Abstract
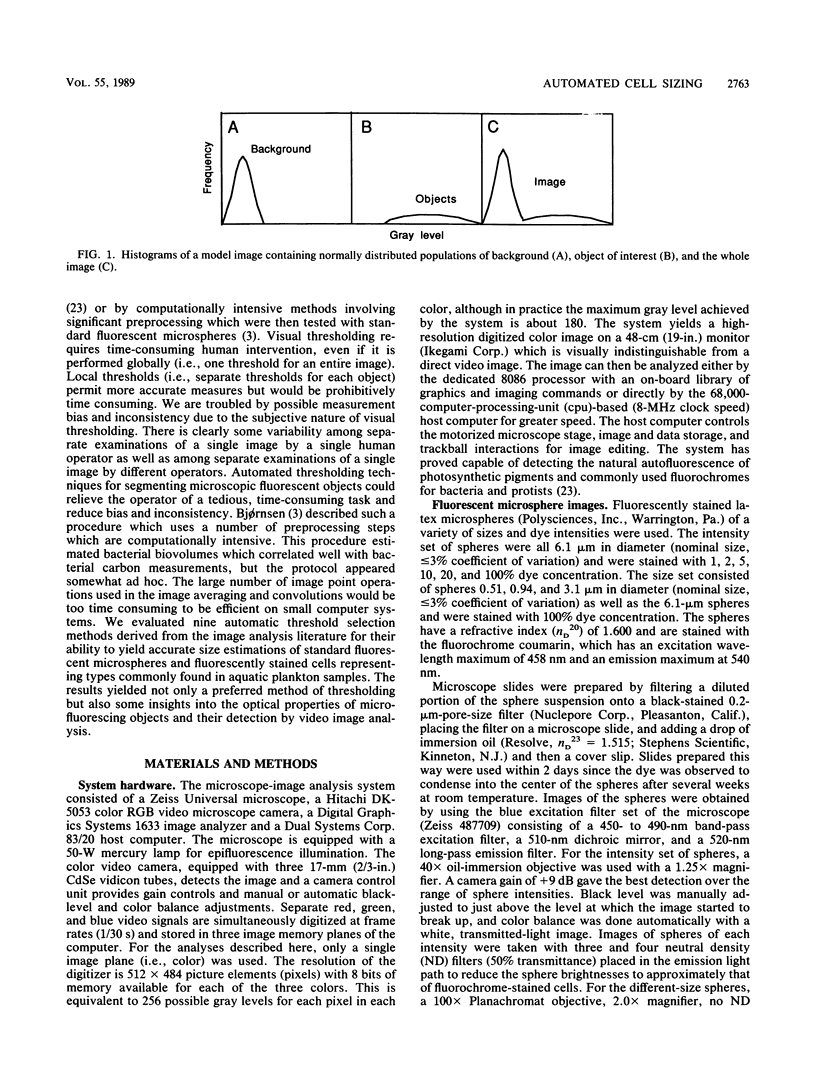
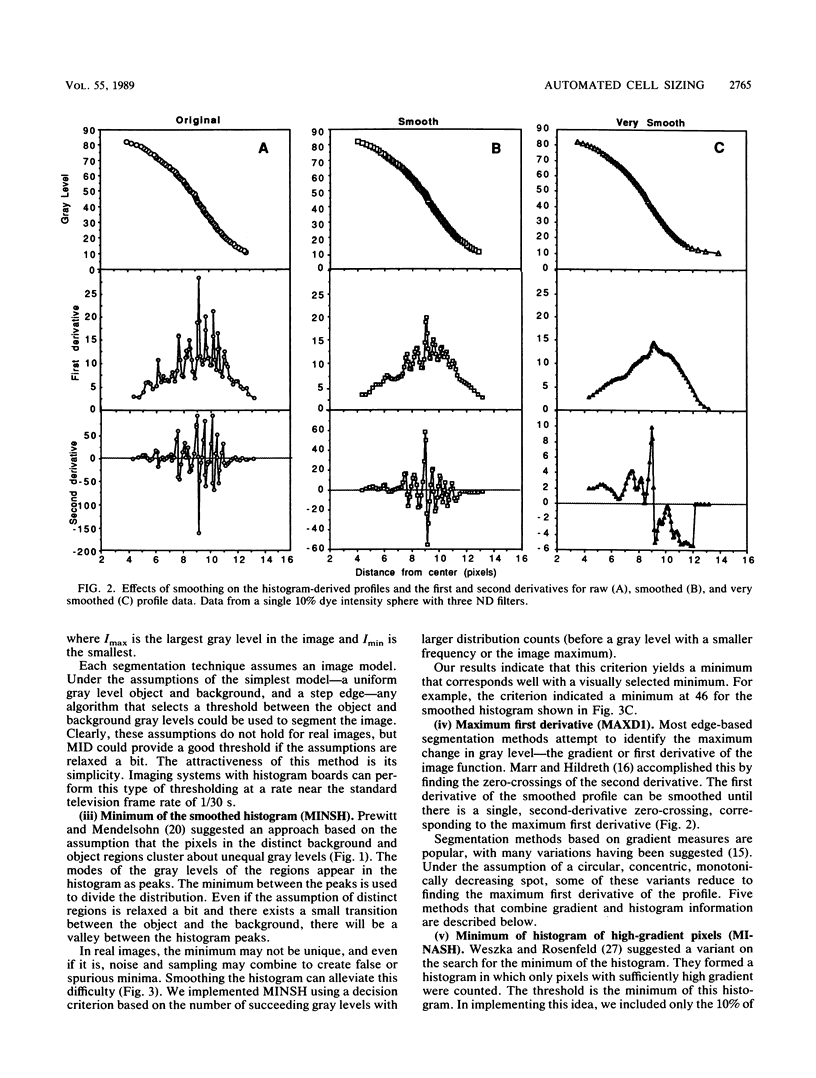
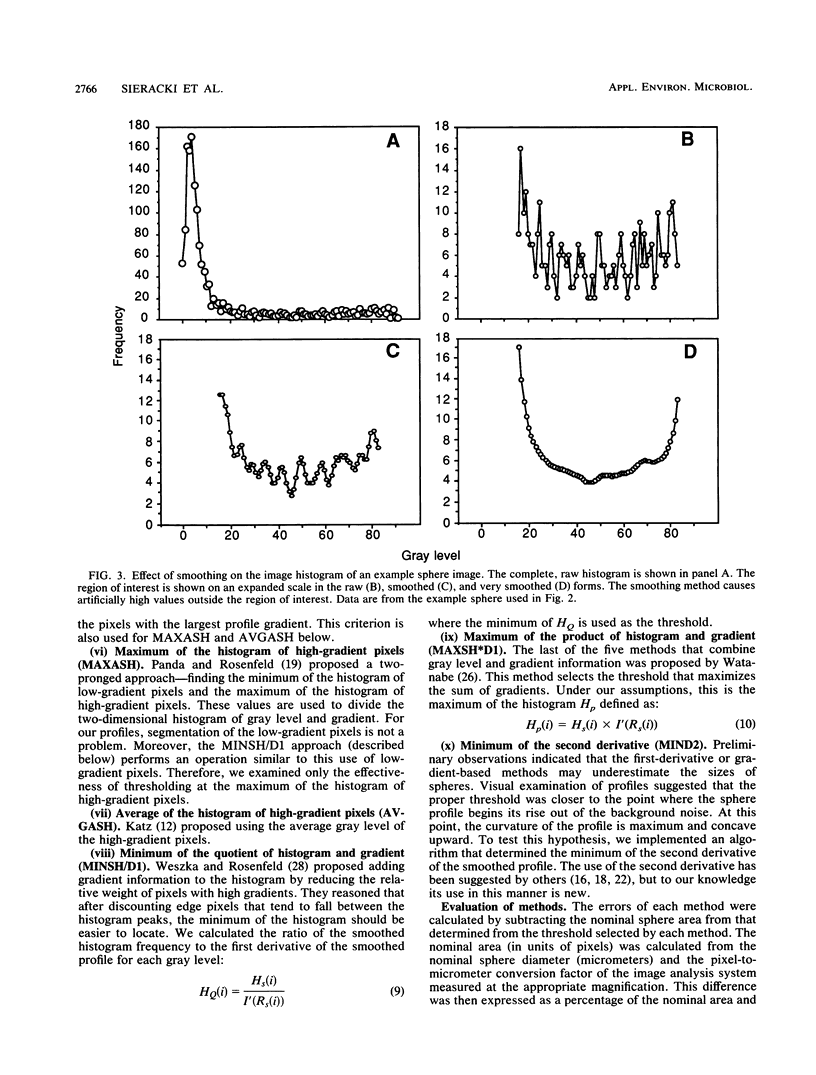
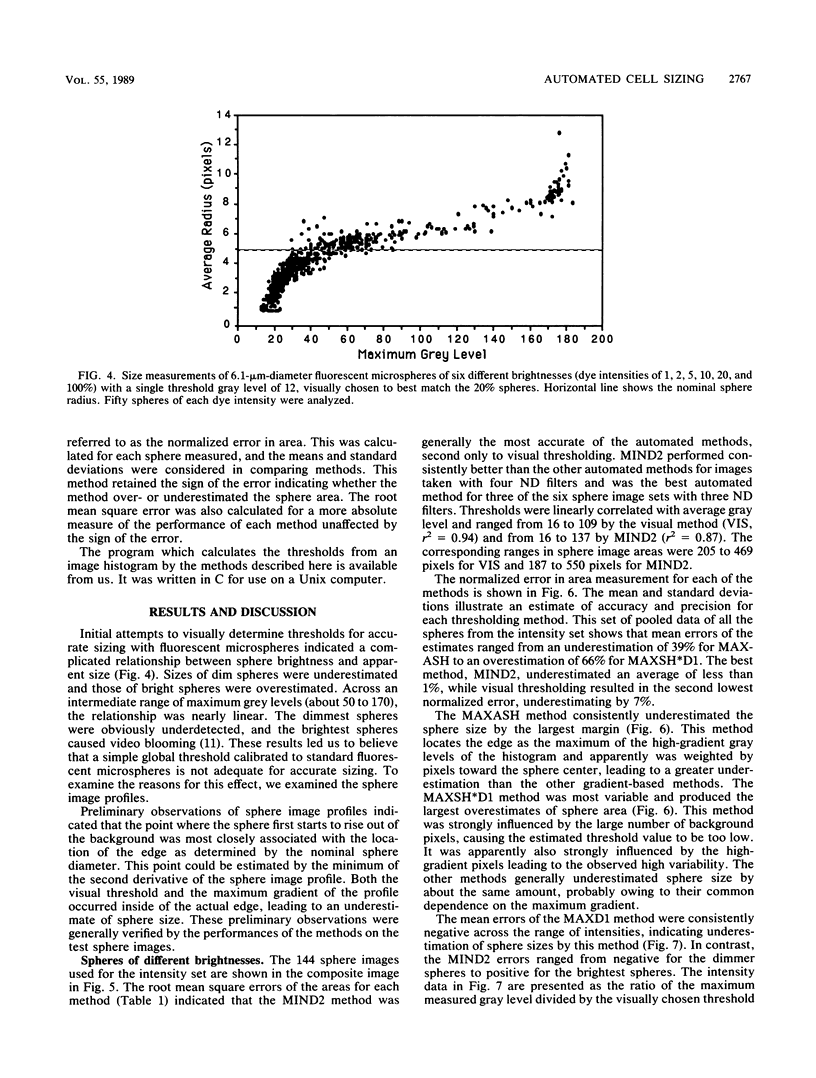
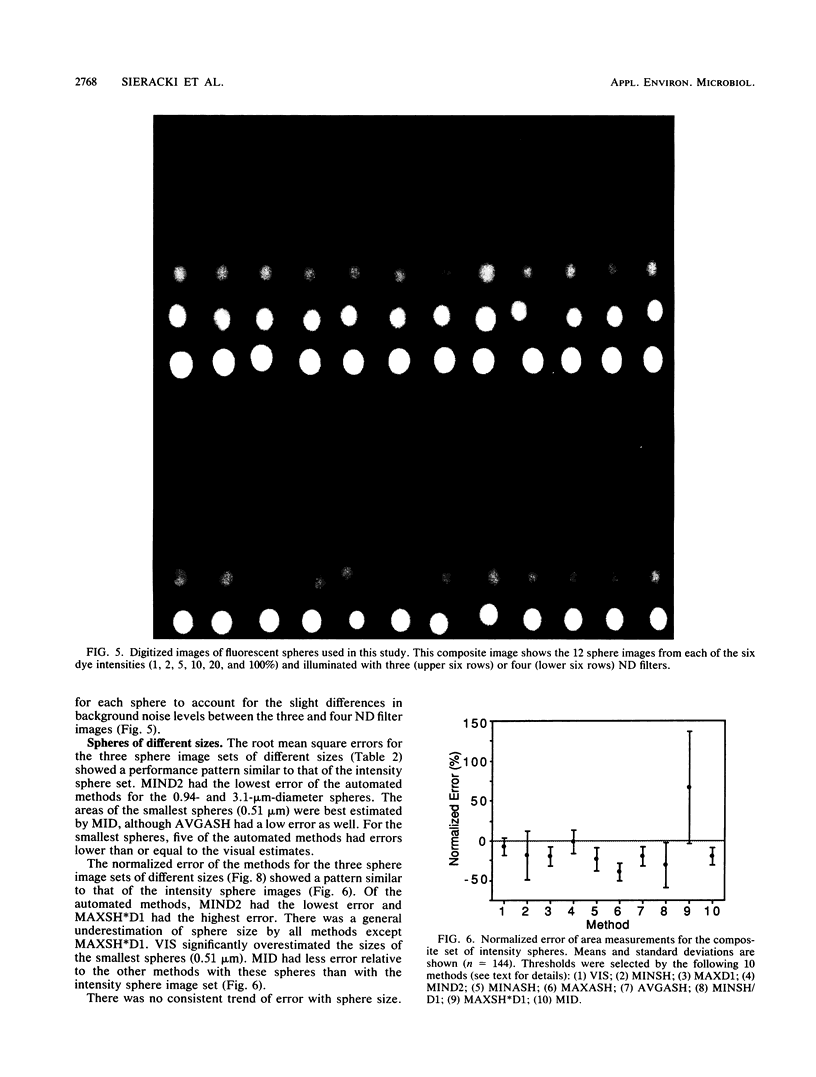
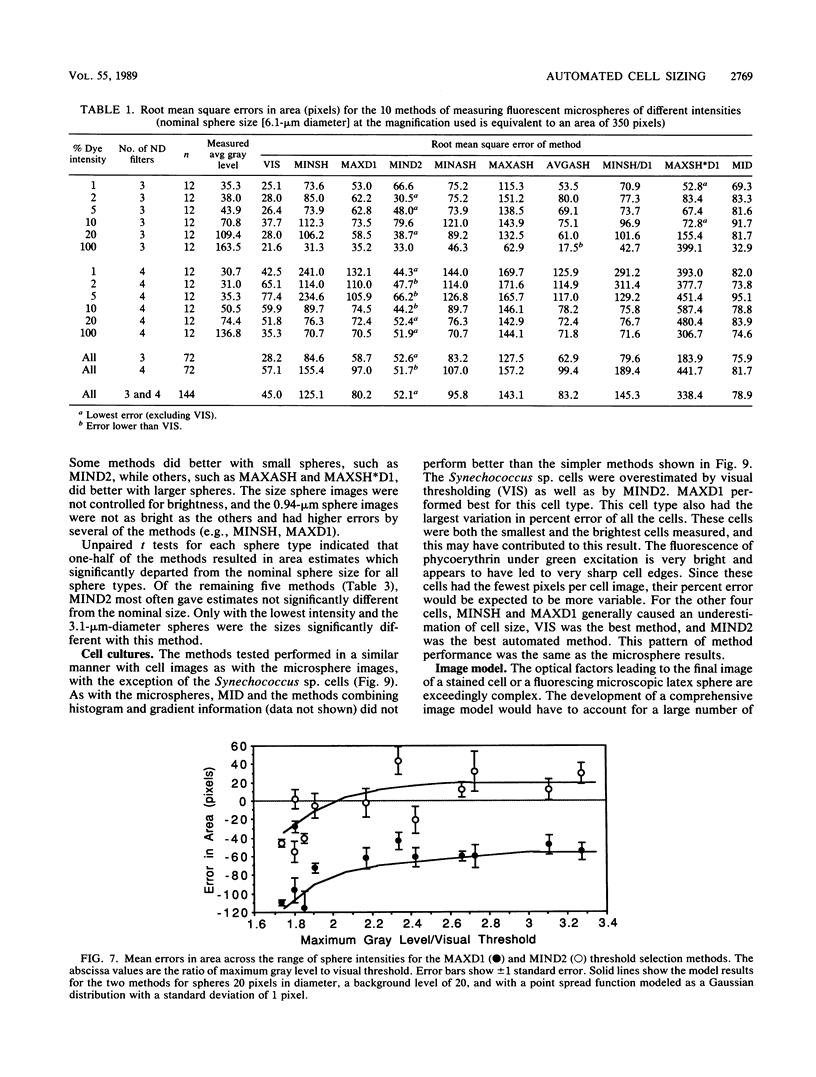
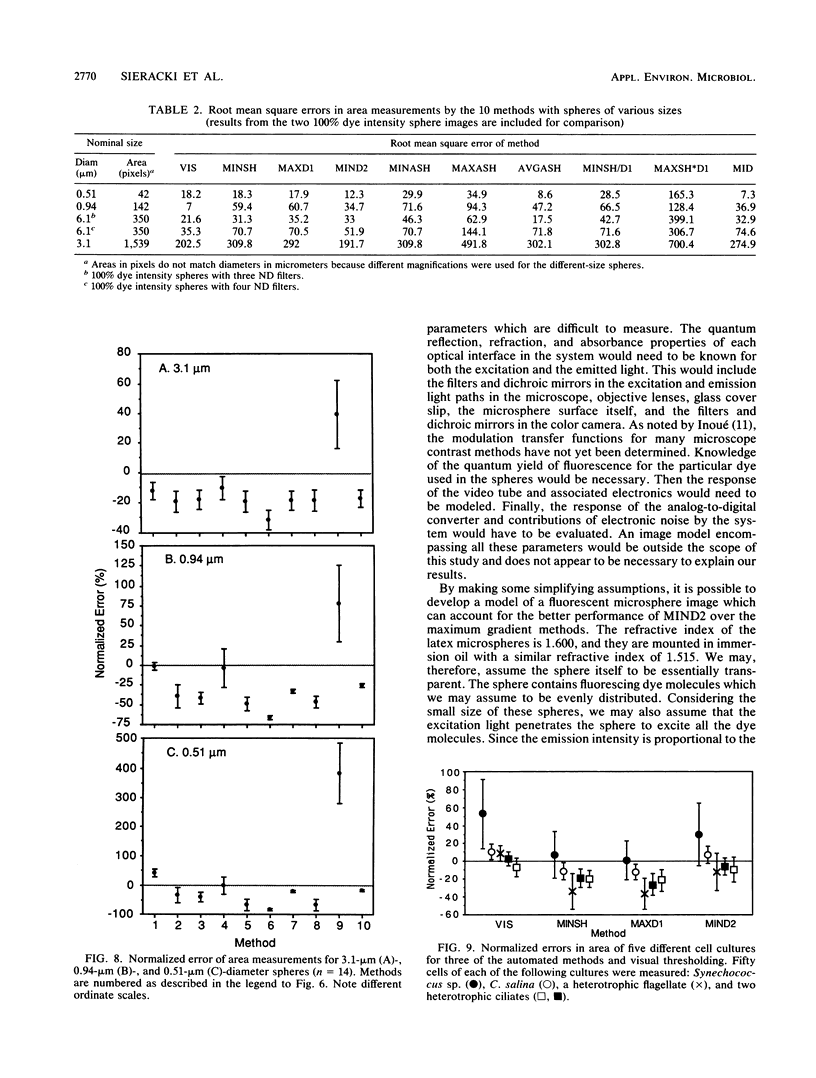
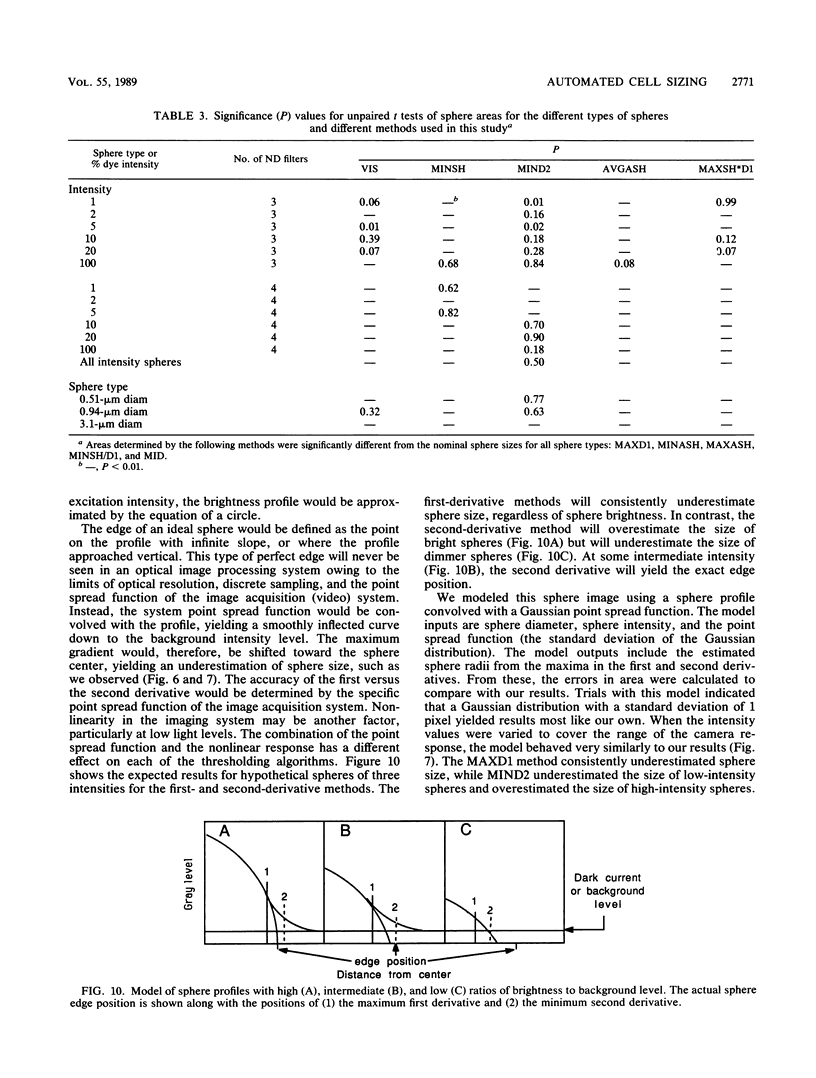
The accurate measurement of bacterial and protistan cell biomass is necessary for understanding their population and trophic dynamics in nature. Direct measurement of fluorescently stained cells is often the method of choice. The tedium of making such