Abstract
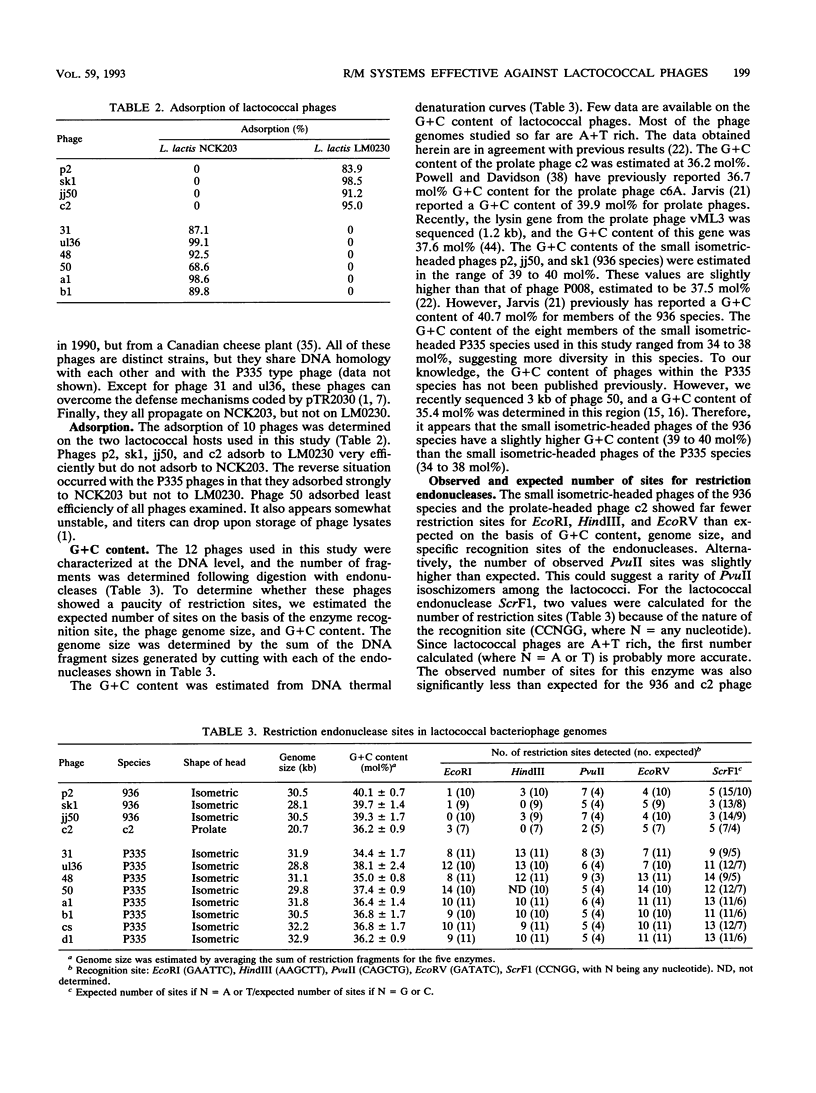
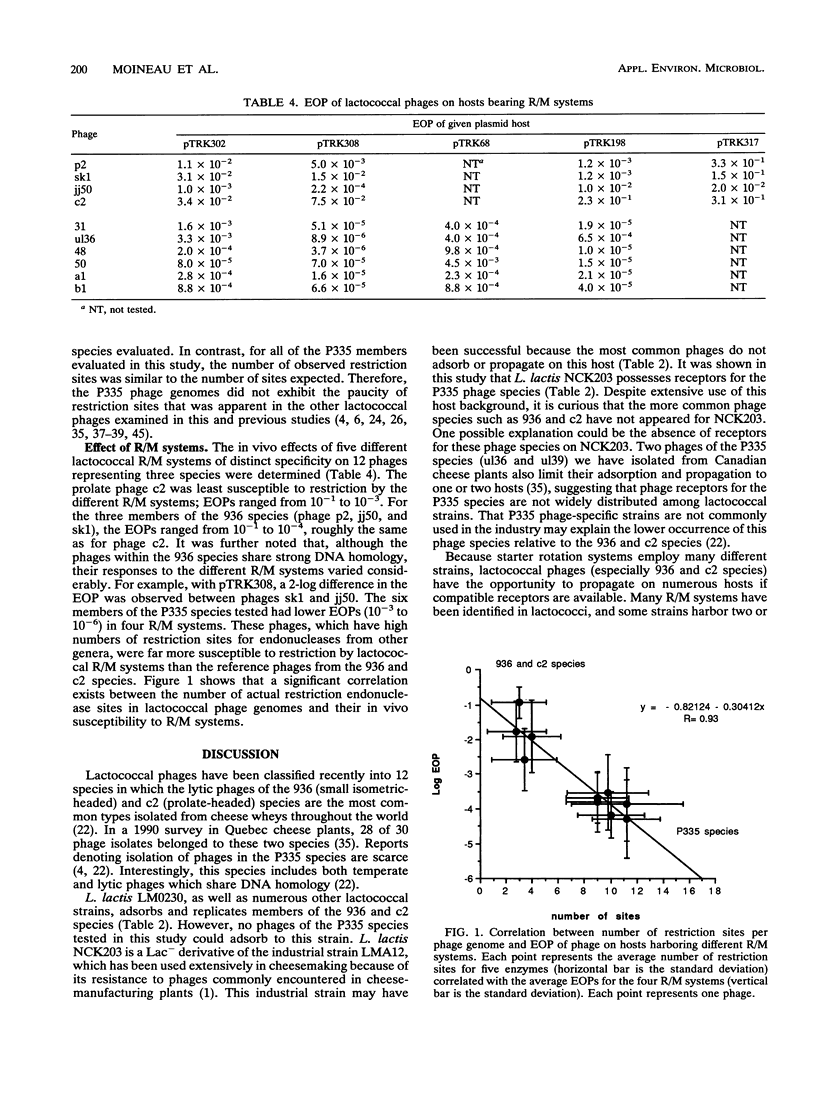
Recently, eight lytic small isometric-headed bacteriophages were isolated from cheese-manufacturing plants throughout North America. The eight phages were different, but all propagated on one strain, Lactococcus lactis NCK203. On the basis of DNA homology, they were classified in the P335 species. Digestion of their genomes in vitro with restriction enzymes resulted in an unusually high number of type II endonuclease sites compared with the more common lytic phages of the 936 (small isometric-headed) and c2 (prolate-headed) species. In vivo, the P335 phages were more sensitive to four distinct lactococcal restriction and modification (R/M) systems than phages belonging to the 936 and c2 species. A significant correlation was found between the number of restriction sites for endonucleases (purified from other bacterial genera) and the relative susceptibility of phages to lactococcal R/M systems. Comparisons among these three phage species indicate that the P335 species may have emerged most recently in the dairy industry.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alatossava T., Klaenhammer T. R. Molecular Characterization of Three Small Isometric-Headed Bacteriophages Which Vary in Their Sensitivity to the Lactococcal Phage Resistance Plasmid pTR2030. Appl Environ Microbiol. 1991 May;57(5):1346–1353. doi: 10.1128/aem.57.5.1346-1353.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussemaer J. P., Schrauwen P. P., Sourrouille J. L., Guy P. Multiple modification/restriction systems in lactic streptococci and their significance in defining a phage-typing system. J Dairy Res. 1980 Oct;47(3):401–409. doi: 10.1017/s0022029900021294. [DOI] [PubMed] [Google Scholar]
- Coveney J. A., Fitzgerald G. F., Daly C. Detailed characterization and comparison of four lactic streptococcal bacteriophages based on morphology, restriction mapping, DNA homology, and structural protein analysis. Appl Environ Microbiol. 1987 Jul;53(7):1439–1447. doi: 10.1128/aem.53.7.1439-1447.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerlad G. F., Daly C., Brown L. R., Gingeras T. R. ScrFI: a new sequence-specific endonuclease from Streptococcus cremoris. Nucleic Acids Res. 1982 Dec 20;10(24):8171–8179. doi: 10.1093/nar/10.24.8171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. L., Sanozky-Dawes R. B., Klaenhammer T. R. Restriction and modification activities from Streptococcus lactis ME2 are encoded by a self-transmissible plasmid, pTN20, that forms cointegrates during mobilization of lactose-fermenting ability. J Bacteriol. 1988 Aug;170(8):3435–3442. doi: 10.1128/jb.170.8.3435-3442.