Abstract
We used high-resolution array analysis to discover a recurrent lung cancer amplicon located at 14q13.3. Low-level gain of this region was detected in 15% of lung cancer samples, and high-level amplification was detected in an additional 4% of samples. High-level focal amplification appears to be specific to lung cancers, because it was not detected in >500 samples of other tumor types. Mapping of the commonly amplified region revealed there are three genes in the core region, all of which encode transcription factors with either established lung developmental function (TTF1/NKX2-1, NKX2-8) or potential lung developmental function (PAX9). All three genes were overexpressed to varying degrees in amplified samples, although TTF1/NKX2-1 was not expressed in the squamous cancer subtype, consistent with previous reports. Remarkably, overexpression of any pairwise combination of these genes showed pronounced synergy in promoting the proliferation of immortalized human lung epithelial cells. Analysis of human lung cancer cell lines by both RNAi and ectopic overexpression further substantiates an oncogenic role for these transcription factors. These results, taken together with previous reports of oncogenic alterations of transcription factors involved in lung development (p63, CEBPA), suggest genetic alterations that directly interfere with transcriptional networks normally regulating lung development may be a more common feature of lung cancer than previously realized.
Keywords: gene amplification, lung development, lung oncogene, TTF1 NKX2-8 PAX9, lineage addiction
Many of the major genetic alterations that drive the development of human carcinomas involve transcription factors. The SMAD4 gene is frequently mutated in pancreatic and colon cancer, thereby blocking the ability of TGF-β to inhibit epithelial cell growth. SMAD4 encodes the common transcription coactivator of the five different Smad factors that are regulated by the TGF family of receptors. Mutational activation of the Wnt pathway protein β-catenin occurs often in colon and hepatocellular carcinomas, leading to its direct activation of TCF transcription factors and increased expression of Wnt target genes. Gene amplification at 8q24 and resultant increased expression of MYC is a common occurrence in carcinomas, leading to the increased formation of Myc:Max heterodimer transcription factors that alter gene expression in large part by recruiting histone-modifying enzymes. p53 is the most widely mutated gene in all carcinomas and encodes a transcription factor that responds to DNA damage and a wide variety of other stress signals and alters the expression of several key mediators of cell cycle progression, apoptosis, and senescence.
Less commonly documented in carcinomas are genetic alterations of transcription factors that play tissue-specific developmental roles. However, in lung cancer, there are a few examples of cancer-associated genetic or epigenetic alteration of lung developmental transcription factors. The first is p63, a p53 homologue at 3q27, which is required for formation of lung basal epithelial cells (1, 2). Cancer-associated gene amplification, which is common in squamous lung carcinomas, produces a p40 splice variant with oncogenic activity (3). The second example is CEBPA, a transcription factor that, when knocked out, causes abnormal proliferation of type II alveolar cells in the lung and is silenced by promoter hypermethylation in >50% of lung tumors (4, 5). Reintroduction of its expression suppresses the growth of lung cancer cell lines (6). The final example is the transcription factor encoded by N-MYC, which is frequently amplified in small-cell lung cancer (SCLC) (7, 8), when partially deleted disrupts branching morphogenesis (9–11), and is required for the proliferation of peripheral progenitor cells (12).
Here we report on the frequent alteration by gene amplification in lung cancer of a locus containing the genes for three lung developmental transcription factors and in addition demonstrate oncogenic activity of these genes. Together with the previous reports reviewed above, these results suggest that disruption of lung developmental pathways could be a common characteristic of lung cancer development.
Results
14q13.3 Amplicon Is One of the Most Frequent High-Amplitude Focal Amplicons in Human Lung Cancer.
We used ROMA, a high-resolution array comparative genomic hybridization (CGH) method (13), to profile the DNA copy number alterations of 77 human cancer cell lines and 184 primary human lung tumors. Supporting information (SI) Table 2 summarizes the overall histological classification of all samples. SI Tables 3 and 4 describe individual samples of cancer cell lines and tumors, respectively. Microarray measurements were converted to copy number estimates by using a segmentation algorithm based on Kolmogorov–Smirnov statistics (14). To restrict subsequent analysis to cancer-associated somatic genetic events, we used an automated procedure to mask common germ-line copy number variations (14). Next, we used an automated method, similar to the minimal common region method developed by Tonon et al. (15), to detect high-level focal amplicons (segmented DNA copy number ≥1.5 and ≤5 Mb) and determined their frequency and region of common overlap (see Materials and Methods). Of the 12 most-frequent focal amplicons, nine contained known oncogenes that are very likely to be the driver gene, and three amplicons did not associate with apparent driver genes (Table 1). The 8p12 amplicon contained FGFR1, although Tonon et al. (15) provided evidence that WHSC1L1 was the more likely candidate driver gene in lung cancer. Importantly, five of the five oncogenes that have been considered to be the best-established oncogenes in lung cancer [MYC, KRAS, EGFR, CCND1, and LMYC; reviewed by Minna et al. (8)] were found among the top 12 amplicons (Table 1). Thus it seems likely that the three recurring amplicons without apparent driver genes contain oncogenes important for lung cancer development. These three amplicons, located at 14q13.3, 5p15.33, and 8p11.21, contained between 5 and 11 known genes. We chose to pursue the 14q13.3 amplicon for further analysis, based on its smaller size, lowest gene content, and status as the second most-frequent high-amplitude amplicon (Table 1).
