Abstract
In Escherichia coli, Salmonella, and related bacteria, the PhoQ–PhoP system regulates the expression of a large collection of genes in response to conditions of low magnesium or to the presence of certain antimicrobial peptides. We measured transcription of four PhoP-regulated promoters in E. coli that have significantly different PhoP-binding sites. Surprisingly, three promoters show identical responses to magnesium concentrations that range over four orders of magnitude. By analyzing and testing a simple model of transcriptional regulation, we find an explanation for this puzzle and show that these promoters are indeed differentially regulated at sufficiently high levels of stimulus. We then use this analysis to infer an effective level of phosphorylated PhoP as a function of magnesium stimulus. Our results demonstrate that differential regulation generally depends on the strength of the stimulus and highlight how quantitative analysis of stimulus–response curves can be used to infer properties of cell regulatory circuits that cannot be easily obtained from in vitro measurements.
Keywords: modeling, response regulator, single cell fluorescence, two-component signaling
In bacteria, a collection of genes controlled by a common regulatory network often show considerable diversity in expression patterns. In many cases, this differential regulation can be explained by combinatorial control by multiple transcription factors or by the action of a single transcription factor that functions as both an activator and a repressor. However, an additional and more subtle form of differential regulation can arise for a group of genes when a transcription factor has significantly different binding affinities for the various promoters. In this case, increasing concentrations of the transcription factor can lead to an ordering or hierarchy of transcriptional activation. Such differential regulation has been suggested to play a critical role in cellular processes that require temporal control, as in the synthesis of complex molecular assemblies, or that depend on the strength or type of input stimulus (e.g., refs. 1–5).
The PhoQ–PhoP two-component signaling system, which is found in Escherichia coli, Salmonella, and related bacteria, is an example of a regulatory circuit that controls expression of a large collection of genes (6–9). PhoQ is a histidine kinase that phosphorylates its cognate response regulator PhoP in response to low extracellular levels of magnesium and to the presence of some antimicrobial peptides (6, 10–13). Phosphorylated PhoP (PhoP-P) functions as a regulator of genes associated with magnesium transport, outer-membrane modification, acid resistance, and pathogenesis (6–9, 14–).
Recent in vitro studies of several promoters regulated by PhoQ–PhoP in E. coli identified specific PhoP-binding sites termed PhoP-boxes (7, 18). Furthermore, PhoP binds some of these promoters with significantly different affinities. As a result, these PhoP-regulated promoters may be differentially regulated in vivo so that with increasing levels of PhoQ stimulation, promoters with high-affinity binding sites are activated first, followed by promoters with progressively lower affinity sites. To explore this possibility, we used two-color fluorescent reporter strains to measure stimulus–response curves of several PhoQ–PhoP regulated genes with a high level of precision. Surprisingly, we did not observe differential regulation over a large range of stimulus (magnesium concentration) for several promoters, despite the variations among their PhoP-box sequences and the significant differences in PhoP-binding affinity in vitro. By analyzing a simple model of transcriptional regulation and testing predictions of this model, we find an explanation for this puzzle and show that these promoters are indeed differentially regulated at sufficiently high levels of stimulus. Interestingly, we find that the order of promoter activation in vivo is qualitatively different from the order expected from in vitro measurements. We also use our analysis to infer an effective level of phosphorylated PhoP as a function of magnesium stimulus.
Results and Discussion
The Transcription Profiles in Response to Mg2+ Stimulus Are Indistinguishable for Several PhoP-Regulated Genes.
To study differential regulation in the PhoQ–PhoP system, we constructed a set of isogenic, two-color fluorescent reporter strains for four promoters (mgtA, phoPQ, mgrB, and hemL) that have markedly different binding affinities for PhoP. Each reporter strain contains yfp, the gene for yellow fluorescent protein (YFP), under the control of a particular PhoP-regulated promoter, and cfp, the gene for cyan fluorescent protein (CFP), under the control of a constitutive promoter (Fig. 1a). In the experiments described below, CFP fluorescence serves as an internal reference level for measuring YFP fluorescence in individual cells. This controls for variations in YFP due to extrinsic variability in gene expression (19, 20), variability in experimental measurements, and variations in total protein levels for different culture conditions (21, 22). As a result, the YFP/CFP fluorescence ratio provides a remarkably precise measurement of transcriptional activity as a function of stimulus.