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Miller L. A., Klaenhammer T. R. Cloning, expression, and sequence determination of a bacteriophage fragment encoding bacteriophage resistance in Lactococcus lactis. J Bacteriol. 1990 Nov;172(11):6419–6426. doi: 10.1128/jb.172.11.6419-6426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Miller L. A., Klaenhammer T. R. In vivo genetic exchange of a functional domain from a type II A methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J Bacteriol. 1991 Jul;173(14):4363–4370. doi: 10.1128/jb.173.14.4363-4370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Pierce K., Klaenhammer T. R. The conjugative plasmid pTR2030 encodes two bacteriophage defense mechanisms in lactococci, restriction modification (R+/M+) and abortive infection (Hsp+). Appl Environ Microbiol. 1989 Sep;55(9):2416–2419. doi: 10.1128/aem.55.9.2416-2419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Romero D. A., McKenney D. S., Finer K. R., Klaenhammer T. R. Localization, cloning, and expression of genetic determinants for bacteriophage resistance (Hsp) from the conjugative plasmid pTR2030. Appl Environ Microbiol. 1989 Jul;55(7):1684–1689. doi: 10.1128/aem.55.7.1684-1689.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holo H., Nes I. F. High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. cremoris Grown with Glycine in Osmotically Stabilized Media. Appl Environ Microbiol. 1989 Dec;55(12):3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W. DNA-DNA Homology Between Lactic Streptococci and Their Temperate and Lytic Phages. Appl Environ Microbiol. 1984 May;47(5):1031–1038. doi: 10.1128/aem.47.5.1031-1038.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl Environ Microbiol. 1984 Feb;47(2):343–349. doi: 10.1128/aem.47.2.343-349.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Fitzgerald G. F., Mata M., Mercenier A., Neve H., Powell I. B., Ronda C., Saxelin M., Teuber M. Species and type phages of lactococcal bacteriophages. Intervirology. 1991;32(1):2–9. doi: 10.1159/000150179. [DOI] [PubMed] [Google Scholar]
- Jarvis A. W., Klaenhammer T. R. Bacteriophage Resistance Conferred on Lactic Streptococci by the Conjugative Plasmid pTR2030: Effects on Small Isometric-, Large Isometric-, and Prolate-Headed Phages. Appl Environ Microbiol. 1986 Jun;51(6):1272–1277. doi: 10.1128/aem.51.6.1272-1277.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Klaenhammer T. R. Bacteriophage Resistance Conferred on Lactic Streptococci by the Conjugative Plasmid pTR2030: Effects on Small Isometric-, Large Isometric-, and Prolate-Headed Phages. Appl Environ Microbiol. 1986 Jun;51(6):1272–1277. doi: 10.1128/aem.51.6.1272-1277.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Burge C., Campbell A. M. Statistical analyses of counts and distributions of restriction sites in DNA sequences. Nucleic Acids Res. 1992 Mar 25;20(6):1363–1370. doi: 10.1093/nar/20.6.