Table 1.
Twelve most-frequent focal amplicons in lung cancer
| High-level amplification frequency, % | Inclusive chromosomal gain and amplification frequency, % | Chromosomal location | Size, Mb | Genes within minimal overlapping amplicon | Likely driver gene |
|---|---|---|---|---|---|
| 5.7 | 36 | 8q24.21 | 1.32 | 3 | MYC |
| 4.2 | 19 | 14q13.3 | 0.53 | 5 | Unknown |
| 3.8 | 18 | 12p12.1 | 2.86 | 12 | KRAS |
| 3.4 | 20 | 7p11.2 | 0.82 | 3 | EGFR |
| 3.1 | 13 | 11q13.3 | 0.54 | 6 | CCND1 |
| 2.7 | 9 | 1p34.2 | 0.59 | 10 | LMYC |
| 2.3 | 25 | 1q21.2 | 1.15 | 36 | MCL1 |
| 2.3 | 33 | 5p15.33 | 0.72 | 11 | Unknown |
| 2.3 | 9 | 8p12 | 0.59 | 14 | FGFR1 |
| 2.3 | 12 | 8p11.21 | 0.80 | 9 | Unknown |
| 1.5 | 7 | 2p24.3 | 0.44 | 2 | NMYC |
| 1.5 | 10 | 12q15 | 1.57 | 13 | MDM2 |
High-level amplification frequency was determined by using a cutoff of segmented DNA copy number >1.5. The frequency of both chromosomal gains and amplification was determined by using a less-stringent cutoff of 1.2. The amplicon locations are based on the March 2006 (hg18) human genome assembly (http://genome.ucsc.edu). The size of the amplicon represents the size of the minimal overlapping region (see Materials and Methods). The genes within the amplicon include all genes from the RefSeq set that are partially or fully contained within its boundaries. The likely driver gene, if present, denotes the commonly accepted driver gene for the previously described amplicon.
The 14q13.3 Amplicon Contains Three Transcription Factor Genes.
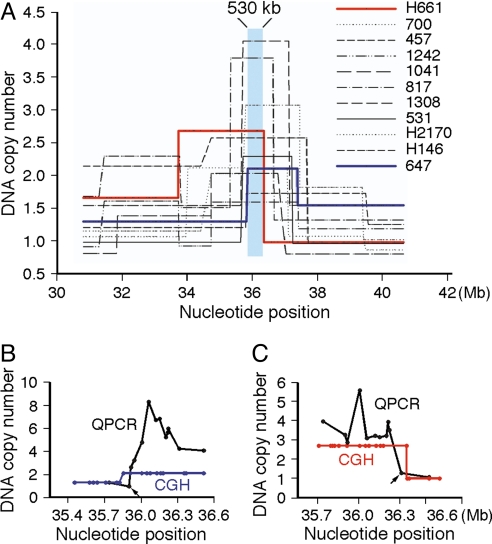
Gain of 14q13.3 was found in 19% of all samples, and high-level amplification of 14q13.3 was found in ≈4% of total samples, including three lung cancer cell lines and eight lung tumors. To visualize the common area of overlap, we plotted the segmented DNA copy number across a 10-Mb region that spanned the 14q13.3 amplicon for each of the highly amplified samples. Each of the individual amplicon structures was different, but two samples (647 and H661) were particularly informative, in that they defined the left and right boundaries of the 480- to 530-kb core amplified region segment shared by all 11 samples (Fig. 1A). The size range of 480–530 kb results from the uncertainty created by gaps in consecutive genome positions of the oligonucleotides on the array. To more precisely map the boundaries of the amplicons in the two informative samples, we used real-time quantitative PCR (QPCR) to measure DNA copy number at ≈15- to 20-kb intervals throughout the two ≈35-Mb boundary regions (Fig. 1 B and C). This analysis refined the boundaries of the commonly amplified core to a 413-kb region containing four protein-coding genes. Three genes, TTF1, NKX2-8, and PAX9, are fully contained within this region and thus are candidate driver genes of 14q13.3 amplification. The fourth gene, SLC25A21, is not a viable candidate, because the common amplified region contains only sequences downstream of its fourth exon.
Fig. 1.
The commonly amplified core region of lung cancer 14q13.3 amplicons. (A) The segmented array CGH DNA copy number values (y axis) for 11 samples containing the 14q13.3 amplicon are plotted against the chromosome 14 nucleotide position (x axis) from the March 2006 (hg18) assembly of the International Human Genome Sequencing Consortium (http://genome.ucsc.edu). The commonly amplified core region (chr14:35,820,860-36,348,636), which maximally spans a 530-kb region, is shaded in blue. The 11 amplified samples consist of eight primary tumors (700, 457, 1242, 1041, 817, 1308, 531, and 647) and three cell lines (NCI-H661, NCI-H2170, and NCI-H146). NCI-H661 is from a large-cell carcinoma, both NCI-H2170 and 647 are from SCC, NCI-H146 is from an SCLC, and the remaining samples are from AC. (B and C) Determination by real-time PCR of the centromeric boundary of the amplicon found in primary tumor sample 647 (B) and of the telomeric boundary of the amplicon found in large-cell carcinoma cell line NCI-H661 (C). The DNA copy numbers measured by both array CGH and real-time PCR are as plotted. This analysis defined the two boundaries (chr14:35,897,833-36,311,043) of the commonly amplified 413-kb region.
