase does generally act nonprocessively in vivo, consistent with conclusions drawn from a previous study (Prescott and Blackburn 1997b). However, we found that the repeat addition processivity was significantly enhanced at extremely short telomeres. We propose that this enhancement of processivity, which is dependent on the ATM ortholog Tel1, allows cells to rapidly elongate critically short telomeres that can arise as a result of a double-strand break caused by DNA damage or DNA replication fork collapse in the telomeric tracts.
Results
Characterization of a tlc1 template mutant
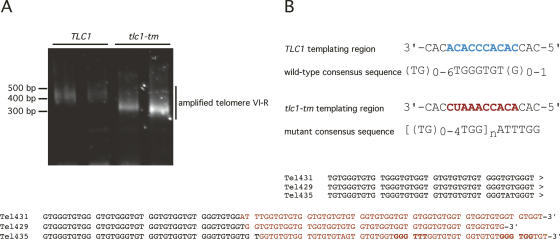
To study telomerase action in cells expressing two different telomerase RNA subunits, we needed a mutant tlc1 that incorporated telomeric repeats that were distinguishable from wild type, yet would still be able to complement a deletion of TLC1 and elongate telomeres to similar levels as wild type. We previously screened an S. cerevisiae tlc1 library, in which 10 of the 16 templating nucleotides were randomized, for complementation of a TLC1 deletion (Forstemann et al. 2003). Out of an estimated 60,000 templates screened, only 40 different mutant templates were able to rescue the senescence phenotype of tlc1Δ (Forstemann et al. 2003). One of the template mutations was isolated three times independently and phenotypically most closely resembled wild type (Forstemann et al. 2003). We called this template mutant tlc1-tm. The template region of tlc1-tm is 3′-CACCUAAACCACACAC-5′, whereas it is 3′-CACAC ACCCACACCAC-5′ for wild-type TLC1.
Wild-type TLC1 was replaced at its endogenous locus by tlc1-tm. After propagating this strain for >100 generations to allow mutant telomeric sequences to be incorporated into telomeres, we performed telomere PCR to amplify telomere VI-R sequences (Fig. 1A). The lengths of the tlc1-tm telomeres were ∼80 base pairs (bp) shorter than those of the isogenic wild-type strain. Despite the modestly shorter telomeres, telomere length homeostasis was maintained, confirming what we reported previously for this mutant when it was expressed from a plasmid in a different genetic background (Forstemann et al. 2003).
Figure 1.
Characterizing the tlc1-tm mutant. (A) Telomere lengths of the indicated strains were determined by telomere PCR of telomere VI-R. The size of the PCR product is the sum of the telomere length, the distance of the subtelomeric primer to the beginning to the telomeric tracts, and the size of the primers used in the telomere PCR (59 bp). Telomere lengths were calculated to be 353 bp and 271 bp for TLC1 and tlc1-tm strains, respectively. The calculated telomere lengths are an average of telomere lengths from two different isolates of each strain. (B) Telomere PCR products from A amplified from the tlc1-tm mutant were cloned and sequenced. Three representative sequences are shown. The complete set of sequences is shown in Supplementary Figure 1. Mutant divergent regions of each sequence (in red) were used to derive a mutant consensus sequence of [(TG)0–4TGG)]nATTTGG. Bolded sequences highlight occurrences of GGG trinucleotide incorporation by the mutant telomerase.
The amplified VI-R telomeres from the tlc1-tm mutant were cloned and sequenced. Analysis of these sequences revealed that telomerase using this template RNA added degenerate repeats with a consensus sequence of [(TG)0–4TGG]nATTTGG, where n indicates that the [(TG)0–4TGG] motif can be repeated for a variable number of times before the incorporation of an ATTTGG sequence (Fig. 1B; Supplementary Fig. S1). Mutant sequences could easily be differentiated from wild-type sequences because of the frequent usage of the GG dinucleotide. Wild-type telomeres, with a consensus sequence of (TG)0–6TGGGTGTG(G)0–1 (Forstemann and Lingner 2001), very rarely contain two consecutive GG dinucleotide sequences without an intervening GGG trinucleotide. GGG trinucleotides are rarely found in mutant repeat sequences, whereas they are present in every wild-type repeat incorporated. Furthermore, the occasional incorporation by the mutant telomerase of ATTTGG is a clear indication of mutant telomeric sequence.
Since the tlc1-tm template region is 3′-CACCUAA ACCACACAC-5′, the mutant telomerase should not be able to incorporate GGG trinucleotides according to canonical Watson-Crick base-pairing. The rare occurrences of GGG trinucleotides in mutant sequences (Fig. 1B; Supplementary Fig. S1) are most likely due to the U residue in the template being able to base-pair with G nucleotides (Crick 1966; Varani and McClain 2000). This potentially poses a problem for differentiating mutant from wild-type sequences. However, some of the GGG trinucleotide occurrences in the mutant sequences were still easily identifiable as mutant (e.g., in cases where the sequence was GGGTTT or GGGTGG, neither of which occurs in wild-type telomeric sequences). The remaining GGG trinucleotide occurrences were very rare (two times in 1373 nt of mutant telomeric sequence, or 217 mutant repeats, from a tlc1-tm mutant). Hence the tlc1-tm mutant added telomeric repeats that were easily distinguishable from wild-type repeats and therefore suitable for use in our coexpression assays.
