Abstract
GTP hydrolysis by the transducin α subunit is stimulated by a membrane-bound protein. The identity of this GTPase-activating protein (GAP) is not yet known, but the recent identification of a new gene family encoding regulator of G protein signaling (RGS) proteins raises the possibility that the transducin GAP is an RGS protein. Biochemical evidence shows that RGS proteins act as GAPs for α subunits of the Gi subfamily of G proteins. To identify an RGS protein that could be a GAP for the transducin α subunit, we investigated the expression of RGS proteins in the retina and identified a new RGS domain, RET-RGS-d, which is specifically expressed in the retina. In situ RNA hybridization analyses revealed that RET-RGS-d is expressed in photoreceptor cells as well as in other cells of the retina. Recombinant RET-RGS-d accelerates single turnover hydrolysis of GTP by transducin. We used RET-RGS-d to isolate a full-length cDNA, RET-RGS1, encoding a new RGS protein with a C terminus that corresponds to RET-RGS-d. The N-terminal half of RET-RGS1 contains a putative transmembrane domain and a string of nine cysteines that are potential substrates for multiple palmitoylation. These findings suggest that RET-RGS1 is an integral membrane protein and that it is a candidate for the membrane-associated protein responsible for the GAP activity detected in photoreceptor membranes.
Transducin is a heterotrimeric G protein that plays a central role in vertebrate phototransduction (1). Photoactivation of rhodopsin stimulates the transducin α subunit (Tα) to bind GTP. This active form of Tα then stimulates a phosphodiesterase (PDE) to hydrolyze cGMP, ultimately causing hyperpolarization of the photoreceptor. Like other G proteins, transducin inactivates itself by hydrolyzing the GTP bound to it.
Current models of vertebrate phototransduction suggest that the rate at which this GTP is hydrolyzed might influence the kinetics and sensitivity of the photoresponse. However, there is a disparity between the results of physiological measurements of the photoresponse and in vitro analyses of GTP hydrolysis. Physiological studies show that rod currents recover from dim flashes within a second (2) whereas biochemical studies show that GTP is hydrolyzed by isolated rod transducin in vitro about 10 times more slowly (3–5). Two possible explanations for this discrepancy have been proposed. One is that recovery is not directly linked to GTP hydrolysis (6, 7); however, the ability of guanosine 5′-[γ-thio]triphosphate to block recovery (8) argues against this idea. Alternatively, a factor that accelerates GTP hydrolysis within intact photoreceptors may be missing or too dilute in transducin preparations (4, 5). Several findings support this second explanation. Calorimetry (9) measurements suggest that GTP hydrolysis and subsequent PDE deactivation occur within a second in concentrated suspensions of photoreceptor outer segments. Biochemical evidence indicates that there is a GTPase-activating protein (GAP) which stimulates GTP hydrolysis by transducin (3–5). Initially, this activity was attributed to the PDE inhibitor subunit (PDEγ) (10). But further studies showed that the GAP for transducin is a protein tightly associated with photoreceptor membranes and distinct from PDEγ (4, 5, 11). Its activity may be enhanced by PDEγ (12, 13), but the GAP for transducin has not been purified and its identity is unknown.
A novel family of proteins that regulate G protein signaling (RGS proteins) has been identified during the past year (14). Genetic and cell-transfection studies have revealed that RGS proteins inhibit G protein signaling (15–17) and directly interact with G protein α subunits (19). About 20 members of this family have been identified, including Sst2p from yeast (18), Egl10 from Caenorhabditis elegans (15), and human Gα interacting protein (GAIP) (19). These proteins share a conserved C-terminal domain of 125 amino acid residues referred to as the RGS domain. Recent reports have shown that RGS proteins accelerate the rate of GTP hydrolysis by α subunits of the Gi subfamily (20–22), and that they do so by stabilizing the transition state of Gα for nucleotide hydrolysis (23).
The discovery of RGS proteins led us to investigate whether the GAP for Tα could be a member of this family. To address this issue, we examined the expression of RGS proteins in the retina. Using degenerate PCR, we amplified a sequence coding for a previously unidentified RGS domain, which we refer to as RET-RGS-d. This sequence is specifically expressed in the retina, including photoreceptor cells. We showed that recombinant RET-RGS-d expressed in bacteria accelerates the GTPase activity of Tα in vitro. Using the sequence of RET-RGS-d, we then screened a bovine retina cDNA library and isolated a cDNA clone that encodes RET-RGS1, a novel RGS protein with structural features suggesting that it is, like the photoreceptor GAP, tightly associated with membranes.