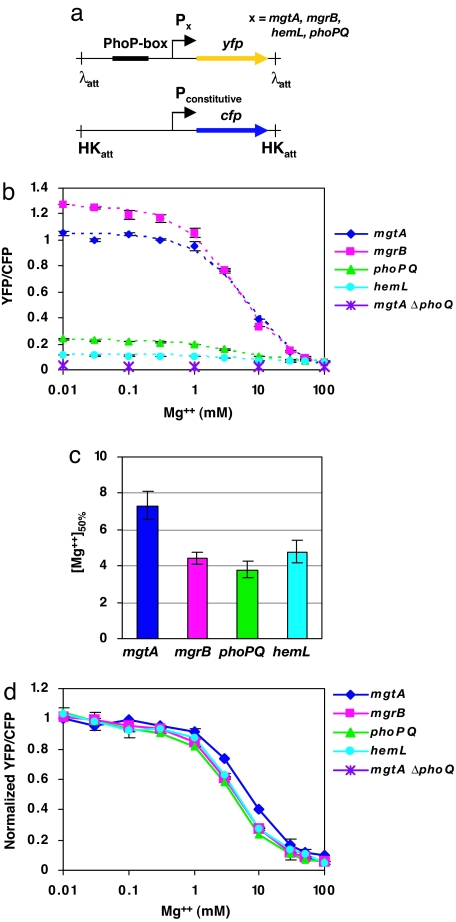
Fig. 1.
Transcription profiles of four PhoP-regulated promoters in response to extracellular [Mg2+]. (a) Each reporter strain contains a chromosomal copy of yfp controlled by a PhoP-regulated promoter, and a copy of cfp, controlled by a constitutive promoter (tetA promoter), at the attachment sites of λ and HK022 phages, respectively. The expression of CFP from the tetA promoter is constant over the range of magnesium concentrations used in our experiments (22). (b) Steady-state transcription of PhoP-regulated promoters as measured by the YFP/CFP fluorescence ratio of single cells. Cultures were grown at 37°C in minimal medium containing the indicated concentrations of MgSO4. Each point indicates the mean of two independent cultures, and each bar indicates the corresponding range. The dotted lines denote fits to saturating curves. (c) Concentration of magnesium at which YFP/CFP is halfway between the maximal and minimal values, determined for each promoter from the fitted curves in a. Bars denote the errors from the nonlinear fits. (d) Normalized curves from b. Each curve was normalized by shifting and rescaling by constants. For details of the analysis, see SI Text.
The PhoQ–PhoP two-component system is stimulated under conditions of low extracellular magnesium (23–25). Indeed, the steady-state YFP/CFP fluorescence ratio of each reporter strain increased with decreasing concentrations of magnesium (Fig. 1b). The magnesium concentration that results in the half-maximal transcriptional response to Mg2+ ([Mg2+]50%) was highest for the mgtA promoter (Fig. 1c), which also has the highest affinity for PhoP in vitro (7). However, the values of [Mg2+]50% for the mgrB, phoPQ, and hemL promoters were not significantly different from each other (Fig. 1c). This suggests that the Mg2+ response curves in Fig. 1b corresponding to these three promoters are the same, after compensating for the basal and maximal levels of transcription at high and low magnesium, respectively. To check this, we normalized the curves by shifting and rescaling so that they coincide at regions of high and low Mg2+ [for details, see supporting information (SI) Text]. After normalization, the Mg2+ response curves of the mgrB, phoPQ, and hemL reporter strains were indistinguishable (Fig. 1d). This result was not due to the particular method of constructing the reporters because an operon fusion of yfp to mgrB at its wild-type locus showed the same normalized curve as the mgrB promoter reporter (SI Fig. 5). In addition, the normalized Mg2+ response curve for the mgtA promoter did not overlap with the corresponding curves for the mgrB, phoPQ, or hemL promoters (Fig. 1d). Under identical growth conditions, the four reporter strains should have the same intracellular concentration of PhoP-P ([PhoP-P]). Therefore, our results suggest that the normalized transcriptional responses of the mgrB, phoPQ, and hemL promoters to [PhoP-P] are indistinguishable in cells grown under the steady-state conditions described in Fig. 1.
