Abstract
A mammalian cytosolic FAD-dependent enzyme that catalyzes the reduction of quinones by N-ribosyl- and N-alkyldihydronicotinamides, but not by NADH, NADPH, or NMNH (reduced nicotinamide mononucleotide), was isolated from bovine kidney more than 30 years ago [S. Liao, J. T. Dulaney and H. G. Williams-Ashman (1962) J. Biol. Chem. 237, 2981–2987]. This enzyme is designated here as quinone reductase type 2 (QR2). Bovine QR2 is a homodimer that migrates on SDS/PAGE at ≈22 kDa. Three tryptic peptides of bovine QR2 (representing 39 amino acids) showed 43% identity to human NAD(P)H:quinone reductase (DT-diaphorase; EC 1.6.99.2), here designated QR1 and 82% identity to a related human cDNA clone [called hNQO2 by A. K. Jaiswal, P. Burnett, M. Adesnik and O. W. McBride (1990) Biochemistry 29, 1899–1906], and designated here as hQR2. The protein encoded by the latter cDNA did not show QR activity when tested with conventional nicotinamide nucleotides. The unexpected high homology between the old flavoenzyme and hQR2 prompted us to clone and overexpress hQR2. The properties of hQR2 were identical to those of the flavoenzyme described by S. Liao and H. G. Williams-Ashman, thus establishing their genetic identity. Recombinant human QR2: (i) reacts with N-ribosyl- and N-alkyldihydronicotinamides, but not with NADH, NADPH, or NMNH; (ii) is very weakly inhibited by dicumarol or Cibacron blue; (iii) is very potently inhibited by benzo[a]pyrene. The x-ray crystal structure of rat QR1 shows that the 43 amino acid C-terminal tail of QR1 provides the binding site for the hydrophilic portions of NADH and NADPH. In the absence of this binding site in QR2, the enzyme retains the essential catalytic machinery, including affinity for FAD, but cannot bind phosphorylated hydride donors.
Keywords: nonphosphorylated dihydronicotinamides, hydride transfer, flavoprotein, polycyclic aromatic hydrocarbons
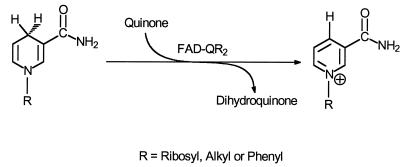
In the early 1960s Liao and Williams-Ashman (1–4) described the purification and properties of a previously unknown mammalian cytosolic FAD-dependent flavoprotein that catalyzes the oxidation of reduced N-ribosyl- and N-alkylnicotinamides by menadione and other quinones (Scheme SI), but not by NADH, NADPH, and NMNH.
Scheme I.
Although this enzyme was highly purified from bovine kidney, and its kinetics, substrate, and inhibitor specificities were determined (4), it has been completely ignored for more than 3 decades, and no information on its structure is available. For reasons to be explained, we here designate this flavoprotein as quinone reductase type 2 (QR2).† Liao et al. (4) pointed out that QR2 has substrate and inhibitor specificities that are strikingly different from those of the more widely known NAD(P)H:(quinone acceptor) oxidoreductase or DT- diaphorase (EC 1.6.99.2)‡ (here designated QR type 1 or QR1). For example, (i) QR2 does not react with NADH, NADPH, or NMNH, all of which are hydride donors for QR1; (ii) QR2 is very weakly inhibited by dicumarol, whereas QR1 is exquisitely sensitive to this anticoagulant; (iii) QR2 is potently inhibited by polycyclic aromatic hydrocarbons and by the estrogens 17β-estradiol, equilenin, and diethylstilbestrol, which only weakly inhibit QR1; and (iv) QR2 is only modestly, if at all, inducible, whereas QR1 is induced by a wide variety of structurally distinct compounds (6, 7). Despite these differences, we show in this paper that QR1 and QR2 are closely related in sequence and structure.
Our laboratory has had a long-standing interest in QR1 because this enzyme is highly inducible by a variety of Michael acceptors and other electrophiles, and its induction is associated with the protection of animals and their cells against electrophile toxicity and chemical carcinogenesis (8). By virtue of its unusual and obligatory 2-electron reduction mechanism (9), QR1 diverts quinones from participating in oxidative cycling that can generate reactive oxygen species and from depleting intracellular glutathione (8). QR1 is also involved in the reductive activation of certain chemotherapeutic quinones such as mitomycins and aziridylbenzoquinones (10). The enzyme is therefore of dual importance for cancer research: it plays roles in both chemical protection against cancer and in the bioactivation of certain cancer chemotherapeutic agents.
The recent report by Friedlos et al. (11) that QR1 could use simple reduced quaternary nicotinamides, such as N-methyldihydronicotinamide and N-ribosyldihydronicotinamide, as hydride donors in place of NADH or NADPH, raised the question whether QR1 and QR2 might be structurally and possibly even functionally related. We examined this possibility in the hope that the three-dimensional crystal structure of rat QR1 (rQR1), recently determined by x-ray diffraction to 2.1-Å resolution (12), might explain the differences in reactivity of the two enzymes with various substituted dihydronicotinamides.
Therefore, we purified bQR2 from bovine kidney by modern improvements of the procedure of Liao et al. (4). We then sequenced three tryptic peptides obtained from bQR2 (consisting of 8, 11, and 20 amino acids) and found that they were highly homologous (82% identical) to the sequence of a cloned human cDNA isolated by Jaiswal and colleagues (5, 13) and designated by these authors as hNQO2. Jaiswal et al. (5, 13) serendipitously isolated the cDNA for hNQO2 during the cloning of human QR1 (hQR1). The deduced amino acid sequences of the two cDNAs coding for hQR1 and hNQO2 were very similar, but no known function was assigned to hNQO2.