In addition to these validated genes, there are four University of California, Santa Cruz, gene predictions within the common amplified region that correspond to EST sequences BX161496, AY102071, BC042093, and BC129834. None of these four candidate genes are predicted to contain coding sequences, nor do they have predicted protein orthologs in six other species, unlike the three validated genes that each clearly defined orthologs in three or more of the six species (http://genome.ucsc.edu). To explore the possibility these four predicted genes may possess oncogenic function but in a noncoding capacity, we examined expression of these four predicted genes by reverse transcriptase-dependent QPCR in a panel of RNA isolated from five normal lung tissues, 20 lung adenocarcinomas (AC), and 20 lung squamous-cell carcinomas (SCC) (which included six amplified tumors). None of the four predicted genes were expressed in any of these samples, effectively ruling out this last possibility for their candidacy as driver oncogenes (data not shown).
The genes TTF1 (also known as NKX2-1 or TITF1 for thyroid transcription factor 1) and NKX2-8 (NK2 transcription factor related locus 8) belong to the same NK2 homeodomain-containing transcription factor gene family (16). PAX9 is a member of the “paired box” (PAX)-containing transcription factor family in which the homeodomain is also present in most family members. However, PAX9 lacks the homeodomain but retains the PAX DNA-binding domain (17).
Correlation of DNA Copy Number and RNA Expression of TTF1, NKX2-8, and PAX9.
A basic premise of gene amplification as a mutational mechanism relevant to cancer is that the gene copy number gain leads to an increased dosage of messenger RNA and protein (18). To determine the impact of DNA copy number on RNA expression of candidate driver genes, we profiled global RNA expression of six amplified AC and 11 nonamplified lung tumors by using expression arrays. This study led to the detection of positive correlations of DNA copy number and RNA expression of TTF1 (0.067, P = 0.02), NKX2-8 (0.431, P = 0.003), and PAX9 (0.101, P = 0.01), demonstrating an influence of DNA copy number on RNA expression of all three genes (SI Fig. 4). Next, we determined by real-time QPCR the expression of these three genes as well as 14q13.13 DNA copy number in an independent set of 50 lung AC tumors. Using 3-fold DNA copy number increase as the cutoff, five AC samples (10%) were found to be amplified for the 14q13.3 region. NKX2-8 was up-regulated in the range of 4- to 15-fold in all five amplified samples (SI Fig. 5A). Clearly, NKX2-8 is a leading candidate driver gene based on its strong correlation of RNA expression and DNA copy number (Pearson correlation coefficient of 0.692, P = 0.0001). However, we could not rule out the driver gene candidacies of PAX9 or TTF1, because both genes also showed positive, albeit weaker, correlations of RNA expression and DNA copy number (Pearson correlation coefficients of 0.343 and 0.241, respectively, with P values = 0.001). Therefore, functional analysis of the three candidate driver genes was essential for identification of the driver gene(s).
Gain-of-Function Studies of TTF1, NKX2-8, and PAX9.
To test the functional consequences of enforced overexpression of candidate driver genes, we tested whether overexpression of any of the candidate driver genes enhanced the tumorigenicity of lung cancer cell lines. We used two lung cancer cell lines, one with the 14q13.3 gain (NCI-H2170, referred to as “H2170” hereafter) and the other without the 14q13.3 amplicon (HCC15). Both of these non-SCLC cancer cell lines are of the SCC subtype. To create stable transfectants, we used retrovirus-mediated gene transfer to enforce expression of individual genes. Overexpression of transfected genes was confirmed by reverse transcriptase-dependent QPCR (data not shown). None of the three candidate genes when overexpressed further enhanced tumor formation of amplified H2170 cells. On the contrary, in nonamplified HCC15 cells, overexpression of either TTF1 or NKX2-8 (but not PAX9) enhanced tumorigenicity by increasing tumor take rate and tumor size (SI Fig. 6), indicating oncogenic function for both TTF1 and NKX2-8 in HCC15 cells. In addition, the observation that the prooncogenic activities of TTF1 and NKX2-8 were manifested only in nonamplified HCC15 cells suggests that functional advantage of increased gene dosages of TTF1 or NKX2-8 may have already been maximized in the amplified H2170 cells.