We did not observe a hierarchy of transcriptional activation among all four PhoP-regulated promoters in response to different levels of magnesium. In fact, three of the promoters showed the same transcriptional profile. One possible explanation for this striking result is that PhoP-P has identical binding affinities at the mgrB, phoPQ, and hemL promoters in vivo. However, this seems unlikely because the sequences of the PhoP-boxes are substantially different (see Fig. 3a) and PhoP binds the promoters with different affinities in vitro (7) (SI Fig. 6). Furthermore, the results described below suggest that the in vivo binding affinities of PhoP-P for the three promoters are indeed different. An alternative explanation is that the level of PhoP-P remains below the in vivo PhoP-P dissociation constants for the mgrB, phoPQ, and hemL promoters throughout the range of [Mg2+] in Fig. 1b. For this range of [PhoP-P], the fractional occupation of these promoters by PhoP-P will be proportional to [PhoP-P] (or proportional to a power of [PhoP-P] for the case of cooperative binding). Furthermore, if we assume that transcriptional activation of a PhoP-regulated promoter is proportional to the fractional occupation by PhoP-P, then the normalized transcription profiles as a function of [PhoP-P] (or equivalently [Mg2+]) will be indistinguishable for the mgrB, phoPQ, and hemL promoters. The distinct behavior of the mgtA promoter can then be attributed to a significantly lower PhoP-P binding constant. These points are illustrated in Fig. 2 and are tested experimentally below.
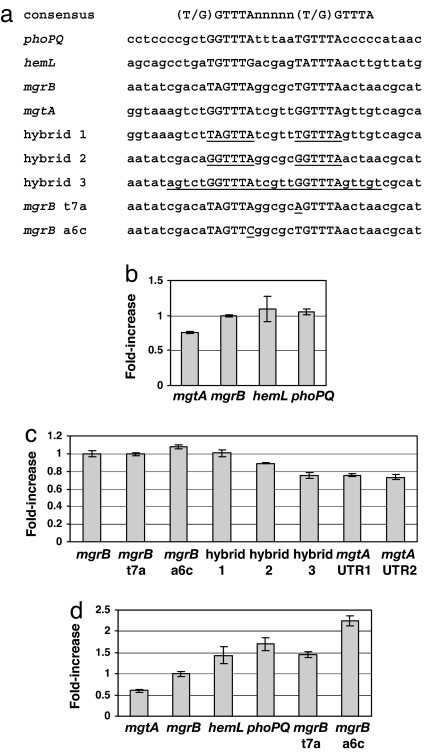
Fig. 3.
Relative transcriptional activation for wild-type and mutated promoters. (a) Sequences of the consensus PhoP-box, the PhoP-boxes of the wild-type promoters, and the PhoP-boxes of the mutated promoters. Hybrid 1 consists of the mgtA promoter with the mgrB PhoP-box. Hybrid 2 and hybrid 3 consist of the mgrB promoter with the mgtA PhoP-box and with portions of flanking mgtA sequence, respectively. (b) Fold increase in transcription of wild-type promoters (relative to the fold increase of mgrB transcription) for cells grown in 100 μM Mg2+ compared with 10 mM Mg2+. Fold increase is defined to be the following: [(YFP/CFP)100 μM Mg2+ − (YFP/CFP)100 μM Mg2+,ΔphoQ]/[YFP/CFP)10 mM Mg2+ − (YFP/CFP)10 mM Mg2+,ΔphoQ]. (c) Fold increase as in b for several mutated promoters listed in a. Two different23 versions of the mgtA reporter are also compared. UTR1 has the native 5′-UTR for mgtA, which includes a potential riboswitch. UTR2 has a truncated 5′-UTR with only 16 base pairs of the original 263 base pairs in the mgtA 5′-UTR. (d) Fold increase in transcription of various promoters (relative to the fold increase of mgrB transcription) for cells exposed to LL-37 in 100 μM Mg2+ for 1 h, compared with cells in 100 μM Mg2+ without LL-37. Fold increases were computed by using YFP/CFP values determined from two independent cultures. The bars denote estimates of the error determined from the corresponding ranges of the YFP/CFP measurements.