The unexpected high homology (82% identity) of the peptides of bQR2 with the amino acid sequence deduced from the human cDNA clone of Jaiswal et al. (5) suggested to us that these enzymes might be genetically identical. Therefore, we cloned the cDNA for hNQO2 and overexpressed and purified the protein that it encodes. This purified protein (which we designate here as QR2) has enzymatic properties identical to those of the flavoprotein isolated by Liao and Williams-Ashman 30 years ago (1–4). The homodimeric subunits of QR2 comprise 231 amino acids, whereas those of QR1 contain 274 amino acids.§ The amino acid sequences of QR1 and QR2 can be aligned without insertions or deletions, but QR2 lacks 43 of the C-terminal residues. Examination of the x-ray crystal structure of rQR1 (12) clarifies the reasons for the differences between the electron donor specificities of QR2 and QR1, since the 43 amino acid carboxyl tail of QR1, which is absent in QR2, provides the binding site for the pyrophosphate–ribose–adenine moiety of NADH and NADPH. This finding also explains the recent observation that QR1 can use not only NADH, NADPH, and NMNH as hydride donors, but also a variety of synthetic reduced N-alkylnicotinamides (11).
EXPERIMENTAL PROCEDURES
Materials.
Protein sequence grade trifluoroacetic acid (TFA) (25%), ammonium sulfate, NADH, NADPH, NMNH, and buffer salts were purchased from Sigma. N-Methylnicotinamide iodide (3-carbamoyl-1-methylpyridinium iodide), sodium hydrosulfite, menadione (vitamin K3), benzo[a]pyrene, and dicumarol were obtained from Aldrich. Cibacron blue was crystallized as the choline salt from methanol (14). All solvents were obtained from J.T. Baker. Centriprep-10 cartridges were from Amicon. Phenyl-Sepharose 6 (fast flow; high substitution), Q-Sepharose Fast Flow, Mono-Q ion exchange column, Superose-12 HR 10/30 gel filtration column, and the fast protein liquid chromatography apparatus were from Pharmacia. The synthesis of oligonucleotides and automatic sequencing of plasmid DNAs were carried out at the Genetic Core Facility of The Johns Hopkins University School of Medicine.
Preparation of N-Methyldihydronicotinamide.
N-Methyldihydronicotinamide was prepared from N-methylnicotinamide iodide according to Rafter and Colowick (15) and stored in 10 mM sodium bicarbonate buffer at pH 10.5 (16). The concentrations of N-methyldihydronicotinamide solutions were determined by use of the molar absorption coefficient (ɛm) of 7000 M−1·cm−1 at 360 nm (15).
Enzyme Assays.
Enzyme activities were determined spectrophotometrically at 25°C, in cuvettes of 1.0-cm light path containing in final volumes of 3.0 ml or 1.0 ml: 60 μM menadione, 80 μM N-methyldihydronicotinamide, 50 mM Tris·HCl buffer (pH 8.50), 0.1% (vol/vol) Triton X-100, and 100 μM dicumarol. The menadione was added as a 30 mM stock solution in ethanol, and the N-methyldihydronicotinamide was added as an 80 mM stock solution (pH 10.5). The absorbance of the reaction mixture was monitored at 360 nm. Minor nonenzymatic decreases in absorbance at 360 nm were subtracted from the total rates to yield the initial enzymatic reaction velocities. The specific activity of the enzyme was calculated on the basis of molar absorbance coefficients (ɛm) of 7000 M−1·cm−1 for N-methyldihydronicotinamide and 1000 M−1·cm−1 for menadione at 360 nm. The oxidized form of N-methylnicotinamide and reduced menadione do not absorb significantly at 360 nm. Specific activities were expressed with respect to protein concentrations measured by the method of Bradford (17) with crystalline BSA as standard.
Purification of QR2 from Bovine Kidney.
The procedures were modified from those described by Liao et al. (4). Bovine kidneys were obtained from the slaughterhouse and quickly frozen. All subsequent operations were carried out at 4°C, unless otherwise noted. One bovine kidney was dissected free from fat and connective tissues and chopped into small pieces. The tissue (420 g) was homogenized in 1 liter of 0.25 M sucrose/3 mM sodium bicarbonate solution. The homogenate was centrifuged at 16,300 × g for 40 min. The supernatant fluid was fractionated with ammonium sulfate, and the fraction precipitating between 40 and 70% saturation was collected, dissolved in 190 ml of 20 mM sodium phosphate buffer (pH 6.8), and dialyzed against water for 24 h. Then, solid KH2PO4 was added to a final concentration of 0.1 M and the pH was adjusted to 4.2 with acetic acid. This solution was heated for 10 min at 38°C to denature most of the QR1 activity, while minimizing the loss of the desired QR2 activity (4). The partially purified material was precipitated with 70% saturated ammonium sulfate, redissolved, and loaded onto a phenyl-Sepharose column in 10 mM Tris acetate (pH 7.5), containing 500 mM sodium chloride, and eluted with a linear gradient from 100% 10 mM Tris acetate/500 mM sodium chloride (pH 7.5) to 100% of a mixture of equal volumes 10 mM Tris acetate (pH 7.5) and ethanol. After concentration of the pooled active fractions (Centriprep-10 cartridges) and dialysis against 5 mM Tris acetate buffer (pH 7.5), the solution was loaded onto a Q-Sepharose Fast Flow column and eluted with a linear gradient from 100% 5 mM Tris acetate (pH 7.5) to 100% 5 mM sodium phosphate/200 mM sodium chloride (pH 6.0). The fractions containing the desired activity were pooled and concentrated by centrifugation on Centriprep-10 cartridges. The proteins were rechromatographed on smaller phenyl-Sepharose and Mono-Q columns, under conditions similar to those described above. Gel filtration was then carried out on a Superose-12 column (10 × 300 mm) in 5 mM sodium phosphate (pH 6.5), containing 200 mM sodium chloride. The column was calibrated with molecular weight markers: BSA (66 kDa), ovalbumin (44 kDa), carbonic anhydrase (29 kDa), trypsinogen (24 kDa).