measurements visually on the large numbers of cells required has prompted the use of automatic image analysis for this purpose. Accurate measurements by image analysis require an accurate, reliable method of segmenting the image, that is, distinguishing the brightly fluorescing cells from a dark background. This is commonly done by visually choosing a threshold intensity value which most closely coincides with the outline of the cells as perceived by the operator. Ideally, an automated method based on the cell image characteristics should be used. Since the optical nature of edges in images of light-emitting, microscopic fluorescent objects is different from that of images generated by transmitted or reflected light, it seemed that automatic segmentation of such images may require special considerations. We tested nine automated threshold selection methods using standard fluorescent microspheres ranging in size and fluorescence intensity and fluorochrome-stained samples of cells from cultures of cyanobacteria, flagellates, and ciliates. The methods included several variations based on the maximum intensity gradient of the sphere profile (first derivative), the minimum in the second derivative of the sphere profile, the minimum of the image histogram, and the midpoint intensity. Our results indicated that thresholds determined visually and by first-derivative methods tended to overestimate the threshold, causing an underestimation of microsphere size. The method based on the minimum of the second derivative of the profile yielded the most accurate area estimates for spheres of different sizes and brightnesses and for four of the five cell types tested. A simple model of the optical properties of fluorescing objects and the video acquisition system is