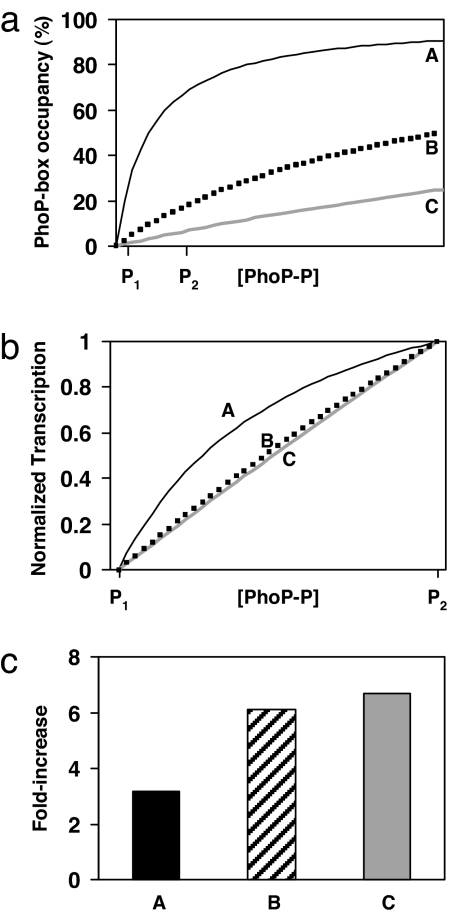
Fig. 2.
A comparison of hypothetical PhoP-P binding curves and normalized transcription profiles for three promoters with different PhoP-P dissociation constants. (a) PhoP-P binding curves for three promoters (A, B, and C) with different dissociation constants. P1 and P2 denote [PhoP-P] at high and low [Mg2+], respectively. In this study, we assume simple binding at these promoters; however, the conclusions of this analysis are the same for the cases of binding by a PhoP-P dimer or of cooperative binding. We also assume in this analysis that the rate of transcription for a particular promoter is a linear function of its fractional occupancy by PhoP-P. (b) The transcription profiles of the promoters in a normalized so that the transcription levels coincide at P1 and P2. The promoters with PhoP-P dissociation constants above P2 (promoters B and C) have normalized transcription profiles that are essentially indistinguishable. In contrast, promoter A, which has a dissociation constant below P2, has a distinct normalized transcription profile. (c) Fold increase in transcription from P1 to P2 for the promoters in a. Note that promoters with different PhoP-P dissociations constants greater than P2 (promoters B and C) show the same fold increase. For details of the analysis, see SI Text.
In the above analysis and in what follows, we have made the simplifying assumption that a single molecule of PhoP-P binds to a PhoP-regulated promoter. However, the conclusions are similar for the cases of binding by a PhoP-P dimer or of cooperative binding.
The Differential Regulation of mgtA Relative to mgrB, phoPQ, and hemL Is Associated with the mgtA PhoP-Binding Site.