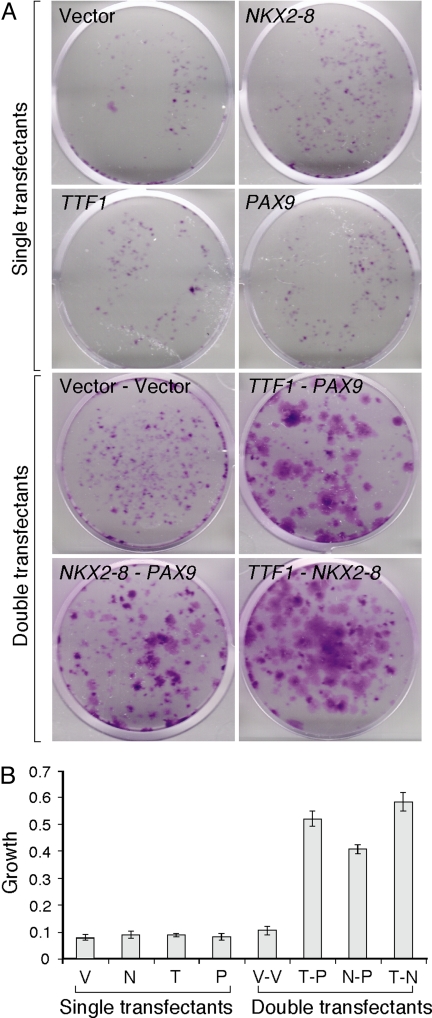
To explore the effects of these three genes on premalignant lung epithelial cells, we performed experiments with normal human lung epithelial cells immortalized by SV40 large T antigen, i.e., BEAS-2B cells (19). Individual and pairwise combinations of the three candidate driver genes were retrovirally transduced into BEAS-2B cells. We found that, when plated at low densities, BEAS-2B cells expressing individual candidate genes did not form colonies differently from empty vector controls (Fig. 2A). However, all three pairwise combinations (i.e., double transfectants) formed much larger colonies, ranked in the order of TTF1-NKX2-8 → TTF1-PAX9 → NKX2-8-PAX9 by visual inspection (Fig. 2A). To quantify the increased colony sizes, the crystal violet in the stained plates was eluted in a 0.1% SDS solution, and the absorbance at 595 nm was measured (20). Consistent with visual inspection, each of the three pairwise combinations showed 4- to 6-fold higher growth than all of the single-transfectant and double-vector control transfectant cells (Fig. 2B). To validate this synergistic growth effect, we repeated the experiment by using different vectors that contained different antibiotic selection marker genes (see Materials and Methods). With this second system, we were able to construct triple transfectants overexpressing all three genes. Significant enhancement of colony growth was detected at a level similar to what was observed with the double transfectants (SI Fig. 7), suggesting that the maximal synergy was reached in the double transfectants. These results confirmed that there is a growth-promoting synergy among the three genes in the background of premalignant lung epithelia.
Fig. 2.
Effects of overexpressing individually or combinatorially the 14q13.3 amplicon genes on the proliferation of premalignant lung epithelial cells. (A) Colony formation of BEAS-2B retroviral transfectants stably expressing individual genes (single transfectant) or pairwise combinations of the three candidate driver genes (double transfectant). The assay was initiated by plating 1,000 cells per well in six-well plates. After 12 days, cells were fixed and stained with crystal violet. Representative pictures of experiments performed in triplicate are shown. (B) The crystal violet stain in A was dissolved in 0.1% SDS solution and quantified by absorbance at 595 nm according to Scragg and Ferreira (20). V, vector only; N, NKX2-8; T, TTF1; P, PAX9; V-V, vector-vector; T-P; TTF1-PAX9; N-P, NKX2-8-PAX9; T-N, TTF1-NKX2-8.
Loss-of-Function Studies of TTF1, NKX2-8, and PAX9.
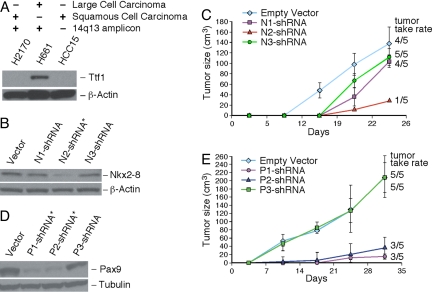
The 14q13.3 amplicon does not appear to be limited to a specific subtype of lung cancer. This is particularly intriguing in that Ttf1 protein is rarely detected in the SCC type of lung cancer based on multiple studies in the literature (21–24). Consistent with the literature, in multiple squamous lung cancer cell lines (including the amplified H2170 cells), we have not been able to detect the protein expression of TTF1 by using a highly specific monoclonal antibody [clone 8G7G3/1 (25)] (Fig. 3A). Notably, the RNA message of TTF1 was undetectable in H2170 by reverse transcriptase-dependent QPCR, suggesting gene silencing at the transcription level (data not shown). Thus, TTF1 is ruled out as a driver gene in the amplified H2170 cells. However, that enforced expression of TTF1 enhanced the tumorigenicity of an amplification-free SCC cells (HCC15) implies that TTF1 has oncogenic activity and SCC cells have the physiological context needed to manifest the oncogenic activity of TTF1.
Fig. 3.