MATERIALS AND METHODS
Degenerate PCR and Isolation of RET-RGS cDNA.
First-strand cDNAs were synthesized from bovine retinal poly(A)+ RNA according to GIBCO/BRL instructions. Degenerate PCR primers corresponding to conserved sequences in the core domain of mammalian RGS proteins were used to amplify related sequences from these retinal cDNAs. The sequence of the 5′ and 3′ primers were the following, respectively: (G/C)(A/T)ITT(C/T)TGG(C/A/T)TIGCITG(C/T)GA and AAIC(T/G)IG(G/C)I(T/A)(A/G)I(G/C)(A/T)(A/G)T(C/T)I(T/C)(T/G)IT(C/T/G)CAT. The-250 bp PCR product was cloned into pCRII.1 using a TA cloning kit (Invitrogen). Ninety percent of the clones obtained coded for a new RGS domain sequence which was used as a probe to screen a λgt10 bovine retinal cDNA library (gift from J. Nathans, Johns Hopkins University, Baltimore).
Sequence Analysis Programs.
On-line blast searches were performed via the National Center for Biotechnology Information at the National Institutes of Health (24). prosite (Geneworks, IntelliGenetics) and tm pred (Institut Suisse de Recherches Experimentales sur le Cancer, Lausanne, Switzerland) were used for protein analysis (25). A dendrogram of RGS family was determined using phylip program (University of Washington, Seattle)
Northern Blot Analysis.
Total RNA was extracted from various bovine tissues using a hot acid phenol protocol (26). A Northern blot carrying 20 μg of each of these total RNA was performed as described (27). Hybridizations were performed at 68°C overnight with 2 × 106 cpm/ml of 32P-labeled DNA probe in 5× SSC (1× SSC = NaCl 150 mM/15 mM sodium citrate, pH 7), 5× Denhart’s solution (0.1% Ficoll, 0.1% BSA, 0.1% polyvinylpyrrolidone), 1% SDS, 100 μg/ml salmon sperm DNA, and 40 μg/ml tRNA. High stringency washes were performed in 0.1 SSC/0.1% SDS at 65°C for 1 hr.
In Situ RNA Hybridizations.
Dissected bovine eye cups were fixed for 5 h at 4°C in 4% paraformaldehyde/0.1 M NaPO4 (pH 7.4) and cryoprotected in 30% sucrose-PBS solution overnight. Samples were then embedded in optimal cutting temperature compound (OCT; Miles), rapidly frozen on dry ice, and store at −70°C until use. Cryosections (10 μm) were thaw mounted onto superfrost plus treated microscope slides (Fisher Scientific), refrozen, and stored at −70°C prior to hybridization.
A fragment corresponding to the RGS domain and the 3′ untranslated region of RET-RGS1 cDNA was subcloned into bluescript KS+ vector (Stratagene). Antisense and sense RNA probes were synthesized using, respectively, T3 and T7 RNA polymerase in presence of digoxigenin-labeled UTP (Boehringer Mannheim). A control antisense RNA probe to rod Tα was synthesized using the same protocol.
Pretreatments of the tissue sections and hybridization were performed as described (28), with the following modifications: hybridization was carried out with 4 μg/ml of probe at 68°C and the last stringent washes were performed in 0.1× SSC/0.1% Tween-20 at 75°C for 40 min.
GTPase Assays. Expression of the RGS domain of RET-RGS and purification.
A PCR fragment coding for the 138 C-terminal amino acid residues of RET-RGS1 corresponding to its RGS domain was subcloned into the His-tagged fusion vector pQE 30 (Qiagen, Chatsworth, CA). The hexahistidine tagged fusion was expressed in Escherichia coli strain XL1 blue and purified on a His Bind resin (Novagen) as recommended by the manufacturer.
Preparation of transducin extract and urea-bleached membranes.
Bovine rod outer segments (ROS) were depleted in the dark of their content of soluble proteins by an isotonic wash with 20 mM Tris·HCl (pH 7.5), 120 mM NaCl, and 1 mM MgCl2. Holotransducin together with PDE were then coextracted by incubating the washed ROS in the dark in hypotonic buffer (5 mM Tris·HCl pH 7.5/100 μM MgCl2). The concentration of Tα was estimated by SDS/PAGE using BSA as a standard. Urea-bleached membranes were prepared in the dark as described in Chen et al. (29).