Two-template single-telomere extension assay (2T-STEX)
Although much work has been done to characterize telomerase activity, it is not known whether a telomere, if extended, is extended by only one telomerase complex once per cell cycle, or whether telomerase complexes can dissociate and reassociate from telomeres while extending a given telomere. To answer this question, we used a modified version of the STEX assay that we developed and described previously (Teixeira et al. 2004; Arneric and Lingner 2007). In the STEX assay, a telomerase-negative strain (referred to as the “recipient”) was created by deleting EST1, which encodes an essential subunit of yeast telomerase (Lundblad and Szostak 1989). Telomerase was added by mating this strain to a telomerase-positive “donor” strain that contains wild-type EST1. Thus, telomeres that had shortened in the recipient strain are extended in the zygote. Telomere elongation was detected by cloning and sequencing of telomeres derived from the recipient parental strain after being amplified by telomere PCR, which is made possible because the recipient chromosome V-R telomere was tagged with the ADE2 gene in the subtelomeric region (Forstemann et al. 2000; Teixeira et al. 2004). Since yeast telomerase adds imperfect repeats, telomere elongation can be detected by the divergence of telomeric sequences in the telomere-distal region relative to the telomeric DNA synthesized by semiconservative DNA replication (Wang and Zakian 1990; Forstemann et al. 2000; Forstemann and Lingner 2001).
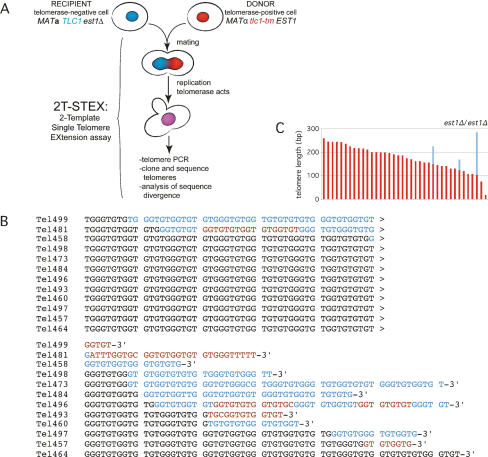
We modified the STEX assay by replacing wild-type TLC1 with tlc1-tm at its endogenous locus in the donor strain. We named this modified assay 2T-STEX (Fig. 2A). This strain was mated to a recipient strain that expresses wild-type TLC1. Telomere extensions were analyzed 3 h after mating (Fig. 2B; Supplementary Fig. S2A), the time required for most cells to mate and complete one S phase (Teixeira et al. 2004). Of 21 telomere elongation events, 11 contained only wild-type sequence and three contained only mutant sequence. Seven contained a mixture of wild-type and mutant sequences.
Figure 2.
2T-STEX analysis. (A) 2T-STEX methodology (see text for details). (B) Example of aligned sequences from a 2T-STEX assay in which the donor strain carried the tlc1-tm allele. Telomere extension events were detected by sequence divergence. Sequences in blue are repeats added by telomerase with wild-type TLC1, while sequences in red are repeats added by telomerase with the mutant tlc1-tm. (C) Analysis of a 2T-STEX assay in which both the recipient and the donor strains were est1Δ. Unlike a standard 2T-STEX assay, the diploids were allowed to undergo ∼30 cell doublings after mating before sequence analysis. Each telomere sequence is represented by a vertical bar where the red and blue parts indicate the nondiverging and diverging regions, respectively. This analysis shows that sequence divergence in the absence of telomerase is very low and not perturbing the 2T-STEX analysis.
We also performed the reciprocal experiment where the donor strain expressed wild-type TLC1 while the recipient expressed the tlc1-tm mutant (Supplementary Fig. S2B). The results were very similar. Of 23 telomere elongation events, 11 contained only wild-type sequence, four contained only mutant sequence, and eight contained a mixture of both wild-type and mutant sequences. In both experiments, the short telomeres were preferentially elongated and long extension events occurred at very short telomeres (data not shown) as expected (Teixeira et al. 2004). Together, these results indicate that telomerase can use both templates to extend the same telomere during one cell cycle.
We also performed a 2T-STEX assay using an est1Δ telomerase-negative donor strain to control for telomerase-independent divergence events (Fig. 2C). Instead of limiting cells to one cell cycle, they were allowed to propagate for ∼30 generations after mating. Three of 38 (7.9%) telomeres sequenced showed telomerase-independent divergence events. This corresponds to an extremely low frequency of divergence after one cell cycle, similar to what we reported previously (Teixeira et al. 2004). Thus, the sequence divergence events we observed in our 2T-STEX assays were due to telomerase-mediated telomere elongation events.
Yeast telomerase is generally nonprocessive for repeat addition except at extremely short telomeres
The average extension length for S. cerevisiae telomerase during one cell cycle is 44 nt for telomeres >200 bp in length (Teixeira et al. 2004). Since the mean telomeric repeat length is 11 nt (Forstemann and Lingner 2001), yeast telomerase in vivo can extend telomeres by about four repeats, which is in contrast with its in vitro biochemical properties, where it can only add one repeat before stopping (Cohn and Blackburn 1995; Prescott and Blackburn 1997b). Therefore, either yeast telomerase has some in vivo processivity or it is able to dissociate from and reassociate to telomeres within a single cell cycle. Our 2T-STEX results suggest that the latter model is possible (Fig. 2A; Supplementary Fig. S2). However, this does not necessary imply that telomerase is completely nonprocessive in vivo.