Homogeneity of the Purified bQR2 and Its Amino Acid Sequence.
Purified bQR2 was subjected to SDS/PAGE on 17% gels according to Laemmli (18). A Waters HPLC system and a Waters Delta Pak 5-μm C4 column (3.9 × 150 mm) were used to analyze the protein and prepare the sample for tryptic digestion and protein sequencing. Two solvent systems were used: (A) 0.1% TFA in water and (B) 0.1% TFA in 85% acetonitrile in water (19). The enzyme was eluted (2.0 ml/min) with a linear gradient from 50% A: 50% B to 30% A: 70% B over a 40-min period at 25°C, monitored at 215 nm. Protein sequencing, amino acid analysis, tryptic digestion, and HPLC separation of peptides, matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) (20) of peptides, and Edman sequencing were carried out at the Harvard MicroChem Facility (Cambridge, MA).
Cloning of hQR2 Gene.
Total cellular RNA was isolated from normal human liver by use of guanidine isothiocyanate-sodium sarkosyl-2-mercaptoethanol, centrifuged through cesium chloride (21), and quantitated by UV absorption. The human liver RNA was reverse transcribed with oligo(dT). The reaction mixture (20 μl) contained: 2 μg RNA, 0.5 μM oligo(dT), 0.5 mM dNTP mixture, 62.5 mM Tris·HCl (pH 8.3), 11.5 mM DTT, 11.25 mM MgCl2, and 12.5 units of reverse transcriptase (avian myeloblastosis virus). The cDNA was then amplified for 30 cycles of 95°C for 1 min, 65°C for 1 min, and 72°C for 1 min in a thermocycler (Ericomp, San Diego), and followed by a final 5-min extension at 72°C. The PCR mixture (50 μl) contained 2 μl of reverse transcriptase cDNA product, 10 mM Tris·HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, 0.2 mM dNTP mixture, 0.2 μM of sense and antisense primers corresponding to the N terminus (primer 1) and the C terminus (primer 2) of the 693 bp open reading frame of the reported cDNA sequence (5), and 2.5 units of Taq DNA polymerase (Boehringer Mannheim).
The primers had the following sequences: Primer 1: 5′-GCATATGGCAGGTAAGAAAGTACTC-3′ (sequence for an added NdeI site and the initiation of translation is underlined) and Primer 2: 5′-CTATTATTGCCCGAAGTGCCAGTG-3′ (antisense sequence of two stop codons TAA and TAG is underlined).
After excision of the ≈700-bp band obtained from reverse transcription–PCR from a low melting point agarose gel, the fragment was ligated into the TA cloning vector (Invitrogen). Colonies with inserts were selected by plating Escherichia coli DH5α cells, which had been transformed with the ligation mixture on an LB (Luria broth) plate containing ampicillin and 5-bromo-4-chloro-3-indolyl β-d-galactoside. Over 90% of the white colonies contained the desired insert. The plasmid DNA was isolated by use of a Qiagen (Chatsworth, CA) Miniprep Kit, and restriction analysis of the fragment with NdeI, EcoRI, and BamHI identified the inserts in the desired orientation. The cDNA coding for the QR2 protein has a single BamHI restriction site. The complete sequence of the insert from both strands was determined by automatic sequencing and was found to be identical with that reported by Jaiswal et al. (5). The hQR2 gene was cut by NdeI/EcoRI digestions and then ligated into a pET-22 expression vector (Novagen), which had been linearized by NdeI/EcoRI digestions. The ligation mixture was then used to transform E. coli DH5α. PCR with primers 1 and 2 of hQR2, with the colonies dispersed in a small amount of LB solution as templates, showed that 9 of 10 colonies carried the desired transformation of the hQR2 gene. The pET-22 vector with hQR2 insert was then transformed into E. coli BL21(DE3) pLys S or BL21(DE3) (Novagen) for overexpression after Miniprep of DNA from the DH5α culture.
Overexpression of Recombinant hQR2 and Its Catalytic Activity.
The E. coli strain carrying the hQR2 gene was grown at 30°C to an absorbance of 0.8–1.0 at 600 nm. Then, 1 mM of isopropyl-β-d-thiogalactopyranoside was added to induce the protein expression, and the bacteria were allowed to grow to the stationary phase. Protein concentrations were determined by the method of Bradford (17) on the cell lysate and on the partially purified hQR2 (40–75% saturated ammonium sulfate).
Inhibition of hQR2 and rQR1 by Benzo(a)pyrene, Dicumarol, and Cibacron Blue.