GTPase assays.
Transducin extract (400 nM Tα final concentration) was incubated at room temperature in the presence of bleached urea-washed membranes (5 μM final concentration) with or without RET-RGS-d, in a buffer containing 20 mM Tris·HCl (pH 7.5), 120 mM NaCl, 1 mM MgCl2, 25 μM AMP-PNP, 1 mM DTT. The reaction was started by the addition [γ-32P]GTP to a final concentration of 50 nM and stopped at different time points by addition of 6% final perchloric acid. The release of 32P was analyzed as described (3).
RESULTS
Amplification of the RET-RGS Domain Sequence and Isolation of RET-RGS1 cDNA.
To investigate expression of RGS proteins in the retina, we performed PCR amplification from bovine retinal cDNA using degenerate primers. We designed these degenerate primers to contain all possible codons for the amino acids found in RGS domains of mammalian RGS1, -2, -3, -4, -5, -6, -13 (16) and GAIP (19) at the positions indicated in Fig. 1. The PCR products from the reaction were of the predicted size. They were cloned, and 40 were sequenced. Four different RGS domain sequences were found. Three correspond to RGS2 (30), -3 (16), and -8 (15). The fourth and most represented encodes a novel RGS domain, which we named RET-RGS-d.
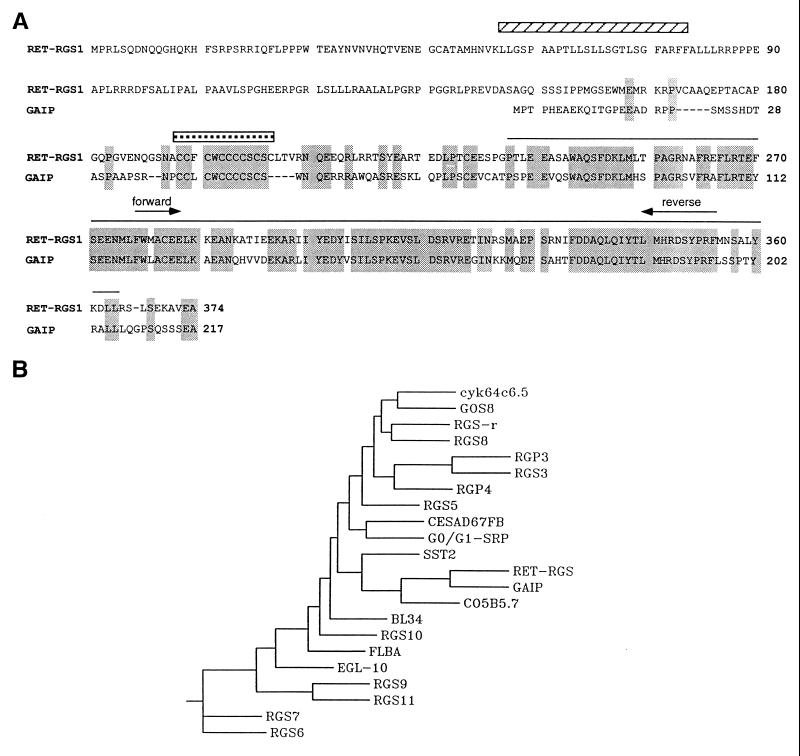
Figure 1.
(A) Sequence alignment between RET-RGS1 and GAIP. Identical amino acid residues are boxed. The sequence corresponding to RET-RGS-d is overlayed by a black line. A striped bar overlays the putative transmembrane domain of RET-RGS1. A stippled bar overlays the cysteine string sequence conserved between RET-RGS1 and GAIP. The PCR degenerate primers used to amplify the conserved RGS domain are indicated by arrows. (B) Phenogram-style tree as determined by phylip analysis of RGS domain sequences available either from the SwissProt database or from recent publications. The RGS domain sequences that were aligned correspond to amino acids 364–430 of the RET-RGS1 sequence. A consensus tree from 40 data sets is shown.
Using this PCR fragment as a probe to screen a λgt10 bovine retina cDNA library, we isolated a 2.4-kb cDNA encoding a new member of the RGS family, RET-RGS1. This cDNA contains a 374 codon ORF preceded by 10 in-frame stop codons upstream of the initiator methionine. The protein encoded by this cDNA has a predicted relative Mr of 44,664. The RGS domain of RET-RGS1, located in the C-terminal part of the protein, is 75% identical to the RGS domain of GAIP (19) (Fig. 1A) and between 30 to 50% identical to RGS domains of other members of this family. A dendrogram showing the relatedness of the RET-RGS-d to RGS domains of other RGS proteins was calculated using the phylip program and is shown in Fig. 1B.