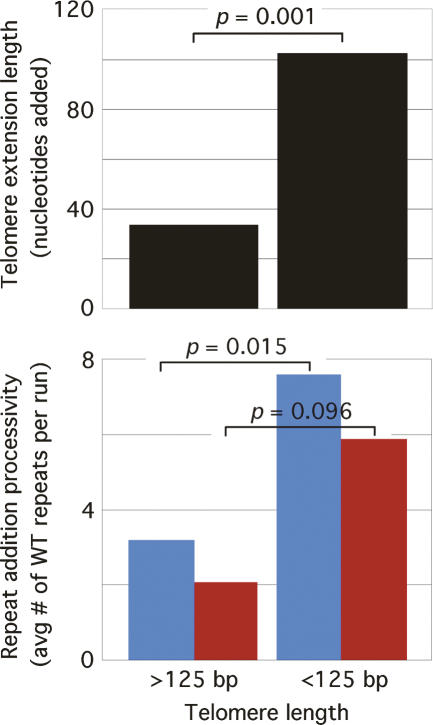
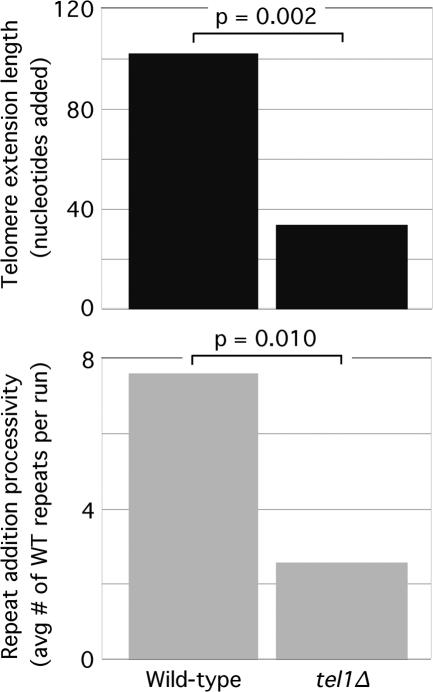
To determine whether telomerase has any repeat addition processivity in vivo, we considered the scenario where telomerase is completely nonprocessive—i.e., if it stops after each repeat added. We counted the total number of repeats added in all extensions from our 2T-STEX assays and determined the fraction of wild-type repeats used (207/295 = 0.702). This fraction did not change significantly as a function of telomere length (data not shown). Given a 70.2% incorporation of wild-type repeats, the expected average number of consecutive wild-type repeats per stretch or run of wild-type repeats should be 3.3, assuming that telomerase is completely nonprocessive in vivo (see Materials and Methods for the mathematical equation used to determine this value). If yeast telomerase is processive, the observed average number of wild-type repeats per run should be higher than 3.3. However, the observed average for telomeres >125 bp in length was 3.2, a value that did not change significantly as a function of telomere lengths >125 bp, indicating that yeast telomerase is indeed nonprocessive in vivo. Strikingly, repeat addition processivity was significantly increased at telomeres <125 bp in length, as the observed average number of wild-type repeats per run was 7.6 (Fig. 3). This effect was also seen for telomerase using the mutant tlc1-tm, where the average number of mutant repeats per run of mutant repeats increased from 2.1 to 5.9 (Fig. 3). Thus, the increased extension length at very short telomeres (Teixeira et al. 2004) can be attributed to the increased telomerase repeat addition processivity (Fig. 3).
Figure 3.
Telomere extension length (top panel) correlates with repeat addition processivity (bottom panel). The average lengths of the telomere extensions from the 2T-STEX assays were determined for telomeres greater than and less than 125 bp in length. Repeat addition processivity is shown as the average number of wild-type repeats added per run of wild-type repeats (blue bars) or the average number of mutant repeats added per run of mutant repeats (red bars) for telomeres greater than and less than 125 bp in length. The P-values associated with two-tailed unpaired t-tests are shown.
Telomeres <125 bp in length do arise in vivo
In the absence of telomerase, S. cerevisiae telomeres shorten at a rate of 3 bp per generation (Marcand et al. 1999). Thus, starting with a wild-type-length telomere of 300 bp in length, it would take ∼58 generations [(300–125)/3 = 58.3] to shorten to a length of 125 bp if telomerase did not act in the intervening time. The frequency of telomere extension by telomerase steadily increases from ∼7% at 300-bp-long telomeres to ∼39% at 125-bp-long telomeres (Teixeira et al. 2004). Consequently, the probability of a telomere shortening from 300 bp to 125 bp without telomerase acting on it in the intervening time is 2.6 × 10−7. Therefore, it is highly improbable that a wild-type-length telomere could shorten to 125 bp in length via normal telomere erosion mechanisms such as incomplete DNA replication or nucleolytic degradation.
