Abstract
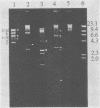
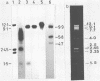
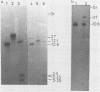
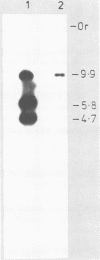
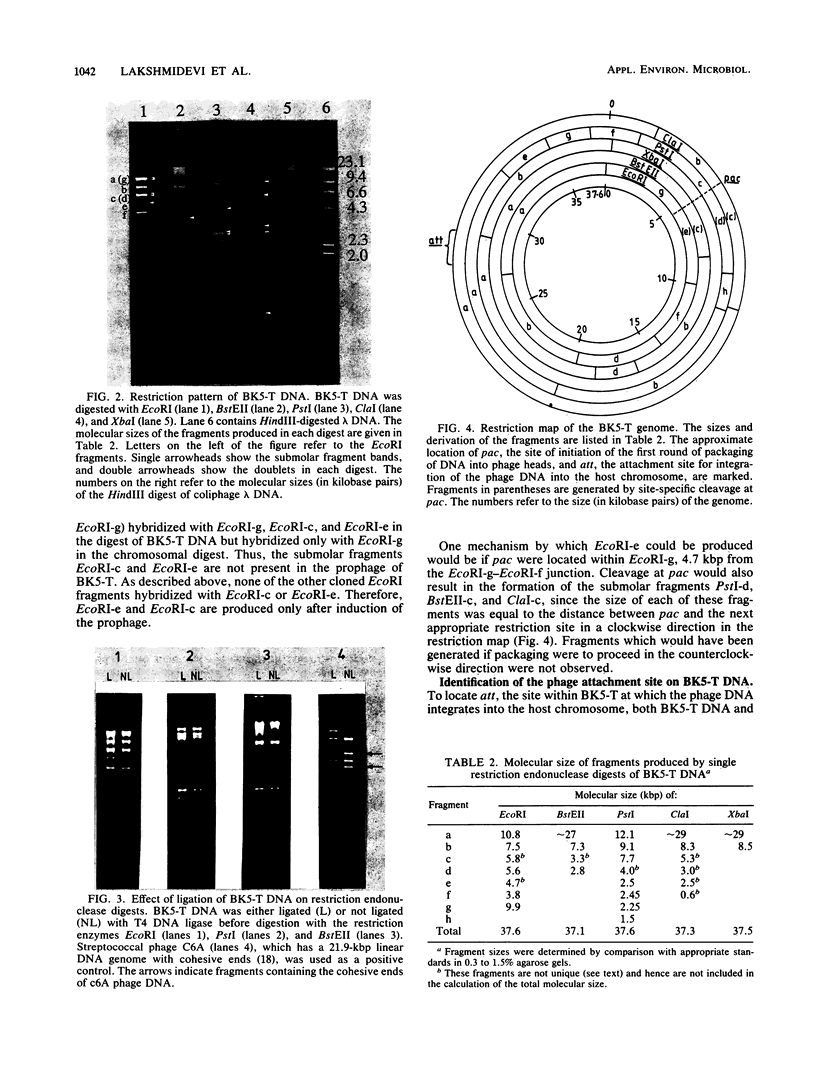
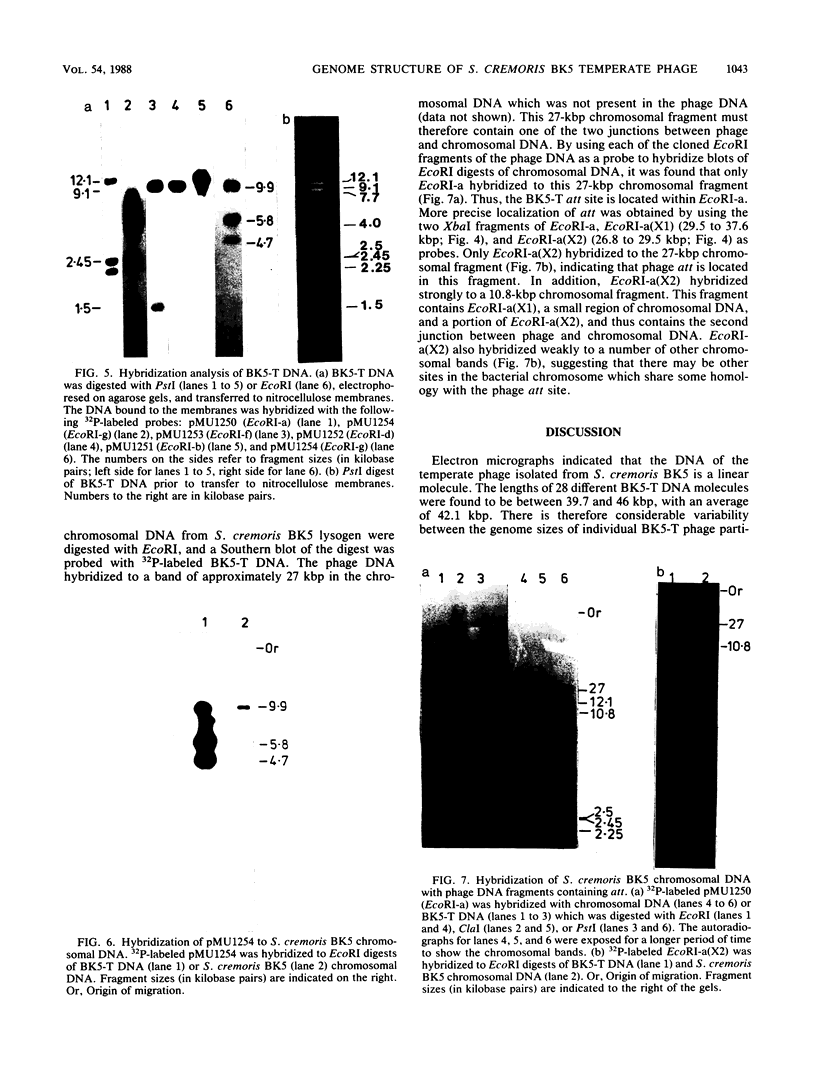
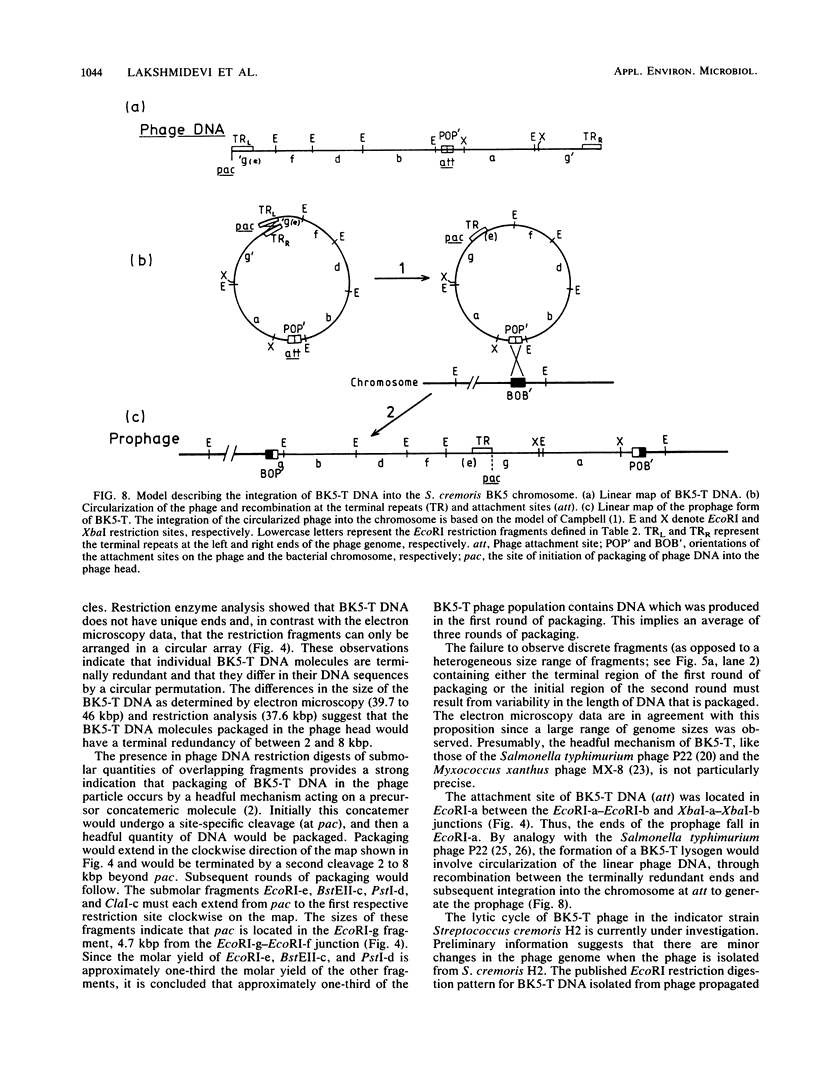
The temperate bacteriophage BK5-T was isolated from Streptococcus cremoris BK5 by induction with mitomycin C. Electron microscopy revealed that BK5-T DNA consists of linear molecules, ranging in size from 39.7 to 46 kilobase pairs. Restriction analysis of self-ligated BK5-T DNA showed that the ends of the DNA were not cohesive. The EcoRI restriction fragments of the phage genome were cloned into pACYC184. Restriction enzyme analysis of both the phage DNA and the cloned EcoRI fragments with EcoRI, BstEII, PstI, ClaI, and XbaI yielded a 37.6-kilobase-pair-long circular restriction map for the phage genome. It was concluded that the BK5-T DNA molecules in the population differ in their sequence by a circular permutation and that individual DNA molecules are terminally redundant. The map location of the sites at which packaging of BK5-T DNA into phage heads is initiated (pac) and at which the phage integrates into the bacterial chromosome (att) were established.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Casjens S., Huang W. M., Hayden M., Parr R. Initiation of bacteriophage P22 DNA packaging series. Analysis of a mutant that alters the DNA target specificity of the packaging apparatus. J Mol Biol. 1987 Apr 5;194(3):411–422. doi: 10.1016/0022-2836(87)90671-1. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins A. R., Sandine W. E. Incidence and properties of temperate bacteriophages induced from lactic streptococci. Appl Environ Microbiol. 1977 Jan;33(1):184–191. doi: 10.1128/aem.33.1.184-191.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson E. N., Jackson D. A., Deans R. J. EcoRI analysis of bacteriophage P22 DNA packaging. J Mol Biol. 1978 Jan 25;118(3):365–388. doi: 10.1016/0022-2836(78)90234-6. [DOI] [PubMed] [Google Scholar]
- Jackson E. N., Miller H. I., Adams M. L. EcoRI restriction endonuclease cleavage site map of bacteriophage P22DNA. J Mol Biol. 1978 Jan 25;118(3):347–363. doi: 10.1016/0022-2836(78)90233-4. [DOI] [PubMed] [Google Scholar]
- Jarvis A. W. DNA-DNA Homology Between Lactic Streptococci and Their Temperate and Lytic Phages. Appl Environ Microbiol. 1984 May;47(5):1031–1038. doi: 10.1128/aem.47.5.1031-1038.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. P., Schlievert P. M. A physical map of the group A streptococcal pyrogenic exotoxin bacteriophage T12 genome. Mol Gen Genet. 1983;189(2):251–255. doi: 10.1007/BF00337813. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Mata M., Trautwetter A., Luthaud G., Ritzenthaler P. Thirteen Virulent and Temperate Bacteriophages of Lactobacillus bulgaricus and Lactobacillus lactis Belong to a Single DNA Homology Group. Appl Environ Microbiol. 1986 Oct;52(4):812–818. doi: 10.1128/aem.52.4.812-818.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Powell I. B., Davidson B. E. Characterization of streptococcal bacteriophage c6A. J Gen Virol. 1985 Dec;66(Pt 12):2737–2741. doi: 10.1099/0022-1317-66-12-2737. [DOI] [PubMed] [Google Scholar]