Interestingly, two sequences in the N-terminal part of RET-RGS1 upstream of the RGS domain are potential sites of membrane interaction. The first is a putative transmembrane domain located between residues 56 and 80 as predicted by a structural analysis program, tm pred (Fig. 1A). The second is a stretch of 9 cysteines located between residues 193 to 205. A similar string of cysteines occurs on GAIP (Fig. 1A). This motif is also found on membrane-bound proteins of the cysteine string protein (csp) family (31), and it is a substrate for multiple palmitoylation (32).
RET-RGS-d Is Specifically Expressed in the Retina.
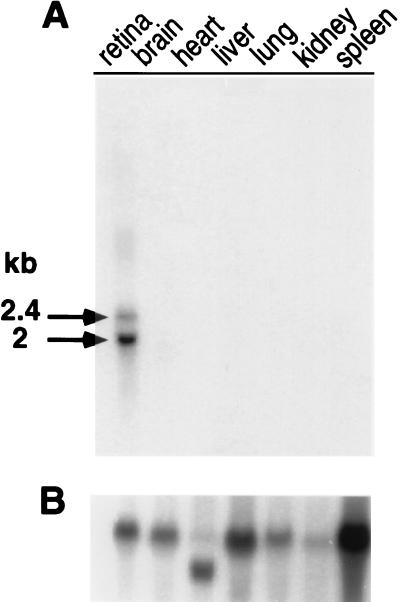
To examine the pattern of RET-RGS gene expression, we hybridized a probe complementary to RET-RGS-d to Northern blots carrying RNA prepared from various bovine tissues. Remarkably, RET-RGS transcripts were detected only in the retina. Two major transcripts of 2.4 and 2 kb were detected (Fig. 2A). Additional bands at 3.6 kb and 3.4 kb and 2.7 kb were detectable following longer exposures (data not shown).
Figure 2.
Northern blot analysis of RET-RGS-d expression in various bovine tissues. Twenty micrograms of total RNA prepared from various bovine tissues (see Materials and Methods) was analyzed in each lane. The blot was serially hybridized with an RET-RGS d-probe (A) and then, as a control for RNA loading and integrity, with a human β-actin probe (B). Autoradiographs were exposed 40 h for A and 20 h for B at −70°C with intensifying screens.
To further analyze the origin of these five transcripts, we hybridized total retinal RNA with a probe corresponding to the 5′ coding region of RET-RGS1. Interestingly, only a subset of the bands detected with the RET-RGS-d probe hybridized with the 5′ coding region of RET-RGS1 (data not shown). We conclude that RET-RGS1 is not the only transcript containing the coding sequence for RET-RGS-d. Therefore, it appears that the RET-RGS gene encodes a family of retina-specific transcripts which have identical RGS domain sequences but distinct 5′ coding sequences. Our preliminary studies of cDNA clones identified using RET-RGS-d as a probe also indicate the existence of multiple types of transcripts.
RET-RGS-d Transcripts Are Expressed Throughout the Retina.
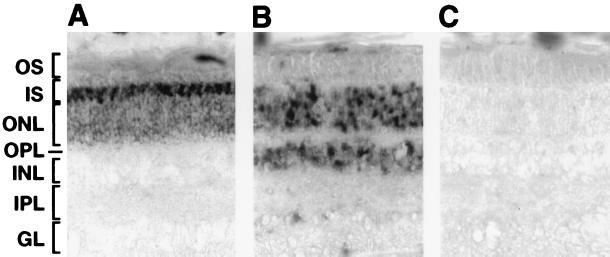
To determine whether RET-RGS proteins are expressed in photoreceptors, we analyzed the pattern of expression of RET-RGS-d by in situ hybridization on sections of bovine retina using digoxigenin-labeled RNA probes. A Tα antisense probe, used as a control, specifically labeled photoreceptors, validating the protocol used (Fig. 3A). Hybridizations were performed at high stringency to reduce nonspecific background (Fig. 3C), as well as cross-hybridization of the RET-RGS-d probe to other RGS sequences. RET-RGS-d antisense probe labeled all the nuclear layers of cells that compose the retina: the outer nuclear layer, which corresponds to the photoreceptors; the inner nuclear layer, corresponding to the bipolar, horizontal, and amacrine cells; and the ganglion cell layer (Fig. 3B). These results show that the transcripts containing RET-RGS-d are expressed in photoreceptors as well as in other cells of the retina.