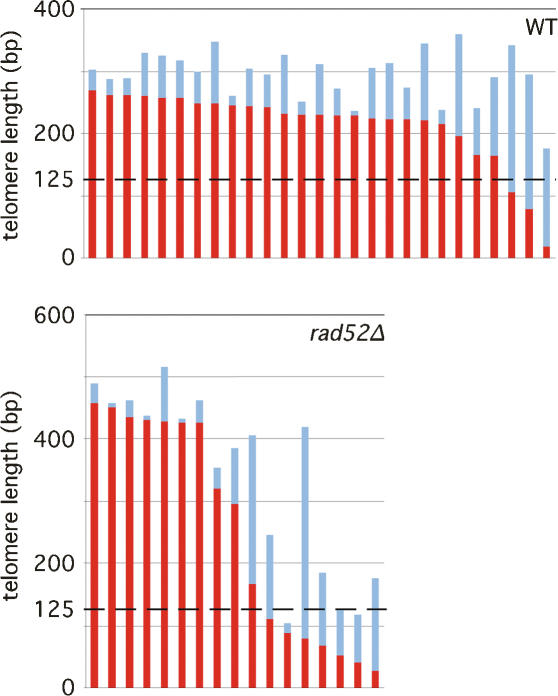
To detect whether or not telomeres <125 bp in length actually occur in vivo, we sequenced telomeres from telomerase-positive cells after ∼40 generations of clonal expansion. While all yeast telomeric repeats are degenerate, only the distal region of telomeres have sequences that are divergent (Wang and Zakian 1990; Forstemann et al. 2000). This sequence divergence is dependent on telomerase activity (Forstemann et al. 2000). The average point of divergence in a wild-type strain is 214 bp into the telomeric tract, counting away from the centromere (Fig. 4). A rad52Δ strain was included in this experiment to eliminate any possibility of recombination-dependent divergence events. The rad52Δ strain had an average point of divergence of 253 bp into the telomeric tract. The slightly greater point of divergence in the rad52Δ strain can be attributed to the slightly longer telomeres seen in this strain (27 wild-type telomeres sequenced with an average length of 291 bp compared with 17 rad52Δ telomeres sequenced with an average length of 348 bp). Although the average points of divergence in both strains were much >125 bp, both strains contained telomere sequences in which the points of divergence were before 125 bp into the telomeric tract (three of 27 sequences for wild type; seven of 17 sequences for rad52Δ). Taken together, this indicates that telomeres <125 bp in length do occur in vivo and most likely do not arise from normal telomere shortening processes.
Figure 4.
Telomeres <125 bp in length do occur in vivo. A RAD52/rad52Δ heterozygous diploid was sporulated and tetrad was dissected. Wild-type or rad52Δ isolates derived from the same tetrad were allowed to propagate for ∼40 generations. Chromosome VI-R telomeres were amplified by telomere PCR, cloned, and sequenced. Each telomere sequence is represented by a vertical bar where the red and blue parts indicate the nondiverging and diverging regions, respectively.
Tel1 is needed for the enhanced repeat addition processivity at extremely short telomeres
The phosphoinositide 3-kinase-related kinases Tel1 and Mec1 (S. cerevisiae orthologs of human ATM and ATR, respectively) are required for maintaining telomere length homeostasis, as simultaneous deletion of both TEL1 and MEC1 results in progressive shortening of telomeres leading to cellular senescence (Ritchie et al. 1999). Cells lacking MEC1 have slightly shortened telomeres, while cells lacking TEL1 have very short but stable telomeres (Greenwell et al. 1995; Ritchie et al. 1999), suggesting that Tel1 has the major role in telomere length regulation. To test if Tel1 is important for making telomerase become more processive at telomeres <125 bp in length, we performed the 2T-STEX assay in a tel1Δ background (Fig. 5). The percentage of wild-type repeats used was 62.5% (25 of 40). For this percentage, the expected average number of wild-type repeats per run of wild-type repeats should be 2.7 if telomerase was completely nonprocessive. At telomeres <125 bp in length, the observed average number of wild-type repeats per run was 2.6, indicating that in cells lacking TEL1, telomerase is nonprocessive even at telomeres <125 bp in length. Accordingly, we found that the increase in telomere extension lengths at telomeres <125 bp in length was not observed in a tel1Δ background (Fig. 5). Regular “one-template” STEX assays (in which both the donor and recipient cells carry wild-type copies of TLC1) performed in a tel1Δ background also lacked long extension events at short telomeres (Arneric and Lingner 2007). The change in repeat addition processivity at short telomeres as measured by the average number of wild-type repeats per run (∼2.9-fold; 7.6 repeats for wild type vs. 2.6 repeats for te1lΔ) mirrored the change in the average telomere extension length (threefold; 102 nt for wild type vs. 34 nt for tel1Δ) (Fig. 5). Thus, the inability of telomerase to add long telomeric extensions on extremely short telomeres in tel1Δ mutants is a result of its inability to act processively at these telomeres.
Figure 5.
Tel1 is needed for enhanced repeat addition processivity at critically short telomeres. (Top panel) The average lengths of the telomere extensions from the 2T-STEX and tel1Δ 2T-STEX assays were determined for telomeres <125 bp in length. (Bottom panel) Repeat addition processivity is shown as the average number of wild-type repeats added per run of wild-type repeats for telomeres <125 bp in length. The P-values associated with two-tailed unpaired t-tests are shown.
Discussion
In this study, we showed that S. cerevisiae telomerase complexes can dissociate and reassociate to extend a given telomere during one cell cycle. We demonstrated that telomerase acts nonprocessively in repeat addition except at extremely short telomeres (i.e., telomeres <125 bp in length). We determined that, although these telomeres do not likely arise from normal telomere erosion events such as incomplete DNA replication or nucleolytic degradation, these telomeres do occur in vivo. Furthermore, we found that the enhancement of repeat addition processivity at extremely short telomeres is mediated by Tel1.
Telomerase can dissociate and reassociate to extend a given telomere during one cell cycle
Previous work (Singer and Gottschling 1994; Prescott and Blackburn 1997b; Henning et al. 1998; Lin et al. 2004) and the work reported here clearly demonstrate that mutations in the templating region of TLC1 can be copied into telomeric DNA in vivo. By coexpressing a tlc1 template mutant along with wild-type TLC1 and analyzing telomere extension events after one cell cycle, we found that some extensions contained both wild-type and mutant repeats, indicating that both RNA templates were used to extend these telomeres. This suggests that telomerase complexes can dissociate and reassociate to extend a given telomere during one cell cycle.