Continuous expression of NKX2-8 and PAX9 is essential to the tumor maintenance of amplified SCC cells. (A) Western blot analysis indicates the absence of Ttf1 protein expression in two lung SCC cell lines (H2170 and HCC15), as expected from the literature (21–24). Two additional SCC cell lines (EPLC-272H and CHAGO-K-1) were also shown by immunoblotting to lack the expression of Ttf1 protein (data not shown). Analysis of the large-cell lung cancer cell line (H661) was included as a positive control. Twenty micrograms of whole-cell extracts were loaded. The 14q13.3 amplicon status and tumor subtype are as indicated. (B and D) Effective shRNAs (indicated by an asterisk) in knocking down the endogenous protein expression of NKX2-8 or PAX9 were identified by immunoblotting of whole-cell extracts prepared from stable transfectants based in the amplified H2170 cells. Immunoblottings of β-actin or tubulin were performed to control for total amounts of proteins analyzed. (C and E) Stable H2170 transfectants expressing individual shRNAs of NKX2-8 or PAX9 were evaluated for tumorigenicity in athymic nude mice. Approximately 24 h before injections, the mice were gamma-irradiated at 400 rad to minimize residual immune responses. Subsequently, five million cells of individual transfectant populations were injected into a group of five athymic nude mice s.c., and these animals were observed weekly for tumor formation. Tumor take rate was calculated as of the last weekly tumor measurement, and the averaged tumor size (y axis) was plotted against days after injection (x axis).
Because amplified SCC cells such as H2170 present a simplified case in which TTF1 is functionally irrelevant because of its lack of protein expression, we wished to use this cell system to address whether continued overexpression of the other two candidate driver genes play a tumor maintenance role. We screened small-hairpin RNA (shRNA) clones to identify the effective ones that could knock down the endogenous protein expression of NKX2-8 or PAX9 in H2170 cells. Preexisting anti-NKX2-8 shRNA clones were obtained from Open Biosystems (Huntsville, AL) (26), and the shRNA inserts were transferred into a RNA polymerase II-based retroviral expression vector (pMLP) that has been shown to consistently induce high suppression of target genes (27). With PAX9, we used the BIOPREDsi webtool (28) and identified three top RNAi sites in the ORF. These three sites were then specifically targeted by using the RNAi Codex webtool (29) to generate 101-mer oligonucleotides that we then cloned into the pMLP vector. After retroviral infection and drug selection to eliminate uninfected H2170 host cells, immunoblotting analysis of cell extracts of stable transfectants identified one anti-NKX2-8 (N2-shRNA; Fig. 3B) and two anti-PAX9 shRNAs (P1- and P2-shRNA; Fig. 3D) as the most potent in suppressing endogenous target protein expression. As judged by scanning densitometry, N2-shRNA conferred nearly 85% reduction of Nkx2-8 protein expression (relative to the empty vector control), and both P1- and P2-shRNA reduced the endogenous Pax9 protein expression to ≈7% of empty-vector control transfectants (Fig. 3 B and D). Each population of stable H2170 shRNA transfectants was evaluated for tumorigenicity, and we found that knocking down NKX2-8 or PAX9 severely compromised the tumorigenicity of H2170 cells. In the case of NKX2-8, the effective N2-shRNA reduced the tumor take rate to only one of five mice, and in the one mouse-forming tumor, the tumor was much smaller (Fig. 3C). With PAX9, the two effective shRNAs also resulted in significantly reduced tumor formation (Fig. 3E). We believe it is unlikely that nonspecific effects of the shRNAs caused the inhibition of tumor formation. First, the degree of tumorigenicity reduction correlates with the severity of knockdowns, with the most effective shRNAs (N2-, P1-, or P2-shRNA) giving the strongest suppression of tumor formation and take rate, whereas other less-effective shRNAs showed milder effects on tumorigenicity. Second, we have assessed the impact of the effective NKX2-8 shRNA (N2-shRNA) in a nonamplified SCC cell line (HCC15). We found that N2-shRNA did not influence the tumorigenicity of HCC15 as it did with H2170 (data not shown), indicating that (i) this shRNA does not have a generalized deleterious off-target effect, and (ii) only a subset of lung cancer cell lines depend on maintaining high levels of NKX2-8 expression. In sum, our studies suggest that both NKX2-8 and PAX9 play a pivotal role in the tumor maintenance of SCC cell with the 14q13.3 amplification.
Association of Focal 14q13.3 Amplicons with Disease Progression.
In an independent genomic DNA copy number data set consisting of 91 clinically annotated lung AC profiled by Agilent's (Austin, TX) 44K platform (a full description of the data set is to be published elsewhere), 17 samples (16%) within this data set were found to harbor focal amplification (≤5 Mb) of 14q13.3 containing the three candidate driver genes (SI Table 5 summarizes the associated clinical and molecular parameters). Eight of these 17 samples (8.8% overall frequency) were considered highly amplified (log2 of segmented DNA copy number >0.8). In addition to these 17 samples with focal amplification, 12 other samples contained low-level wide gain at 14q13 (SI Table 5). We analyzed the data set for potential correlations of the 14q13.3 amplicon status with the 17 clinical parameters by Spearman rank order correlation calculations and Fisher's exact tests. By stratifying the samples into two groups [early-stage lung cancer (stages I and II) vs. late stage (stage III cancers)], a significant positive correlation was observed between focal amplification of 14q13.3 and late-stage cancer (SI Table 6). Also, focal amplification of 14q13.3 correlates with recurrence (SI Table 5). Thus, the 14q13.3 focal amplification appears to confer more advanced malignant properties to lung cancer cells. In contrast, wide DNA copy number gain involving 14q13.3 did not correlate with late-stage lung cancer or recurrence, reinforcing the basic premise of this study that DNA copy number increases should be considered in the context of whether they are focal or wide.