Inhibition of partially purified recombinant hQR2 (specific activity 15 μmol per min/mg of protein) was compared with rQR1 purified from livers of azo-dye-induced rats with a specific activity 1875 μmol per min/mg of protein (22). The stock solutions of the inhibitors were prepared in ethanol. Appropriate concentrations of inhibitors were obtained by diluting stock solutions prepared in ethanol and adding 10 μl of these solutions to the reaction mixture containing 50 mM Tris·HCl buffer at pH 8.5, 30 μM menadione, and 80 μM N-methyldihydronicotinamide in a total volume of 1.0 ml. The reactions were carried out at 25°C and monitored at 360 nm for both QR1 and QR2, since N-alkyldihydronicotinamides are also good hydride donors for QR1 (11).
RESULTS AND DISCUSSION
Purification of Bovine Kidney QR2.
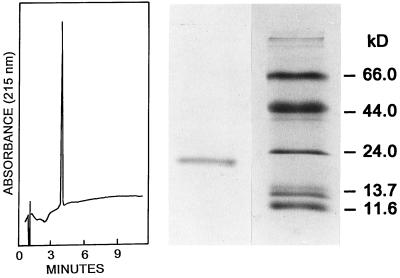
The enzyme was purified by successive ammonium sulfate fractionation, heat treatment, and hydrophobic (phenyl-Sepharose) and ion-exchange chromatographies, as described in Experimental Procedures. These steps resulted in a 3100-fold increase in specific activity to give a final product that catalyzed the oxidation of 30 μmol of N-methyldihydronicotinamide per min/mg of protein with menadione as electron acceptor, which is comparable to the specific activity reported by Liao et al. (4) with N-propyldihydronicotinamide as substrate. The rates of reaction of bQR2 with N-methyl- and N-propyldihydronicotinamide are similar (4). SDS/PAGE of the purified enzyme displayed a single band migrating at the position corresponding to ≈22 kDa (Fig. 1). We believe that this value underestimates the true molecular weight, since QR2 is very hydrophobic, as indicated by its tight binding to phenyl-Sepharose (this work) and its exceedingly potent inhibition by polycyclic aromatic hydrocarbons and phenolic steroids (2, 4). It is well known that hydrophobic proteins tend to bind higher quantities of SDS and consequently migrate more rapidly than do most common proteins (23).
Figure 1.
Reverse-phase HPLC (Left) and SDS/PAGE (Right) of purified bovine kidney QR2. The single band observed on the SDS/PAGE migrates at 22.0 kDa. The molecular weight standards are β-galactosidase (11.6 kDa), RNase (13.7 kDa), trypsinogen (24 kDa), ovalbumin (44 kDa), and BSA (66 kDa). The HPLC of purified bQR2 shows a single absorption peak at 215 nm.
The homogeneity of the purified enzyme was further corroborated by the presence of a single sharp peak on reverse-phase HPLC in 0.1% TFA in water/acetonitrile (Fig. 1). The bright yellow solutions of the purified enzyme showed an absorption spectrum in the range of 350–550 nm, which is characteristic of flavoproteins, as reported earlier by Liao et al. (4), who identified the flavin as FAD. The flavin appears to be very tightly bound to the protein, since extensive chromatographic procedures (ion-exchange and hydrophobic chromatography in the presence of small amounts of ethanol, as well as gel filtration) failed to remove FAD from the active site, and the activity of the purified enzyme was not further increased by added FAD.
Amino Acid Sequence of Tryptic Peptides of bQR2.
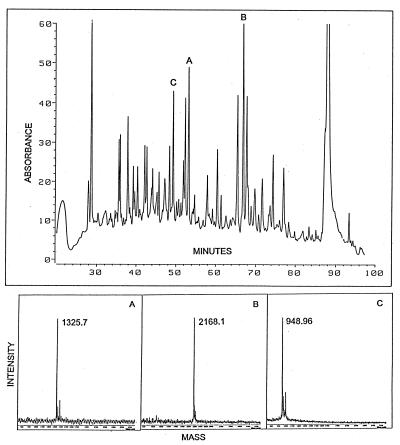
The fractions containing the purified bQR2 that had been subjected to reverse-phase HPLC in TFA/acetonitrile were evaporated to a small volume in a vacuum centrifuge. The material was then digested with trypsin and chromatographed on a Zorbax C18 column. UV absorption showed nearly 50 well-resolved peaks (Fig. 2). Three peaks (designated A, B, and C) were subjected to MALDI-MS (Fig. 2) and Edman sequencing (Table 1). There was excellent agreement between the calculated weights of the peptides (containing 11, 20, and 8 amino acids, respectively) and the corresponding MALDI-MS M/z (M + 1) determinations (Table 1). The three peptides derived from bQR2 had a high degree of identity (75–91%) with the deduced sequence of hQR2 cDNA, and a lesser yet highly significant homology to hQR1 (5) (Table 1 and Fig. 3). On gel filtration, bQR2 migrated at 40 kDa (not shown) indicating that this enzyme is a homodimer like QR1.
Figure 2.
Tryptic mapping of homogeneous beef kidney QR2. (Upper) HPLC trace from a Zorbax C18 column monitored at 215 nm is shown and three peptides selected for MALDI-MS and Edman sequencing are identified as A, B, and C (Table 1). Simultaneous monitoring at 277 nm and 292 nm (not shown) indicated the presence of tyrosine residues in A and B, and a tryptophan residue in C. (Lower) MALDI-MS (20) of peptide fragments A, B, and C. The molecular weights and Edman sequences of the peptides are given in Table 1.
Table 1.