Est2, the reverse transcriptase protein subunit of telomerase (Lingner et al. 1997; Counter et al. 1997), is found at telomeres throughout the cell cycle, whereas Est1 only associates with telomeres in late S and G2 phases, when telomeres are lengthened by telomerase (Taggart et al. 2002). Since our data show that telomerase can dissociate and reassociate from telomeric 3′ ends, the localization of Est2 to telomeres is likely dynamic. In other words, telomerase core complexes are unlikely to be sitting statically at telomeres waiting for Est1 to arrive and activate them in late S phase. Indeed, the observation from our 2T-STEX assays that some telomere extension events began with the incorporation of mutant sequence in an assay using a wild-type TLC1 recipient or wild-type sequence in an assay using a mutant tlc1-tm recipient support the argument that telomerase complexes can and do dissociate and reassociate from telomeres, both before they act on telomeres and while telomeres are being extended.
As it has been shown that yeast telomerase is likely a dimer or multimer (Prescott and Blackburn 1997a, b), it is formally possible that the telomere extensions containing a mixture of wild-type and mutant sequences are the result dimeric or multimeric telomerase complexes, with each telomere extension event carried out by only one telomerase complex that is capable of using different RNA templates present in the complex. We are not in favor of this model because the extensions containing a mixture of wild-type and mutant sequences do not reveal any discernable pattern that would be indicative of a regular intracomplex template switching mechanism (Fig. 2; Supplementary Fig. S2).
Telomerase is nonprocessive except at extremely short telomeres
Previous work by Prescott and Blackburn (1997b) also analyzed telomerase processivity by coexpressing wild-type TLC1 and various template-mutated tlc1. Sequencing telomeres from the TLC1/tlc1 heterozygous diploids after extensive clonal expansion, Prescott and Blackburn (1997b) found that the mutant repeats were not present as long stretches; instead, they occurred most often either singly or in pairs. Prescott and Blackburn (1997b) suggested that the most likely interpretation for this pattern is that the mutant enzymes acted nonprocessively. Furthermore, some of mutant repeats were separated by single wild-type repeats, indicating that the wild-type telomerase was likely similarly nonprocessive. However, since their study did not analyze STEX events after one cell cycle, Prescott and Blackburn (1997b) did not exclude the possibility that the mixed wild-type and mutant repeat patterns were the result of processive elongation by telomerase, with successive bursts of elongation separated by degradation of newly added telomeric repeats, generated over many cell cycles. Since our study analyzes extension events during one cell cycle, we conclusively showed that telomerase does indeed act nonprocessively in vivo at telomeres that are >125 bp in length.
The Prescott and Blackburn (1997b) study was also not able to assess telomerase processivity as a function of telomere length. Our work demonstrated that while telomerase acts nonprocessively on telomeres >125 bp in length, it possesses significant repeat addition processivity when acting on telomeres <125 bp in length. We propose that this enhancement of processivity is a mechanism that cells use to rapidly elongate critically short telomeres. We showed previously that the frequency of telomere extension decreases steadily as a function of telomere length. Thus, cells have two ways to respond to critically short telomeres—an increase in frequency of extension coupled with an increase in telomerase processivity. While the increase in repeat addition processivity is only approximately two- to threefold, this would correspond to extension lengths of ∼88–132 nt, given that the average extension length during one cell cycle is 44 nt for telomeres >200 bp in length (Teixeira et al. 2004), which is quite significant considering that wild-type yeast telomeres are only ∼300 bp in length.
Although both the frequency of extension and telomerase processivity change with telomere length, the relationship of each to telomere length is different. While the extension frequency steadily increases as telomeres get shorter, repeat addition processivity is bimodal with respect to telomere length. Telomerase is generally nonprocessive at all telomeres greater than ∼125 bp in length but processive on telomeres <125 bp in length. This agrees with the bimodal distribution seen for telomere length extension, where much longer extensions are observed at very short telomeres (Teixeira et al. 2004). Telomerase repeat addition processivity, unlike telomere extension frequency, does not steadily increase with decreasing telomere length. Hence, although an increase in both extension frequency and telomerase processivity occurs at critically short telomeres, the increase in processivity may occur specifically to rapidly elongate these telomeres.
One potential caveat of this study is that it assumes that there is an excess of telomerase of both template sequences available at every telomere. If this assumption is incorrect, there may not be free competition of the two telomerases in our 2T-STEX assays to extend a given telomere. Consequently, the same telomerase would have an advantage to rebind and extend the same telomere. As a result, the calculated processivity, as measured by the average number of wild-type repeats per run of wild-type repeats, could be viewed as only a maximum estimate. Indeed, it has been reported that telomere length homeostasis requires that telomerase levels are limiting in cultured human cells (Cristofari and Lingner 2006), and that telomerase is present in low abundance in yeast (Mozdy and Cech 2006). Our calculated processivity for telomerase is extremely low at telomeres >125 bp in length, so even this “maximum” estimate suggests a nonprocessive enzyme. At a telomere <125 bp in length, it is formally possible that one telomerase enzyme undergoes several cycles of single repeat additions separated by periods where the enzyme has “dissociated” from the telomeric 3′ end, but staying within its vicinity, yielding a pattern of repeats that resembles processive repeat addition. However, since the concept of repeat addition processivity is unique to the telomerase enzyme, and given that the molecular mechanisms behind processive repeat addition by telomerase from any organism has not yet been elucidated, it is not possible to discount this mode of action as processive repeat addition, especially considering that in any model for processive repeat addition, after the extension of the telomeric 3′ end, the new 3′ end must be “dissociated” from the active site of the telomerase enzyme in order for it to be translocated into a position where the enzyme can once again extend it. Repeat addition processivity is the ability of one telomerase enzyme to synthesize more than one repeat at one telomere, an ability that we have shown in this study that yeast telomerase possesses at extremely short telomeres.