Figure 3.
Analysis of the pattern of expression of RET-RGS-d in the retina by in situ RNA hybridizations. Bovine retina sections were hybridized as described in Materials and Methods with antisense rod Tα (A), antisense RET-RGS-d (B), and sense RET-RGS-d (C) RNA probes. OS, outer segments; IS, inner segments; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GL, ganglion layer.
RET-RGS-d Accelerates GTP Hydrolysis by Transducin α in Vitro.
RGS proteins inhibit G protein-dependent signaling pathways and interact with α subunits of G proteins. A straightforward explanation for these observations is that these proteins stimulate the return of Gα proteins to their inactivated GDP bound form by accelerating hydrolysis of bound GTP. We therefore investigated the capacity of RET-RGS-d to affect GTP hydrolysis by the α subunit of transducin in vitro. A His-tagged recombinant protein corresponding to the 138 C-terminal amino acid residues of RET-RGS1 (Fig. 1A) was expressed in E. coli and purified by Ni2+ resin chromatography. The RGS domain was expressed alone rather than in the context of RET-RGS1 for three reasons. (i) We were concerned that the putative membrane interaction sites of RET-RGS1 would interfere with efficient expression and/or purification of the recombinant protein. (ii) Our Northern blot analyses suggest that several isoforms containing the RET-RGS domain are present in the retina. (iii) It is important to establish whether or not the RGS domain itself is sufficient to stimulate GTP hydrolysis by transducin.
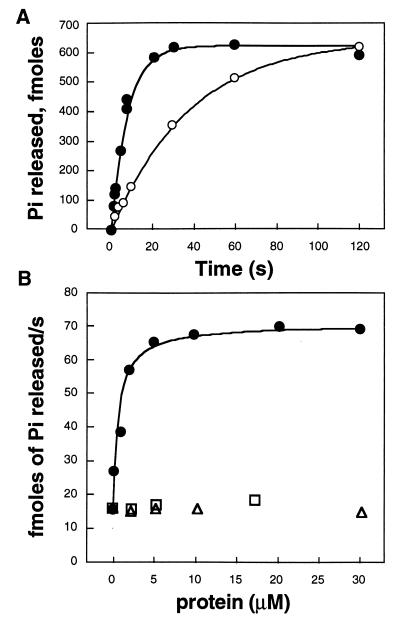
The effect of RET-RGS-d on Tα GTPase was tested using a single turn-over assay with [γ-32P]GTP as substrate (3). Bovine ROS membranes were depleted of their endogenous GAP activity by washing in 6 M urea (11). These membranes were then bleached and reconstituted with an extract of transducin and holoPDE. The experiment was started by addition of [γ-32P]GTP, and Pi release was analyzed in the presence or absence of RET-RGS-d. The rate of GTP hydrolysis was measured at saturating concentrations of photoactivated rhodopsin and Tα, 5 μM and 400 nM, respectively, conditions where neither binding of transducin to photoactivated rhodopsin nor GDP/GTP exchange are rate limiting. Therefore, the kinetics were solely dependent on the rate of hydrolysis of GTP bound to Tα.
In the absence of RET-RGS-d or in the presence of boiled RET-RGS-d, the kcat for hydrolysis of GTP was 0.026/s corresponding to a characteristic time τ of 38 s. The addition of 5 μM RET-RGS-d accelerated GTP hydrolysis 5-fold to a kcat of 0.127/s equivalent to a τ of 8 s (Fig. 4A). A similar experiment performed in the dark showed significantly reduced Pi release, confirming that the GTP hydrolysis accelerated by RET-RGS-d is derived from the light-dependent GTPase activity of Tα and not from a different type of nucleotidase that might be present in the preparation (data not shown).
Figure 4.
GTPase accelerating activity of RET-RGS-d on transducin. (A) Kinetics of GTP hydrolysis by transducin reconstituted with bleached urea-stripped ROS membranes in the absence (○) or presence (•) of 5 μM RET-RGS-d. (B) Titration of the GAP activity of RET-RGS-d (•), using his-tagged neurocalcin (□) and BSA (▵) as controls. The rate of hydrolysis of GTP by Tα was measured at 5 μM final concentration of bleached urea-stripped ROS membranes and 400 nM final concentration of transducin, concentrations that we determined to be saturating under these conditions. These experiments were repeated three times with similar results.