Discussion
Previous high-resolution oligonucleotide array-based CGH studies of human lung cancer have been reported (15, 30). Using bacterial artificial chromosome arrays, Garnis et al. (31) determined the whole-genome copy number alterations in 28 non-SCLC cell lines. Here, we have used high-resolution genome scanning by ROMA of 261 lung cancer samples to determine the most frequently occurring high-amplitude focal amplicons in lung cancer. The second most frequently recurring high-level amplicon is at 14q13.3. We note that both Zhao et al. (30) and Garnis et al. (31) also detected the 14q13.3 amplicon in their studies. Although the high-level amplification of 14q13.3 amplicon as judged by ROMA in lung cancer is only 4.2%, by relaxing the amplification cutoff from 1.5 (high level) to 1.2 (lower level), our CGH data implicate 19% of analyzed samples are amplified at 14q13.3. Collectively, these observations suggest that the 14q13.3 amplicon is a highly recurrent event in human lung cancer.
A convergence of recent observations suggests a cell lineage dependency (or lineage addiction) model of human cancer (32). In the lineage addiction model, tumor cells are highly dependent on the survival mechanisms that are built into the lineage precursor cells during development, which might be genetically altered to provide a survival advantage. In contrast to the well publicized oncogene addiction model that invokes a tumor-specific gain-of-function event (33), lineage dependency relies on the deregulation of key developmental survival mechanisms to promote tumorigenesis. Interestingly, given its essential function in lung development (34, 35) and restrictive expression in lung tissue (36), TTF1 was speculated to be an appealing candidate cell lineage-survival oncogene in lung cancer (32). Very recently, Tanaka et al. (37) provided evidence that sustained TTF1 expression may be crucial for a subset of lung AC. However, in light of the findings of our study, we believe there are several lines of evidence suggesting that TTF1 as well as NKX28 and PAX9 are lung cell lineage addiction oncogenes. In addition to lung cancer, by using the same ROMA platform of array CGH, we have profiled other cancer types (see SI Table 7). Other than the amplified lung cancer samples, the only samples showing DNA gain at 14q13 were six breast and two colon cancer DNAs. However, in each case, the DNA gain was extremely wide (>20 Mb), and it is thus unlikely that the 14q13.3 locus is the specific target of amplification. Based on these observations and other published CGH studies (reviewed in refs. 38 and 39), it appears that the focal 14q13.3 amplicon takes place preferentially in lung cancer, consistent with a lineage connection to the underlining driver genes.
Classically, each amplicon is presumed to target one driver gene for its functional advantage. However, data emerging from recent studies suggest that multiple coamplified genes within a given amplicon could be functionally relevant. Several reports in the literature lend support to the thesis of “multiple driver genes” per amplicon. Amplicons that have been shown functionally involving multiple driver genes include the 11q22 amplicon in liver cancer (40), the HER2 amplicon (41), and the 8p11-12 amplicon (42) in breast cancer, the 3q26.2 amplicon in ovarian cancer (43), and finally the 19q13 amplicon in pancreatic cancer (44). Based on our findings, the focal point of this study, 14q13.3 amplicon, is very likely to also involve multiple coamplified driver genes. Functionally, both NKX2-8 and TTF1 were able to enhance the tumorigenicity of a nonamplified lung SCC cell line, establishing that both genes are capable of oncogenic activity. Furthermore, substantial synergistic growth effects of all pairwise combinations of the three genes was observed when introduced into immortalized premalignant lung epithelial cells. This surprising finding highlights the possibility that these candidate oncogenes functionally cooperate to promote lung cancer cell growth.
In sum, we conclude that we have discovered an important recurrent amplicon in lung cancer. The three genes affected by this lesion are potentially novel lung oncogenes that cooperate to promote lung cancer cell proliferation. Our findings have strengthened the growing body of evidence that many amplification events select multiple cooperating oncogenes. This has important implications for the large functional genomic projects aimed at identifying cancer-relevant genes. Large genome-scale cDNA or RNAi screens that assay one gene at a time will not uncover activities that rely on multiple collaborating genes. Only through direct genomic analysis of human cancer will we be able to discover these important interactions.
Materials and Methods
Primary Lung Tumors and Lung Cancer Cell Lines.