Critically short telomeres do not occur due to normal telomere erosion
As discussed in the Results section, it is highly improbable for a yeast telomere to shorten to 125 bp or less in length through telomere erosion caused by incomplete DNA replication or nucleolytic degradation. However, we showed that these extremely short telomeres do occur in vivo. Such telomeres likely arise from double-strand DNA breaks caused by DNA damage or the collapse of DNA replication forks. Indeed, it is well established that telomeric repeats are difficult to replicate and result in increased replication fork pausing events (Cooper et al. 1997; Ivessa et al. 2002; Crabbe et al. 2004; Makovets et al. 2004; Fouche et al. 2006). If a replication fork were to collapse while traversing the telomeric tracts, there would be no replication origin distal to the site of fork collapse to generate a fork to finish replication of the telomere resulting in a truncated telomere. A processive telomerase may then be essential to rapidly elongate a critically shortened truncated telomere.
The function of Tel1 in telomere length homeostasis
Cells lacking TEL1 have very short, but stable, telomeres (Greenwell et al. 1995; Ritchie et al. 1999). Tel1 is able to in vitro phosphorylate Cdc13 (Tseng et al. 2006), a single-stranded telomere-binding protein involved in both telomere protection and telomerase recruitment (Garvik et al. 1995; Lin and Zakian 1996; Nugent et al. 1996; Chandra et al. 2001; Pennock et al. 2001). Furthermore, Tel1 has been shown recently to specifically associate with short telomeres (Bianchi and Shore 2007; Sabourin et al. 2007) and is needed for the preferential binding of Est1 and Est2 to short telomeres (Goudsouzian et al. 2006; Sabourin et al. 2007). Thus, Tel1 appears to be important for the recruitment of telomerase to telomeres. Consistent with this view, we recently demonstrated that cells lacking TEL1 have a greatly decreased frequency of telomerase-mediated telomere elongation (Arneric and Lingner 2007).
In addition, at an artificial telomere lacking subtelomeric repeats, telomerase is unable to mediate the preferential elongation of short telomeres by telomerase in a strain lacking TEL1 (Arneric and Lingner 2007). In this study, we found that Tel1 is also important for increasing telomerase processivity at critically short telomeres. Thus, Tel1 is a major regulator in telomere length homeostasis, affecting multiple aspects of telomerase activity including telomerase recruitment, telomere length determination, and telomerase repeat addition processivity. Since occurrences of telomeres that are <125 bp in length should be relatively infrequent, the reduced frequency of elongation, rather than the reduced processivity, is likely the primary cause of the shortened telomeres of tel1Δ cells.
Interestingly, the frequency of telomere extension at a natural telomere is still regulated in a length-dependent manner even in the absence of Tel1 (Arneric and Lingner 2007). This can be attributed to a redundant mechanism for telomerase regulation that is dependent on the subtelomeric region. Curiously, although our tel1Δ 2T-STEX assayed extension events at an artificial telomere lacking subtelomeric repeats, the extension length defect at short telomeres in tel1Δ mutants is also present at a natural telomere (Arneric and Lingner 2007), suggesting that repeat addition processivity is not affected by the presence or absence of a subtelomeric region. Hence, this redundant mechanism for telomerase regulation seems to only affect the frequency of telomere extension, not extension length and telomerase processivity.
It will be of considerable interest to know if these multiple functions of Tel1 are separable through the phosphorylation of different substrates. Although Cdc13 is the only known substrate for the Tel1 kinase so far (Tseng et al. 2006), it is likely that other substrates exist and phosphorylation of these different substrates may mediate the different aspects of Tel1 function. Since Tel1 has been found recently to be enriched at short telomeres (Bianchi and Shore 2007; Sabourin et al. 2007), it is likely that the phosphorylation of these substrates occur specifically at short telomeres. Some of these substrates may have been identified by a recent proteomic study to identify Mec1 and Tel1 targets (Smolka et al. 2007). In particular, the telomeric protein Rif1 was among the substrates identified (Smolka et al. 2007). Phosphorylation of Rif1 by Tel1 may mediate some of the functions of Tel1 at telomeres. Clearly, much work still needs to be done to elucidate the mechanisms behind telomere length homeostasis and the role of Tel1 in these processes.
Materials and methods
Yeast strains and media
Yeast strains used in this study are listed in Table 1. Standard yeast media and growth conditions were used (Sherman 1991). Mating of yeast cells was performed essentially as described (Teixeira et al. 2004).
Table 1.
Yeast strains used in this study
Telomere sequencing and analysis
Telomere V-R PCR, cloning, and sequence analysis were performed as described (Forstemann et al. 2000; Teixeira et al. 2004). For amplifying telomeres of chromosome VI-R, a primer specific for the subtelomeric sequence of this chromosome was used (5′-ACGTGTGCGTACGCCATATCAATATGC-3′). Sequencing was performed by Fasteris SA.