A titration experiment showed that acceleration of Tα GTPase activity by RET-RGS-d is specific and saturable, with a half-maximal effect at 1 μM RET-RGS-d (Fig. 4B). His-tagged neurocalcin and BSA taken as controls did not stimulate the GTPase activity of Tα (Fig. 4B).
These findings show that RET-RGS-d acts as a GAP for Tα in vitro, confirming our initial hypothesis that an RGS protein can accelerate the GTPase of transducin. While our study was in progress other groups reported that different members of the RGS family are also GAPs for α subunits of the Gi subfamily (20–22).
DISCUSSION
The existence in photoreceptors of a GTPase accelerating activity for transducin, distinct from its effector, was first reported in 1993 (5). Attempts to purify this factor have been complicated by the fact that this protein is tightly bound to membranes and rapidly loses its activity when solubilized in detergent (33). We are taking an alternative route to identify this protein based on a molecular approach. Genetic analyses from yeast and nematodes led to the recent discovery of a superfamily of proteins, named RGS proteins, involved in negative regulation of G protein signaling (15, 17). Recent studies have shown that RGS proteins stimulate GTP hydrolysis by G protein α subunits (20–22). We therefore performed a screen to identify RGS proteins that are candidates for the GAP that regulates transducin in photoreceptors.
By PCR amplification from bovine retinal cDNA using degenerate primers, we identified a new RGS domain, RET-RGS-d, specifically expressed in the retina. Recombinant RET-RGS-d stimulates the GTPase activity of transducin in vitro. A previous study showed that the core domain of GAIP is sufficient for the interaction of the protein with Gαi3 (19), but we report here evidence that an RGS domain by itself is sufficient for GAP activity. These data suggest that RGS proteins are composed of two modules; a conserved C-terminal RGS domain responsible for the GAP activity and a variable N-terminal domain involved in other functions such as localization or interaction with other proteins.
The GAP for transducin was first recognized as a membrane-associated protein whose activity became more pronounced as the concentration of bovine ROS was raised in a single-turnover GTPase assay (5). At low ROS concentrations the kcat for GTP hydrolysis was ≈0.028/s (5) and at saturating ROS concentrations the kcat increased to ≈0.12/s. These values agree well with the single-turnover GTP hydrolysis rates we report here, 0.026/s in the absence and 0.127/s in the presence of saturating RET-RGS-d. This suggests that RET-RGS-d and the GAP in bovine ROS membranes accelerate GTP hydrolysis by similar mechanisms. Nevertheless, our rates are slower than those (0.7–1/s) measured at higher concentrations of ROS supplemented with PDEγ (12, 13). Biophysical measurements of GTP hydrolysis, including calorimetry (>1/s) (9) and light-scattering (0.7–4/s) (34), also reported faster rates. The slower rates we observe might be attributable to PDEγ or some other important factor being less abundant in our transducin preparations (12, 13). However, in preliminary experiments (data not shown) we did not detect further acceleration of GTP hydrolysis by supplementing our assays with exogenous PDEγ. This suggests that the RET-RGS domain alone lacks structures needed for PDEγ to accelerate GTP hydrolysis, but further studies are required to resolve this issue.
Using RET-RGS-d as a probe to screen a bovine retinal cDNA library, we isolated a cDNA encoding a new RGS protein, RET-RGS1. Domains outside of the RGS domain in RET-RGS1 might also play a specific role in GAP function or perhaps in some other type of activity. It has been shown that both the N- and C-terminal domains of the yeast RGS protein, Sst2, are required for its function in vivo (35). However, the N-terminal sequence of RET-RGS1 upstream of its core domain is unrelated to any known RGS proteins except for a stretch of cysteines also present on GAIP. Homologous stretches of cysteines found in proteins of the cysteine string family are heavily palmitoylated (32). Interestingly, a recent study has shown that GAIP expressed in mammalian cells is palmitoylated and tightly bound to membranes (36). This suggests that the cysteine string sequence on RET-RGS1 may also serve as a membrane anchor. Moreover, in contrast to GAIP or any known RGS proteins, the N-terminal domain of RET-RGS1 also contains a putative transmembrane domain. RET-RGS1 is therefore the only RGS protein identified to this date that is likely to be an integral membrane protein. Immunological analyses of the subcellular localization of native and full-length recombinant RET-RGS1 will be required to definitively address this issue.