described which explains how the second derivative best approximates the position of the edge.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chow C. K., Kaneko T. Automatic boundary detection of the left ventricle from cineangiograms. Comput Biomed Res. 1972 Aug;5(4):388–410. doi: 10.1016/0010-4809(72)90070-5. [DOI] [PubMed] [Google Scholar]
- Kirsch R. A. Computer determination of the constituent structure of biological images. Comput Biomed Res. 1971 Jun;4(3):315–328. doi: 10.1016/0010-4809(71)90034-6. [DOI] [PubMed] [Google Scholar]
- Marr D., Hildreth E. Theory of edge detection. Proc R Soc Lond B Biol Sci. 1980 Feb 29;207(1167):187–217. doi: 10.1098/rspb.1980.0020. [DOI] [PubMed] [Google Scholar]
- Prewitt J. M., Mendelsohn M. L. The analysis of cell images. Ann N Y Acad Sci. 1966 Jan 31;128(3):1035–1053. doi: 10.1111/j.1749-6632.1965.tb11715.x. [DOI] [PubMed] [Google Scholar]
- Sieracki M. E., Johnson P. W., Sieburth J. M. Detection, enumeration, and sizing of planktonic bacteria by image-analyzed epifluorescence microscopy. Appl Environ Microbiol. 1985 Apr;49(4):799–810. doi: 10.1128/aem.49.4.799-810.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]