Primary fresh-frozen lung tumors were obtained from the Cooperative Human Tissue Network (CHTN, Eastern Division, Philadelphia, PA) and from the laboratory of M. Wigler at Cold Spring Harbor Laboratory. Tumor samples, coded anonymously, were examined histologically to ensure at least 70% malignant tissue before CGH study. The majority of cancer cell lines were acquired from either American Type Culture Collection (Manassas, VA) or Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). Genomic DNA were prepared by using the proteinase-K method with at least three rounds of phenol/phenol-chloroform extraction to minimize protein contaminants (45). RNA of cell lines and tumors was prepared by using Qiagen (Valencia, CA) RNeasy mini kit.
CGH and Gene Expression Profiling.
CGH microarrays bearing 85,000 oligonucleotides designed to hybridize BglII restriction fragments of human genome were custom synthesized by Nimblegen Systems (Madison, WI) (13). The microarray hybridization was performed essentially as described (46). A more detailed description is presented in SI Text.
QPCR, RNAi, and Cell Culture Assay.
QPCR experiments to determine DNA copy number and RNA expression were conducted essentially as described by Mu et al. (47). The QPCR probes were designed by using Primer Express 2.0 software (Applied Biosystems, Foster City, CA), and the sequences are listed in SI Text. shRNA selection and creation of stable shRNA transfectants were as described (26, 27). The actual shRNA inserts cloned into the pMLP retroviral expression vector (27) are listed in SI Text. Retrovirus-mediated gene transfer was achieved with the murine stem cell virus-based expression vectors (see SI Text).
Supplementary Material
Acknowledgments
We thank L. Rodgers (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) and M. Wigler for providing normal human DNA and various cancer cell lines, S. Lowe and members of his laboratory for interesting discussions, and J. Zuber for informing us of the BIOPREDsi webtool. D.M. was supported in part by a grant from Joan's Legacy Foundation, New York. This work was supported by Cold Spring Harbor Laboratory Cancer Center Grant 5P30CA45508.
Abbreviations
- AC
adenocarcinoma
- CGH
comparative genomic hybridization
- SCLC
small-cell lung cancer
- SCC
squamous-cell carcinoma
- QPCR
quantitative PCR
- shRNA
small-hairpin RNA.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708286104/DC1.
References
- 1.Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 2.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 3.Hibi K, Trink B, Patturajan M, Westra WH, Caballero OL, Hill DE, Ratovitski EA, Jen J, Sidransky D. Proc Natl Acad Sci USA. 2000;97:5462–5467. doi: 10.1073/pnas.97.10.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugahara K, Iyama KI, Kimura T, Sano K, Darlington GJ, Akiba T, Takiguchi M. Cell Tissue Res. 2001;306:57–63. doi: 10.1007/s004410100420. [DOI] [PubMed] [Google Scholar]
- 5.Halmos B, Huettner CS, Kocher O, Ferenczi K, Karp DD, Tenen DG. Cancer Res. 2002;62:528–534. [PubMed] [Google Scholar]
- 6.Shim M, Powers KL, Ewing SJ, Zhu S, Smart RC. Cancer Res. 2005;65:861–867. [PubMed] [Google Scholar]
- 7.Wistuba II, Gazdar AF, Minna JD. Semin Oncol. 2001;28:3–13. [PubMed] [Google Scholar]
- 8.Minna JD, Roth JA, Gazdar AF. Cancer Cell. 2002;1:49–52. doi: 10.1016/s1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- 9.Moens CB, Auerbach AB, Conlon RA, Joyner AL, Rossant J. Genes Dev. 1992;6:691–704. doi: 10.1101/gad.6.5.691. [DOI] [PubMed] [Google Scholar]
- 10.Charron J, Malynn BA, Fisher P, Stewart V, Jeannotte L, Goff SP, Robertson EJ, Alt FW. Genes Dev. 1992;6:2248–2257. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- 11.Stanton BR, Perkins AS, Tessarollo L, Sassoon DA, Parada LF. Genes Dev. 1992;6:2235–2247. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- 12.Knoepfler PS, Cheng PF, Eisenman RN. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lucito R, Healy J, Alexander J, Reiner A, Esposito D, Chi M, Rodgers L, Brady A, Sebat J, Troge J, et al. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E, Esposito D, Alexander J, Troge J, Grubor V, et al. Genome Res. 2006;16:1465–1479. doi: 10.1101/gr.5460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, Khatry DB, Protopopov A, You MJ, Aguirre AJ, et al. Proc Natl Acad Sci USA. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanfel MN, Moses KA, Schwartz RJ, Zimmer WE. Cell Mol Biol (Noisy-le-grand) 2005;(Suppl 51):OL785–OL799. [PubMed] [Google Scholar]