Besides RET-RGS1, we also detected other retina-specific transcripts containing the sequence encoding RET-RGS-d. Our preliminary studies suggest that these transcripts are splice variants of the same RET-RGS gene and that they encode a family of RGS proteins with a common RGS domain expressed throughout the retina. Their expression throughout the retina suggests that RET-RGS proteins act as GAPs for several different types of G proteins. Since RET-RGS proteins contain identical RGS domains, it is possible that their specificity toward a particular G protein is restricted in part by expression in distinct cell types or by localization within specific subcellular compartments. This raises two important issues that must be addressed in identifying the GAP or GAPs that have a physiological association with transducin. First, is RET-RGS1 or any other member of the RET-RGS family specifically expressed in photoreceptors? Most proteins known to be involved in phototransduction are expressed specifically in photoreceptors. This suggests that the GAP for transducin is likely to be a photoreceptor-specific protein. However, the possibility that the GAP for transducin is a more ubiquitous protein cannot be excluded. Second, is any RET-RGS protein present in the outer segments of photoreceptors? Immunocytochemical analyses will be required to answer these questions and will be performed as antibodies become available. Particular attention will be brought to RET-RGS1, which, because of its putative membrane attachment, is an excellent candidate for the GAP for transducin.
During the preparation of this manuscript, Chen et al. (37) reported cloning a new RGS protein (RGS-r) that is expressed in the retina and which accelerates multiple turn-over GTP hydrolysis by Tα in vitro. Sequence comparisons show that RET-RGS1 and RGS-r encode different proteins. The RGS domains of RET-RGS1 and of RGS-r are mostly related to GAIP and RGS8 respectively (Fig. 1B) and are only 37% identical to each other. The N-terminal domain of RGS-r is also much shorter than that of RET-RGS1 (43 amino acids vs. 236 amino acids) and there is no homology between the N-terminal domains of RET-RGS1 and RGS-r. Precise localization studies will be required to determine the functions of these RGS proteins in the retina.
Acknowledgments
We thank Matthew Czislowsky for help with DNA sequencing, Glenda Froelick for tissue sectioning, and Tony Scotti for excellent technical assistance. We thank Robert Hughes for providing His-tagged neurocalcin. We are grateful to Thierry Lepage, Luc De Vries, and the members of our laboratory for helpful discussions. These studies were supported by National Institutes of Health Grant EY06641 to J.B.H.
ABBREVIATIONS
- Tα
α subunit of transducin
- GAP
GTPase-activating protein
- ROS
rod outer segments
- PDE
phosphodiesterase
- GAIP
Gα interacting protein
- RGS
regulator of G protein signaling.
Footnotes
References
- 1.Yarfitz S, Hurley J B. J Biol Chem. 1994;269:14329–14322. [PubMed] [Google Scholar]
- 2.Baylor D A, Nunn B J, Schnapf J L. J Physiol (London) 1984;357:575–607. doi: 10.1113/jphysiol.1984.sp015518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arshavsky V Yu, Gray Keller M P, Bownds M D. J Biol Chem. 1991;266:18530–18537. [PubMed] [Google Scholar]
- 4.Antonny B, Otto Bruc A, Chabre M, Vuong T M. Biochemistry. 1993;32:8646–8653. doi: 10.1021/bi00084a036. [DOI] [PubMed] [Google Scholar]
- 5.Angleson J K, Wensel T G. Neuron. 1993;11:939–949. doi: 10.1016/0896-6273(93)90123-9. [DOI] [PubMed] [Google Scholar]
- 6.Erickson M A, Robinson P, Lisman J. Science. 1992;257:1255–1258. doi: 10.1126/science.1519062. [DOI] [PubMed] [Google Scholar]
- 7.Tsuboi S, Matsumoto H, Yamazaki A. J Biol Chem. 1994;269:15016–15023. [PubMed] [Google Scholar]