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger D. H., Bickle T. A. Bacteriophage survival: multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts. Microbiol Rev. 1983 Sep;47(3):345–360. doi: 10.1128/mr.47.3.345-360.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski R. E. Coevolution of bacteria and phage: are there endless cycles of bacterial defenses and phage counterdefenses? J Theor Biol. 1984 Jun 7;108(3):319–325. doi: 10.1016/s0022-5193(84)80035-1. [DOI] [PubMed] [Google Scholar]
- Powell I. B., Davidson B. E. Resistance to In Vitro Restriction of DNA from Lactic Streptococcal Bacteriophage c6A. Appl Environ Microbiol. 1986 Jun;51(6):1358–1360. doi: 10.1128/aem.51.6.1358-1360.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevots F., Mata M., Ritzenthaler P. Taxonomic differentiation of 101 lactococcal bacteriophages and characterization of bacteriophages with unusually large genomes. Appl Environ Microbiol. 1990 Jul;56(7):2180–2185. doi: 10.1128/aem.56.7.2180-2185.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Klaenhammer T. R. Evidence for Plasmid Linkage of Restriction and Modification in Streptococcus cremoris KH. Appl Environ Microbiol. 1981 Dec;42(6):944–950. doi: 10.1128/aem.42.6.944-950.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Klaenhammer T. R. Restriction and modification in group N streptococci: effect of heat on development of modified lytic bacteriophage. Appl Environ Microbiol. 1980 Sep;40(3):500–506. doi: 10.1128/aem.40.3.500-506.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Leonhard P. J., Sing W. D., Klaenhammer T. R. Conjugal strategy for construction of fast Acid-producing, bacteriophage-resistant lactic streptococci for use in dairy fermentations. Appl Environ Microbiol. 1986 Nov;52(5):1001–1007. doi: 10.1128/aem.52.5.1001-1007.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M. Molecular evolution of bacteriophages: evidence of selection against the recognition sites of host restriction enzymes. Mol Biol Evol. 1986 Jan;3(1):75–83. doi: 10.1093/oxfordjournals.molbev.a040377. [DOI] [PubMed] [Google Scholar]
- Shearman C., Underwood H., Jury K., Gasson M. Cloning and DNA sequence analysis of a Lactococcus bacteriophage lysin gene. Mol Gen Genet. 1989 Aug;218(2):214–221. doi: 10.1007/BF00331271. [DOI] [PubMed] [Google Scholar]
- Sijtsma L., Wouters J. T., Hellingwerf K. J. Isolation and characterization of lipoteichoic acid, a cell envelope component involved in preventing phage adsorption, from Lactococcus lactis subsp. cremoris SK110. J Bacteriol. 1990 Dec;172(12):7126–7130. doi: 10.1128/jb.172.12.7126-7130.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]