Comparison of sequences of tryptic peptides of bQR2 with hQR1 and hQR2
| Peptide (position) | Amino acid
|
Mol. wt. MALDI (calculated) | |
|---|---|---|---|
| Sequence | Identity with bQR2 | ||
| A | bQR2 VLIVYAHQEPR | – | 1325.7 |
| (6–16) | hQR2 ∗∗∗∗∗∗∗∗∗∗K | 10/11 | (1324.5) |
| (hQR1 A∗∗∗L∗∗S∗RT) | 6/11 | ||
| B | bQR2 DITGTLSNPGFFNYGVEAHK | 2168.1 | |
| (55–74) | hQR2 ∗∗∗∗∗∗∗∗∗EV∗∗∗∗∗∗T∗E | 16/20 | (2167.3) |
| (hQR1 ∗∗∗∗K∗KD∗AN∗Q∗PA∗SVL) | 9/20 | ||
| C | bQR2 MVASWAQR | 949.0 | |
| (204–211) | hQR2 ∗∗∗A∗S∗∗ | 6/8 | (948.1) |
| (hQR1 ILEG∗KK∗) | 2/8 | ||
, amino acids identical with those of peptides of bQR2. Mol. wt., molecular weight.
Figure 3.
Sequence and location of the three tryptic peptides (A, B, and C) obtained from purified bQR2 compared with the deduced protein sequences of hQR2 (231 amino acids; Mr 25,952) and hQR1 (274 amino acids; Mr 30,832). The tryptic peptides correspond to the QR sequence as follows: A, residues 6–16; B, residues 55–74; C, residues 204–211. Among the 39 amino acids of the tryptic peptides sequenced there is 82% (32 of 39) identity between hQR2 and the purified bQR2. Note that this similarity is much closer than 49% identity between hQR1 and hQR2 in the overlapping region (231 amino acids from the N terminus).
Cloning, Sequencing, and Overexpression of hQR2.
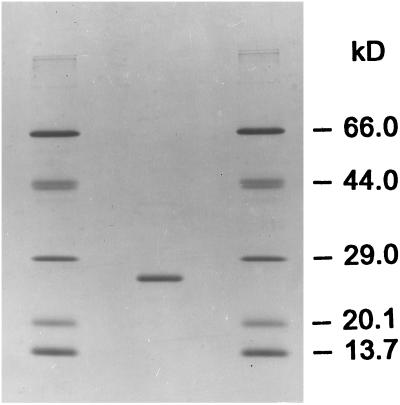
The hQR2 gene was amplified from total RNA of liver by reverse transcription–PCR with the use of two primers corresponding to the N-terminal and C-terminal sequences (see Experimental Procedures and ref. 5). The crude E. coli lysate of an overnight culture, induced with isopropyl-β-d-thiogalactopyranoside when it had reached an absorbance at 0.8–1.0 at 600 nm, had a specific activity of 1.6 μmol per min/mg of protein with N-methyldihydronicotinamide as hydride donor and menadione as acceptor. The enzyme was purified from the crude lysate as follows: nucleic acids were precipitated with 0.1% polyethyleneimine, and the supernatant fluid was then fractionated with solid ammonium sulfate. The 35–65% fraction, which contained most of the activity, was applied to a phenyl-Superose Fast Flow column and eluted with an increasing glycerol gradient. Pooled fractions were concentrated and applied to a Q Sepharose Fast Flow column, and eluted with a 0–200 mM NaCl gradient. Fractions with activity were combined and applied to a Mono Q column and eluted with a 0–175 mM NaCl gradient, and then subjected to gel filtration on a Superose 12 column. SDS/PAGE of the purified hQR2 showed one major band with an apparent molecular weight of 24.6 kDa (calculated 25,951) and one minor higher molecular weight contaminant (Fig. 4). The specific activity of the purified enzyme was 242 μmol per min/mg of protein.
Figure 4.
SDS/PAGE of purified hQR2. The cloned, overexpressed, and purified hQR2 migrated with a Mr 24.6 kDa. There is a trace contaminant of higher molecular weight. The standards are: RNase (13.7 kDa), soybean trypsin inhibitor (20.1 kDa), carbonic anhydrase (29 kDa), ovalbumin (44 kDa), and BSA (66 kDa).
Properties of Recombinant hQR2.
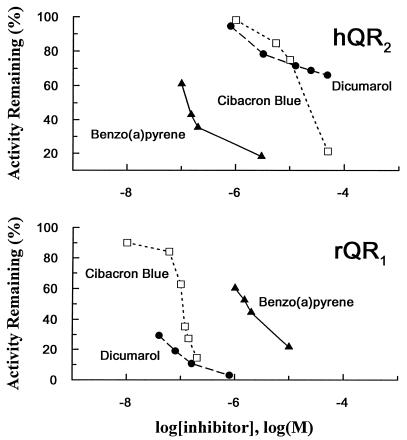
In sharp contrast to the high sensitivity of QR1 to inhibition by dicumarol (Ki = 1–2 nM; ref. 24) and Cibacron blue (Ki = 0.2–0.5 μM) (14, 22, 25), only marginal inhibition of QR2 was observed with 10 μM dicumarol or Cibacron blue, whereas very potent inhibition (≈75%) was observed with 0.1 μM benzo(a)pyrene (Fig. 5). The high affinity of hQR2 for hydrophobic ligands is in accord with the truncation of the C-terminal portion of QR2, which binds the pyrophosphate moiety of NAD(P)H in QR1 (see below and Fig. 6). For both types of enzymes, the affinity for FAD is very high, because the FAD is not removed by a variety of chromatographic steps. The spectrum of purified hQR2 showed absorbances characteristic of flavins in the 350–500 nm range. However, the peak absorbances of FAD are significantly red shifted when bound to hQR2, i.e., from 450 to 456 nm and from 375 to 392 nm for the two major peaks. Thus, the truncation does not appear to affect FAD binding. The affinity for dicumarol appears to reside in major part in the C-terminal tail, since truncation of this tail in QR2 significantly reduces the affinity for this inhibitor.