- 8.Sather W A, Detwiler P B. Proc Natl Acad Sci USA. 1987;84:9290–9294. doi: 10.1073/pnas.84.24.9290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vuong T M, Chabre M. Proc Natl Acad Sci USA. 1991;88:9813–9817. doi: 10.1073/pnas.88.21.9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arshavsky V Y, Bownds M D. Nature (London) 1992;357:416–417. doi: 10.1038/357416a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otto-Bruc A, Antonny B, Vuong T M. Biochemistry. 1994;33:15215–15222. doi: 10.1021/bi00255a001. [DOI] [PubMed] [Google Scholar]
- 12.Arshavsky V Y, Dumke C L, Zhu Y, Artemyev N O, Skiba N P, Hamm H E, Bownds M D. J Biol Chem. 1994;269:19882–19887. [PubMed] [Google Scholar]
- 13.Angleson J K, Wensel T G. J Biol Chem. 1994;269:16290–16296. [PubMed] [Google Scholar]
- 14.Roush W. Science. 1996;271:1056–1058. doi: 10.1126/science.271.5252.1056. [DOI] [PubMed] [Google Scholar]
- 15.Koelle M R, Horvitz H R. Cell. 1996;84:115–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- 16.Druey K M, Blumer K J, Kang V H, Kehrl J H. Nature (London) 1996;379:742–746. doi: 10.1038/379742a0. [DOI] [PubMed] [Google Scholar]
- 17.Dohlman H G, Apaniesk D, Chen Y, Song J, Nusskern D. Mol Cell Biol. 1995;15:3635–3643. doi: 10.1128/mcb.15.7.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dietzel C, Kurjan J. Mol Cell Biol. 1987;7:4169–4177. doi: 10.1128/mcb.7.12.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Vries L, Mousli M, Wurmser A, Farquhar M G. Proc Natl Acad Sci USA. 1995;92:11916–11920. doi: 10.1073/pnas.92.25.11916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson N, Linder M E, Druey K M, Kehrl J H, Blumer K J. Nature (London) 1996;383:172–175. doi: 10.1038/383172a0. [DOI] [PubMed] [Google Scholar]
- 21.Hunt T W, Fields T A, Casey P J, Peralta E G. Nature (London) 1996;383:175–177. doi: 10.1038/383175a0. [DOI] [PubMed] [Google Scholar]
- 22.Berman D M, Wilkie T M, Gilman A G. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- 23.Berman D M, Kozasa T, Gilman A G. J Biol Chem. 1996;271:27209–27212. doi: 10.1074/jbc.271.44.27209. [DOI] [PubMed] [Google Scholar]
- 24.Altschul S F, Gish W, Miller W, Myers W W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann K, Stoffel W. Biol Chem Hoppe-Seyler. 1993;347:166. doi: 10.1515/bchm3.1992.373.1.187. [DOI] [PubMed] [Google Scholar]
- 26.Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) Eugene: Univ. of Oregon Press; 1995. [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Harland R M. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 29.Chen C-K, Inglese J, Lefkowitz R J, Hurley J B. J Biol Chem. 1995;270:18060–18066. doi: 10.1074/jbc.270.30.18060. [DOI] [PubMed] [Google Scholar]
- 30.Siderovski D P, Heximer S P, Forsdyke D R. DNA Cell Biol. 1994;13:125–147. doi: 10.1089/dna.1994.13.125. [DOI] [PubMed] [Google Scholar]
- 31.Umbach J A, Mastrogiacomo A, Gundersen C B. J Physiol (Paris) 1995;89:95–101. doi: 10.1016/0928-4257(96)80556-0. [DOI] [PubMed] [Google Scholar]
- 32.Gundersen C B, Mastrogiacomo A, Faull K, Umbach J A. J Biol Chem. 1994;269:19197–19199. [PubMed] [Google Scholar]
- 33.Angleson J K, Wensel T G. Biophys J. 1995;68:A18. (abstr.). [Google Scholar]
- 34.Dratz E A, Lewis J A, Schaechter L E, Parker K R, Kliger D S. Biochem Biophys Res Commun. 1987;146:379–386. doi: 10.1016/0006-291x(87)90540-7. [DOI] [PubMed] [Google Scholar]
- 35.Dohlman H G, Song J, Ma D, Courchesne W E, Thorner J. Mol Cell Biol. 1996;16:5194–5209. doi: 10.1128/mcb.16.9.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Vries L, Elenko E, Hubler L, Jones T L Z, Gist Farquhar M. Proc Natl Acad Sci USA. 1996;93:15203–15208. doi: 10.1073/pnas.93.26.15203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen C-K, Wieland T, Simon M I. Proc Natl Acad Sci USA. 1996;93:12885–12889. doi: 10.1073/pnas.93.23.12885. [DOI] [PMC free article] [PubMed] [Google Scholar]