- Reyrolle J., Chopin M. C., Letellier F., Novel G. Lysogenic strains of lactic Acid streptococci and lytic spectra of their temperate bacteriophages. Appl Environ Microbiol. 1982 Feb;43(2):349–356. doi: 10.1128/aem.43.2.349-356.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M., MacHattie L. A., Thomas C. A., Jr The P22 bacteriophage DNA molecule. I. The mature form. J Mol Biol. 1968 Oct 14;37(1):21–40. doi: 10.1016/0022-2836(68)90071-5. [DOI] [PubMed] [Google Scholar]
- Shimizu-Kadota M., Kiwaki M., Hirokawa H., Tsuchida N. ISL1: a new transposable element in Lactobacillus casei. Mol Gen Genet. 1985;200(2):193–198. doi: 10.1007/BF00425423. [DOI] [PubMed] [Google Scholar]
- Shimizu-Kadota M., Sakurai T., Tsuchida N. Prophage Origin of a Virulent Phage Appearing on Fermentations of Lactobacillus casei S-1. Appl Environ Microbiol. 1983 Feb;45(2):669–674. doi: 10.1128/aem.45.2.669-674.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwag E., Fink J. M., Zissler J. Physical characterization of the genome of the Myxococcus xanthus bacteriophage MX-8. Mol Gen Genet. 1985;199(1):123–132. doi: 10.1007/BF00327521. [DOI] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver S., Levine M. Recombinational circularization of Salmonella phage P22 DNA. Virology. 1977 Jan;76(1):29–38. doi: 10.1016/0042-6822(77)90278-1. [DOI] [PubMed] [Google Scholar]
- Weaver S., Levine M. The timing of erf-mediated recombination in replication, lysogenization, and the formation of recombinant progeny by Salmonella phage P22. Virology. 1977 Jan;76(1):19–28. doi: 10.1016/0042-6822(77)90277-x. [DOI] [PubMed] [Google Scholar]