Figure 5.
Striking differences between the inhibitor specificities of the two types of QR. (Upper) hQR2. (Lower) rQR1. Activity was measured with 30 μM menadione/80 μM N-methyldihydronicotinamide in 50 mM Tris·HCl buffer (pH 8.5) in the presence of 1% ethanol. Benzo(a)pyrene is a potent inhibitor of hQR2, consistent with the results with rQR2 and bQR2 (1–4), whereas QR1 is known to be very sensitive to inhibition by dicumarol and Cibacron blue (22, 24).
Figure 6.
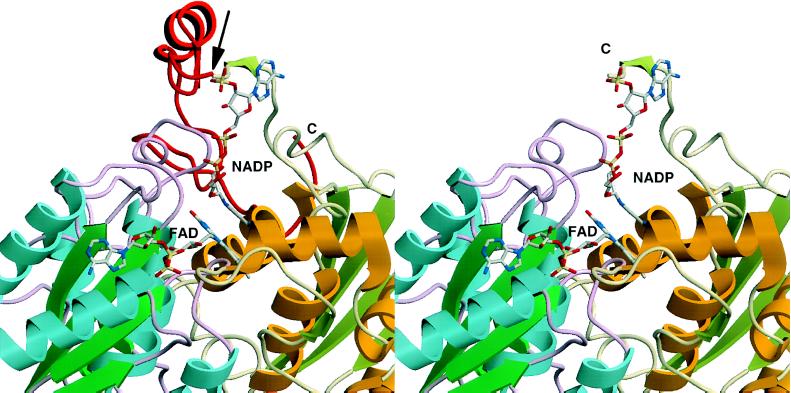
Three-dimensional structure of rQR1 in complex with FAD and NADP+ (12). The C-terminal 43 amino acids (232–274) are shown in red (Left), and have been truncated at the arrow (Right). Different colors of the backbone indicate the two different subunits. It can be seen that the truncated portion of QR2 is critical for binding of the pyrophosphate moiety of the NAD(P)H cofactor. Note that the contact regions between the enzyme and FAD are presumably preserved in QR2 (Right), consistent with the tight binding of FAD. The structural comparison between rQR1 and hQR2 is appropriate since the homology between rQR1 and hQR1 is very high (85% identity).
As reported for the mouse, rat, and bQR2, recombinant hQR2 is also completely inert toward NADPH, NADH, and NMNH. That NMNH is neither a substrate nor an inhibitor of hQR2 suggests that the introduction of only one negatively charged phosphate group abolishes binding of dihydronicotinamide derivatives to QR2.
Comparison of Structures and Function of QR1 and QR2.
The similarities and differences of kinetic properties, hydride donor specificities, and inhibitor profiles of QR1 and QR2 are consistent with the amino acid sequences of the two enzymes and the x-ray crystal structure of QR1 (12). The high degree of sequence homology between hQR2 and hQR1 legitimizes such comparisons. The crystal structure of rQR1 (12) and proteolytic digestion experiments (26) have indicated that this enzyme consists of two domains: a major catalytic N-terminal domain (amino acids 1–220) and a much smaller C-terminal domain (residues 221–274). Truncation of QR2 at residue 231 occurs in a loop near the juncture of the two domains between β-sheets 8 and 9 (figure 1 of ref. 12). The two-domain structure is also consistent with the observations (26) that trypsin, Staphylococcus aureus protease, and chymotrypsin cleaved rQR1 in the same region and produced similar major truncations at K239–K240 (27.0 kDa), D229–L230 (25.9 kDa), and F228–D229 (25.7 kDa),‡ respectively. Moreover, the proteolytic N-terminal fragments retained the dimeric form and the binding affinity for FAD (26), similar to QR2. The proximity to the active site of the C-terminus of QR2, which stabilizes the adenine ring of NAD(P)H in QR1, could explain the greater hydrophobicity of the active site, which favors binding of hydrophobic substrates and inhibitors.
In analogy to QR1 (12, 27), we anticipate that QR2 will also promote a ping-pong mechanism resulting in a two-electron reduction, because the basic enzymatic machinery appears to be the same for the two enzymes. Furthermore, (i) the inhibition of QR1 by dicumarol is linearly competitive with respect to NADH, but not to quinones (25), consistent with the observation that QR2 lacks affinity for dicumarol and is unable to use NADH or NADPH, which are competitors with dicumarol; (ii) the x-ray crystal structure of rat liver QR2 showed that Cibacron blue, a potent and competitive inhibitor of QR1 with respect to NAD(P)H (22), occupied the same site as the nucleotides (12). This observation is also consistent with the ping-pong enzyme kinetics. Moreover, Cibacron blue is only a weak inhibitor of QR2, and this enzyme does not bind to a Cibacron blue affinity column, in striking contrast to QR1, which binds to Cibacron blue very tightly and was purified in a single step on this type of column (22); and (iii) the adenine ring of NADP+ and the anthraquinone ring of Cibacron blue interact with the C-terminal domain (residues 232 and higher), especially the main chain of the loop connecting strands 8 and 9 (L230–F236) (12). The binding site for the pyrophosphate moiety of NAD(P)H resides in this C-terminal tail. It is therefore of interest that the C-terminal 43 amino acids are exceedingly rich in the basic amino acid lysine (8 of 43 residues), whereas the remainder of the chain contains only 8 lysines among 231 amino acid residues. These observations suggest that QR2 may be viewed as a C-terminal truncation analogue of QR1, and that the absence of the carboxyl-terminal tail should favor the participation of smaller hydride donors as cofactors in the oxidoreduction reactions (Fig. 6).