Expected average number of repeats per run
Assuming that telomerase is completely nonprocessive at adding repeats, the expected average number of wild-type repeats per run of wild-type repeats from the 2T-STEX events was determined using the following equation:
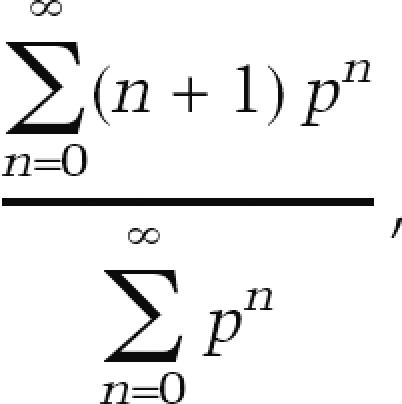 |
where p is the probability of telomerase adding a wild-type repeat, which, for a nonprocessive telomerase, would be equivalent to the fraction of added repeats that have wild-type sequence.
Acknowledgments
We thank Alessandro Bianchi for providing the telomere VI-R primer sequence for telomere PCR and Brian Luke for constructive comments on the manuscript. M.C. was supported by an EMBO Long-term Fellowship and a Human Frontiers Science Program Long-term Fellowship Award. This work was supported by the Swiss National Science Foundation, the Swiss Cancer League, the Human Frontier Science Program, and the EU Sixth Framework Programme.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1588807
References
- Arneric M., Lingner J., Lingner J. Tel1p kinase and subtelomere bound Tbf1p mediate preferential elongation of short telomeres by telomerase in yeast. EMBO Rep. 2007 doi: 10.1038/sj.embor.7401082. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A., Shore D., Shore D. Increased association of telomerase with short telomeres in yeast. Genes & Dev. 2007;21:1726–1730. doi: 10.1101/gad.438907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D., Caputo E., Li J., Hieter P., Boeke J.D., Li J., Hieter P., Boeke J.D., Hieter P., Boeke J.D., Boeke J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Chandra A., Hughes T.R., Nugent C.I., Lundblad V., Hughes T.R., Nugent C.I., Lundblad V., Nugent C.I., Lundblad V., Lundblad V. Cdc13 both positively and negatively regulates telomere replication. Genes & Dev. 2001;15:404–414. doi: 10.1101/gad.861001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M., Blackburn E.H., Blackburn E.H. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- Cooper J.P., Nimmo E.R., Allshire R.C., Cech T.R., Nimmo E.R., Allshire R.C., Cech T.R., Allshire R.C., Cech T.R., Cech T.R. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- Counter C.M., Meyerson M., Eaton E.N., Weinberg R.A., Meyerson M., Eaton E.N., Weinberg R.A., Eaton E.N., Weinberg R.A., Weinberg R.A. The catalytic subunit of yeast telomerase. Proc. Natl. Acad. Sci. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe L., Verdun R.E., Haggblom C.I., Karlseder J., Verdun R.E., Haggblom C.I., Karlseder J., Haggblom C.I., Karlseder J., Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- Crick F.H. Codon–anticodon pairing: The wobble hypothesis. J. Mol. Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Cristofari G., Lingner J., Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M.G., Miller K.M., Cooper J.P., Miller K.M., Cooper J.P., Cooper J.P. Indecent exposure: When telomeres become uncapped. Mol. Cell. 2004;13:7–18. doi: 10.1016/s1097-2765(03)00531-8. [DOI] [PubMed] [Google Scholar]
- Forstemann K., Lingner J., Lingner J. Molecular basis for telomere repeat divergence in budding yeast. Mol. Cell. Biol. 2001;21:7277–7286. doi: 10.1128/MCB.21.21.7277-7286.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann K., Hoss M., Lingner J., Hoss M., Lingner J., Lingner J. Telomerase-dependent repeat divergence at the 3′ ends of yeast telomeres. Nucleic Acids Res. 2000;28:2690–2694. doi: 10.1093/nar/28.14.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann K., Zaug A.J., Cech T.R., Lingner J., Zaug A.J., Cech T.R., Lingner J., Cech T.R., Lingner J., Lingner J. Yeast telomerase is specialized for C/A-rich RNA templates. Nucleic Acids Res. 2003;31:1646–1655. doi: 10.1093/nar/gkg261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouche N., Ozgur S., Roy D., Griffith J.D., Ozgur S., Roy D., Griffith J.D., Roy D., Griffith J.D., Griffith J.D. Replication fork regression in repetitive DNAs. Nucleic Acids Res. 2006;34:6044–6050. doi: 10.1093/nar/gkl757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton T.B., Blackburn E.H., Blackburn E.H. Identification of Kluyveromyces lactistelomerase: Discontinuous synthesis along the 30-nucleotide-long templating domain. Mol. Cell. Biol. 1998;18:4961–4970. doi: 10.1128/mcb.18.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B., Carson M., Hartwell L., Carson M., Hartwell L., Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudsouzian L.K., Tuzon C.T., Zakian V.A., Tuzon C.T., Zakian V.A., Zakian V.A. S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol. Cell. 2006;24:603–610. doi: 10.1016/j.molcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Greenwell P.W., Kronmal S.L., Porter S.E., Gassenhuber J., Obermaier B., Petes T.D., Kronmal S.L., Porter S.E., Gassenhuber J., Obermaier B., Petes T.D., Porter S.E., Gassenhuber J., Obermaier B., Petes T.D., Gassenhuber J., Obermaier B., Petes T.D., Obermaier B., Petes T.D., Petes T.D. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- Greider C.W. Telomerase is processive. Mol. Cell. Biol. 1991;11:4572–4580. doi: 10.1128/mcb.11.9.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider C.W., Blackburn E.H., Blackburn E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Greider C.W., Blackburn E.H., Blackburn E.H. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- Greider C.W., Blackburn E.H., Blackburn E.H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- Harley C.B., Futcher A.B., Greider C.W., Futcher A.B., Greider C.W., Greider C.W. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Henning K.A., Moskowitz N., Ashlock M.A., Liu P.P., Moskowitz N., Ashlock M.A., Liu P.P., Ashlock M.A., Liu P.P., Liu P.P. Humanizing the yeast telomerase template. Proc. Natl. Acad. Sci. 1998;95:5667–5671. doi: 10.1073/pnas.95.10.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug N., Lingner J., Lingner J. Telomere length homeostasis. Chromosoma. 2006;115:413–425. doi: 10.1007/s00412-006-0067-3. [DOI] [PubMed] [Google Scholar]