- 17.Robson EJ, He SJ, Eccles MR. Nat Rev Cancer. 2006;6:52–62. doi: 10.1038/nrc1778. [DOI] [PubMed] [Google Scholar]
- 18.Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO. Proc Natl Acad Sci USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ke Y, Reddel RR, Gerwin BI, Miyashita M, McMenamin M, Lechner JF, Harris CC. Differentiation. 1988;38:60–66. doi: 10.1111/j.1432-0436.1988.tb00592.x. [DOI] [PubMed] [Google Scholar]
- 20.Scragg MA, Ferreira LR. Anal Biochem. 1991;198:80–85. doi: 10.1016/0003-2697(91)90509-r. [DOI] [PubMed] [Google Scholar]
- 21.Bejarano PA, Baughman RP, Biddinger PW, Miller MA, Fenoglio-Preiser C, al-Kafaji B, Di Lauro R, Whitsett JA. Mod Pathol. 1996;9:445–452. [PubMed] [Google Scholar]
- 22.Fabbro D, Di Loreto C, Stamerra O, Beltrami CA, Lonigro R, Damante G. Eur J Cancer. 1996;32A:512–517. doi: 10.1016/0959-8049(95)00560-9. [DOI] [PubMed] [Google Scholar]
- 23.Di Loreto C, Di Lauro V, Puglisi F, Damante G, Fabbro D, Beltrami CA. J Clin Pathol. 1997;50:30–32. doi: 10.1136/jcp.50.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khoor A, Whitsett JA, Stahlman MT, Olson SJ, Cagle PT. Hum Pathol. 1999;30:695–700. doi: 10.1016/s0046-8177(99)90096-5. [DOI] [PubMed] [Google Scholar]
- 25.Holzinger A, Dingle S, Bejarano PA, Miller MA, Weaver TE, DiLauro R, Whitsett JA. Hybridoma. 1996;15:49–53. doi: 10.1089/hyb.1996.15.49. [DOI] [PubMed] [Google Scholar]
- 26.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 27.Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 28.Huesken D, Lange J, Mickanin C, Weiler J, Asselbergs F, Warner J, Meloon B, Engel S, Rosenberg A, Cohen D, et al. Nat Biotechnol. 2005;23:995–1001. doi: 10.1038/nbt1118. [DOI] [PubMed] [Google Scholar]
- 29.Olson A, Sheth N, Lee JS, Hannon G, Sachidanandam R. Nucleic Acids Res. 2006;34:D153–D157. doi: 10.1093/nar/gkj051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao X, Weir B. A., LaFramboise T, Lin M, Beroukhim R, Garraway L, Beheshti J, Lee JC, Naoki K, Richards WG, et al. Cancer Res. 2005;65:5561–5570. doi: 10.1158/0008-5472.CAN-04-4603. [DOI] [PubMed] [Google Scholar]
- 31.Garnis C, Lockwood WW, Vucic E, Ge Y, Girard L, Minna JD, Gazdar AF, Lam S, MacAulay C, Lam WL. Int J Cancer. 2006;118:1556–1564. doi: 10.1002/ijc.21491. [DOI] [PubMed] [Google Scholar]
- 32.Garraway LA, Sellers WR. Nat Rev Cancer. 2006;6:593–602. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 33.Weinstein IB. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 34.Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- 35.DeFelice M, Silberschmidt D, DiLauro R, Xu Y, Wert SE, Weaver TE, Bachurski CJ, Clark JC, Whitsett JA. J Biol Chem. 2003;278:35574–35583. doi: 10.1074/jbc.M304885200. [DOI] [PubMed] [Google Scholar]
- 36.Ordonez NG. Adv Anat Pathol. 2000;7:123–127. doi: 10.1097/00125480-200007020-00007. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S, Shimada Y, Osada H, Kosaka T, Matsubara H, et al. Cancer Res. 2007;67:6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- 38.Myllykangas S, Knuutila S. Cancer Lett. 2006;232:79–89. doi: 10.1016/j.canlet.2005.07.045. [DOI] [PubMed] [Google Scholar]
- 39.Knuutila S, Bjorkqvist AM, Autio K, Tarkkanen M, Wolf M, Monni O, Szymanska J, Larramendy ML, Tapper J, Pere H, et al. Am J Pathol. 1998;152:1107–1123. [PMC free article] [PubMed] [Google Scholar]
- 40.Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan S-T, Luk J, Wigler M, Hannon GJ, et al. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kao J, Pollack JR. Genes Chromosomes Cancer. 2006;45:761–769. doi: 10.1002/gcc.20339. [DOI] [PubMed] [Google Scholar]
- 42.Yang ZQ, Streicher KL, Ray ME, Abrams J, Ethier SP. Cancer Res. 2006;66:11632–11643. doi: 10.1158/0008-5472.CAN-06-2946. [DOI] [PubMed] [Google Scholar]
- 43.Nanjundan M, Nakayama Y, Cheng KW, Lahad J, Liu J, Lu K, Kuo W. L., Smith-McCune K, Fishman D, Gray JW, et al. Cancer Res. 2007;67:3074–3084. doi: 10.1158/0008-5472.CAN-06-2366. [DOI] [PubMed] [Google Scholar]
- 44.Kuuselo R, Savinainen K, Azorsa DO, Basu GD, Karhu R, Tuzmen S, Mousses S, Kallioniemi A. Cancer Res. 2007;67:1943–1949. doi: 10.1158/0008-5472.CAN-06-3387. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 46.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 47.Mu D, Chen L, Zhang X, See LH, Koch CM, Yen C, Tong JJ, Spiegel L, Nguyen KC, Servoss A, et al. Cancer Cell. 2003;3:297–302. doi: 10.1016/s1535-6108(03)00054-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.