There is substantial evidence that, by virtue of its obligatory two-electron reduction mechanism, QR1 plays an important role in protecting cells against the toxicities of quinones and the reactive oxygen species that arise from oxidative cycling of quinones (8). QR1 is highly induced in mammalian cells coordinately with other Phase 2 enzymes, resulting in protection against the toxic and neoplastic effects of electrophiles (7, 28). The role of QR2 is much more obscure. It is only modestly inducible (5, 13) and its natural hydride donor is presumably N-ribosyldihydronicotinamide, which may arise from the hydrolytic breakdown of NADH or NADPH that occurs to varying degrees in a number of tissue extracts (3, 4). The quantitative importance of these reactions in intact tissues is not known. NMNH can be hydrolyzed by phosphatases to N-ribosyldihydronicotinamide. Significant quantities of oxidized N-ribosylnicotinamide can also arise from the enzymatic breakdown of NAD and NADP in mammalian tissues (3, 29), or by enzymatic synthesis from nicotinamide in the presence of phosphoribosylpyrophosphate (for review see ref. 30). To our knowledge, however, no enzymes promoting reduction of N-ribosylnicotinamide have been described in animal tissues, although Kaplan and colleagues (31) have described this reaction in Pseudomonas fluorescens. A search for such enzymes in animal tissues may therefore be rewarding.
Nicotinamide is released from NAD by a number of important cellular reactions, including nicotinamide nucleotide hydrolases, ADP ribosyltransferases transferases, the extensive ADP ribosylations of proteins, and reactions involved in the formation of poly(ADP) ribose (30). Both endogenous and exogenous nicotinamide are methylated to N-methylnicotinamide in tissues, and this is a normal excretory product found in the urine. Nevertheless, there is no evidence for the occurrence of N-alkylnicotinamide reducing enzymes in animal tissues. Although the physiological significance of QR2 is unclear, the catalytic properties of QR1 and QR2 differ mostly in hydride donor specificity. Because QR1 is believed to play an important role in the reductive bioactivation of quinone and nitro group-containing cytotoxic agents (10, 11, 13, 32), QR2 may play a similar role provided the appropriate hydride donors are available.
In conclusion, an old flavoenzyme discovered more than 30 years ago, which was not even officially recognized by the International Committee on Enzyme Nomenclature, was identified as type 2 QR. Both types of QR appear to have the same catalytic machinery for FAD-mediated hydride transfer, whereas their substrate and inhibitor specificities are strikingly different as a result of truncation of the C-terminal 43 amino acid residues in QR2 (corresponding to the C-terminal domain of QR1).
Acknowledgments
We are grateful to Profs. S. Liao and H. G. Williams-Ashman (University of Chicago) for helpful insight into the discovery and properties of QR2, which they described in the early 1960s. Dr. Mario A. Bianchet kindly prepared the color figures of the QR1 structure. Mindan Huang provided helpful technical assistance. We also thank Dr. William S. Lane, Harvard MicroChem Facility (Cambridge, MA) for the sequence determinations of the purified bQR2, and Gale Doremus for preparing the manuscript and illustrations. These studies were supported by Grant PO1 CA 44530 from the National Cancer Institute, Department of Health and Human Services and by The Burroughs Wellcome Fund (Morrisville, NC). Q.Z. was supported by a fellowship from the National Cancer Institute (5T32 CA09243).
ABBREVIATIONS
- MALDI-MS
matrix-assisted laser desorption ionization mass spectrometry
- NMNH
reduced nicotinamide mononucleotide
- QR1
quinone reductase type 1
- QR2
quinone reductase type 2
- bQR
bovine QR
- hQR
human QR
- rQR
rat QR
- TFA
trifluoroacetic acid
Footnotes
Our interest in the old flavoenzyme described more than 30 years ago by Liao et al. (4) arose from the fact that it catalyzed the same QR reactions as the well-known NAD(P)H:quinone reductase (DT-diaphorase), whereas it has distinct hydride donor requirements and inhibitor specificities. To our surprise and satisfaction, high homology (82% identity) between this old flavoenzyme from bovine kidney and a human cDNA clone, serendipitously cloned by Jaiswal et al. (5) during the cloning of hQR1, unequivocally established the genetic relationship between these two types of oxidoreductases. Therefore, we have named this old flavoenzyme QR2.
This enzyme, which is officially designated NAD(P)H:(quinone acceptor) oxidoreductase (EC 1.6.99.2), has been referred to by a bewildering array of pseudonyms and acronyms, including: DT-diaphorase; QR, quinone reductase; NMOR, NAD(P)H:menadione oxidoreductase; QAO, quinone acceptor oxidoreductase; NQO, nicotinamide nucleotide-quinone oxidoreductase; menadione reductase; phylloquinone reductase; and vitamin K reductase. Because we have consistently used the term QR for this enzyme, we have designated EC 1.6.99.2 as QR1. The prefixes h, r, and b refer to the species of origin of the enzymes human, rat, and bovine, respectively.