- Ivessa A.S., Zhou J.Q., Schulz V.P., Monson E.K., Zakian V.A., Zhou J.Q., Schulz V.P., Monson E.K., Zakian V.A., Schulz V.P., Monson E.K., Zakian V.A., Monson E.K., Zakian V.A., Zakian V.A. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes & Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.J., Zakian V.A., Zakian V.A. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl. Acad. Sci. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Smith D.L., Blackburn E.H., Smith D.L., Blackburn E.H., Blackburn E.H. Mutant telomere sequences lead to impaired chromosome separation and a unique checkpoint response. Mol. Biol. Cell. 2004;15:1623–1634. doi: 10.1091/mbc.E03-10-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Hughes T.R., Shevchenko A., Mann M., Lundblad V., Cech T.R., Hughes T.R., Shevchenko A., Mann M., Lundblad V., Cech T.R., Shevchenko A., Mann M., Lundblad V., Cech T.R., Mann M., Lundblad V., Cech T.R., Lundblad V., Cech T.R., Cech T.R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Lue N.F., Peng Y., Peng Y. Identification and characterization of a telomerase activity from Schizosaccharomyces pombe. Nucleic Acids Res. 1997;25:4331–4337. doi: 10.1093/nar/25.21.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue N.F., Peng Y., Peng Y. Negative regulation of yeast telomerase activity through an interaction with an upstream region of the DNA primer. Nucleic Acids Res. 1998;26:1487–1494. doi: 10.1093/nar/26.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V., Szostak J.W., Szostak J.W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- Makovets S., Herskowitz I., Blackburn E.H., Herskowitz I., Blackburn E.H., Blackburn E.H. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol. Cell. Biol. 2004;24:4019–4031. doi: 10.1128/MCB.24.9.4019-4031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S., Brevet V., Gilson E., Brevet V., Gilson E., Gilson E. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 1999;18:3509–3519. doi: 10.1093/emboj/18.12.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin G.B. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- Mozdy A.D., Cech T.R., Cech T.R. Low abundance of telomerase in yeast: Implications for telomerase haploinsufficiency. RNA. 2006;12:1721–1737. doi: 10.1261/rna.134706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent C.I., Hughes T.R., Lue N.F., Lundblad V., Hughes T.R., Lue N.F., Lundblad V., Lue N.F., Lundblad V., Lundblad V. Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- Pennock E., Buckley K., Lundblad V., Buckley K., Lundblad V., Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- Prescott J., Blackburn E.H., Blackburn E.H. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes & Dev. 1997a;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott J., Blackburn E.H., Blackburn E.H. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes & Dev. 1997b;11:528–540. doi: 10.1101/gad.11.4.528. [DOI] [PubMed] [Google Scholar]
- Prowse K.R., Avilion A.A., Greider C.W., Avilion A.A., Greider C.W., Greider C.W. Identification of a nonprocessive telomerase activity from mouse cells. Proc. Natl. Acad. Sci. 1993;90:1493–1497. doi: 10.1073/pnas.90.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K.B., Mallory J.C., Petes T.D., Mallory J.C., Petes T.D., Petes T.D. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin M., Tuzon C.T., Zakian V.A., Tuzon C.T., Zakian V.A., Zakian V.A. Telomerase and Tel1p preferentially associate with short telomeres in S. cerevisiae. Mol. Cell. 2007;27:550–561. doi: 10.1016/j.molcel.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Singer M.S., Gottschling D.E., Gottschling D.E. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- Smolka M.B., Albuquerque C.P., Chen S.H., Zhou H., Albuquerque C.P., Chen S.H., Zhou H., Chen S.H., Zhou H., Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc. Natl. Acad. Sci. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart A.K., Teng S.C., Zakian V.A., Teng S.C., Zakian V.A., Zakian V.A. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]
- Teixeira M.T., Arneric M., Sperisen P., Lingner J., Arneric M., Sperisen P., Lingner J., Sperisen P., Lingner J., Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- Tseng S.F., Lin J.J., Teng S.C., Lin J.J., Teng S.C., Teng S.C. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 2006;34:6327–6336. doi: 10.1093/nar/gkl786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G., McClain W.H., McClain W.H. The G × U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.S., Zakian V.A., Zakian V.A. Sequencing of Saccharomyces telomeres cloned using T4 DNA polymerase reveals two domains. Mol. Cell. Biol. 1990;10:4415–4419. doi: 10.1128/mcb.10.8.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G.L., Bradley J.D., Attardi L.D., Blackburn E.H., Bradley J.D., Attardi L.D., Blackburn E.H., Attardi L.D., Blackburn E.H., Blackburn E.H. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature. 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]