Both rQR1 and hQR1 contain 274 amino acids according to their cDNA sequence. The purified rQR1, however, contains 273 aminoacids; the N-terminal methionine residue is absent (see ref. 12).
References
- 1.Liao S, Williams-Ashman H G. Biochem Biophys Res Commun. 1961;4:208–213. doi: 10.1016/0006-291x(61)90272-8. [DOI] [PubMed] [Google Scholar]
- 2.Liao S, Williams-Ashman H G. Biochem Pharmacol. 1961;6:53–54. [Google Scholar]
- 3.Liao, S. (1961) Ph.D. dissertation (University of Chicago, Chicago).
- 4.Liao S, Dulaney J T, Williams-Ashman H G. J Biol Chem. 1962;237:2981–2987. [PubMed] [Google Scholar]
- 5.Jaiswal A K, Burnett P, Adesnik M, McBride O W. Biochemistry. 1990;29:1899–1906. doi: 10.1021/bi00459a034. [DOI] [PubMed] [Google Scholar]
- 6.Talalay P, De Long M J, Prochaska H J. Proc Natl Acad Sci USA. 1988;85:8261–8265. doi: 10.1073/pnas.85.21.8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prestera T, Zhang Y, Spencer S R, Wilczak C, Talalay P. Adv Enzyme Regul. 1993;33:281–296. doi: 10.1016/0065-2571(93)90024-8. [DOI] [PubMed] [Google Scholar]
- 8.Prochaska H J, Talalay P. In: Oxidative Stress: Oxidants and Antioxidants. Sies H, editor. London: Academic; 1991. pp. 195–211. [Google Scholar]
- 9.Iyanagi T, Yamazaki I. Biochim Biophys Acta. 1970;216:282–294. doi: 10.1016/0005-2728(70)90220-3. [DOI] [PubMed] [Google Scholar]
- 10.Ross D, Siegel D, Beall H, Prakash A S, Mulcahy R T, Gibson N W. Cancer Metastasis Rev. 1993;12:83–101. doi: 10.1007/BF00689803. [DOI] [PubMed] [Google Scholar]
- 11.Friedlos F, Jarman M, Davies L C, Boland M P, Knox R J. Biochem Pharmacol. 1992;44:25–31. doi: 10.1016/0006-2952(92)90033-f. [DOI] [PubMed] [Google Scholar]
- 12.Li R, Bianchet M A, Talalay P, Amzel L M. Proc Natl Acad Sci USA. 1995;92:8846–8850. doi: 10.1073/pnas.92.19.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaiswal A K. J Biol Chem. 1994;269:14502–14508. [PubMed] [Google Scholar]
- 14.Prestera T, Prochaska H J, Talalay P. Biochemistry. 1992;31:824–833. doi: 10.1021/bi00118a027. [DOI] [PubMed] [Google Scholar]
- 15.Rafter G W, Colowick S P. J Biol Chem. 1954;209:773–777. [PubMed] [Google Scholar]
- 16.Oppenheimer N J. Mol Cell Biochem. 1994;138:245–251. doi: 10.1007/BF00928468. [DOI] [PubMed] [Google Scholar]
- 17.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Matsudaira P T. A Practical Guide to Protein and Peptide Purification for Microsequencing. San Diego: Academic; 1989. pp. 43–47. [Google Scholar]
- 20.Chait B T, Kent S B H. Science. 1992;257:1885–1894. doi: 10.1126/science.1411504. [DOI] [PubMed] [Google Scholar]
- 21.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 22.Prochaska H J. Arch Biochem Biophys. 1988;267:529–538. doi: 10.1016/0003-9861(88)90060-4. [DOI] [PubMed] [Google Scholar]
- 23.Tung J-S, Knight C A. Biochem Biophys Res Commun. 1971;42:1117–1121. doi: 10.1016/0006-291x(71)90020-9. [DOI] [PubMed] [Google Scholar]
- 24.Hollander P M, Ernster L. Biochim Biophys Acta. 1975;169:560–567. doi: 10.1016/0003-9861(75)90200-3. [DOI] [PubMed] [Google Scholar]
- 25.Prochaska H J, Talalay P. J Biol Chem. 1986;261:1372–1378. [PubMed] [Google Scholar]
- 26.Chen S, Deng P S K, Bailey J M, Swiederek K M. Protein Sci. 1994;3:51–57. doi: 10.1002/pro.5560030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosoda S, Nakamura W, Hayashi K. J Biol Chem. 1974;249:6416–6423. [PubMed] [Google Scholar]
- 28.Talalay P. In: Cellular and Molecular Targets of Chemoprevention. Steele V E, Stoner G D, Boone C W, Kelloff G J, editors. Boca Raton, FL: CRC; 1992. pp. 193–205. [Google Scholar]
- 29.Baum C L, Selhub J, Rosenberg I H. Biochem J. 1982;204:203–207. doi: 10.1042/bj2040203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cervantes-Laurean D, McElvaney N, Moss J. In: Modern Nutrition in Health and Disease. 9th Ed. Shils M E, Olson J A, Shike M, Ross A C, editors. Baltimore: Williams & Wilkins; 1997. in press. [Google Scholar]
- 31.Kaplan N O, Colowick S P, Neufeld E F, Ciotti M M. J Biol Chem. 1953;205:17–22. [PubMed] [Google Scholar]
- 32.Beall H D, Murphy A M, Siegel D, Hargreaves R H J, Butler J, Ross D. Mol Pharmacol. 1995;48:499–504. [PubMed] [Google Scholar]