If PhoP-P levels in cells grown under the Mg2+ conditions described in Fig. 1 are well below the dissociation constants of the mgrB, phoPQ, and hemL promoters, then our model predicts that the fold change in transcription corresponding to a change in [Mg2+] within this range will be the same for these promoters (see Fig. 2c). Consistent with the results in Fig. 1 and our model, we found that the fold increase in transcription from 10 mM to 100 μM [Mg2+] was comparable for mgrB, phoPQ, and hemL (Fig. 3b). In contrast, mgtA showed a significantly lower fold increase (Fig. 3b), which suggests that [PhoP-P] at 100 μM [Mg2+] is comparable to or greater than the dissociation constant for the mgtA PhoP-box, Kd,mgtA. To determine whether the distinct Mg2+ response of the mgtA promoter is associated with the PhoP-binding site, we similarly measured the fold increase in transcription for hybrid promoters in which the PhoP-boxes of the mgrB and mgtA promoters were swapped (Fig. 3 a and c). The hybrid promoter constructed by replacing the PhoP-box in the mgtA promoter with the mgrB PhoP-box (hybrid 1) showed a fold increase in transcription that was identical with that of the mgrB promoter. The reciprocal hybrid, with the mgtA PhoP-box replacing the mgrB PhoP-box in the mgrB promoter (hybrid 2), showed a fold increase in transcription that was intermediate between those of the mgrB and mgtA promoters. A second hybrid mgrB promoter, which has a larger substitution of the PhoP-binding region from the mgtA promoter (hybrid 3), showed a fold increase in transcription that was identical with that of mgtA. These results provide further evidence that the differential regulation of the mgtA promoter, relative to the mgrB, phoPQ, and hemL promoters, is due to a lower PhoP-P dissociation constant for the mgtA PhoP-binding site. The results also suggest that additional sequences adjacent to the PhoP-box consensus sequence may play a role in determining PhoP-P binding affinity in vivo at some promoters.
A recent study has shown that a Mg2+-responsive riboswitch within the 5′-UTR of the mgtA transcript contributes to magnesium regulation of mgtA expression in Salmonella typhimurium (26). Computer analysis suggested that the 5′-UTR of the E. coli mgtA transcript may also function as a riboswitch (26). Our E. coli mgtA reporter construct contains the native mgtA 5′-UTR (263 base pairs) upstream of the yfp start codon. However, a different mgtA reporter strain, which contains only 16 base pairs from the native mgtA 5′-UTR, exhibited the same response to Mg2+ (Fig. 3c). In addition, the hybrid 3 construct described above, which produces a transcript that completely lacks the mgtA 5′-UTR, showed the same response to Mg2+ as that of the mgtA promoter (Fig. 3b). If there is a riboswitch in the mgtA 5′-UTR in E. coli, our results suggest that it is not sensitive to the range of [Mg2+] in our experiments.
The Mg2+ Response of the mgrB Promoter Is Insensitive to PhoP-Box Mutations That Weaken Transcription.
If the levels of PhoP-P associated with the range of [Mg2+] used in our experiments are well below the dissociation constant of a particular promoter, then mutations that decrease the PhoP binding affinity, i.e., raise the dissociation constant, should not affect the normalized transcriptional response to a change in [Mg2+]. To test this, we weakened the mgrB promoter by mutating its PhoP-box. Two bases within the PhoP-box of the mgrB promoter that are also present within the consensus sequence were individually mutated (Fig. 3a). The YFP expression levels from the two mutated promoters were markedly attenuated compared with the corresponding level for the mgrB promoter (SI Fig. 7 Left). Nevertheless, for both promoters, the fold increases in transcription and the normalized Mg2+ response curves were comparable with the corresponding data for the wild-type mgrB promoter (Fig. 3c and SI Fig. 7 Right). Thus, consistent with the predictions of our model, weakening the mgrB PhoP-box does not affect the relative transcriptional response for [Mg2+] over the range 100 mM to 10 μM.
Stimulation of PhoQ with the Antimicrobial Peptide LL-37 Reveals Differential Regulation of mgrB, phoPQ, and hemL.
The above results suggest that if the mgrB, phoPQ, and hemL promoters have significantly different PhoP-P dissociation constants, then differential regulation will emerge at sufficiently high levels of PhoP-P. However, such levels must be higher than those obtained for growth in 10 μM Mg2+. We found that it was difficult to achieve steady-state growth conditions for magnesium concentrations <10 μM, presumably because the cells continually deplete the magnesium in the culture medium. We therefore used the antimicrobial peptide LL-37 to further stimulate PhoQ. LL-37 has been shown to activate the PhoQ–PhoP system in S.typhimurium (13). Similarly, for E. coli cells growing in 100 μM Mg2+, the addition of LL-37 resulted in increased PhoP-regulated transcription (data not shown). Notably, the mgrB, phoPQ, and hemL promoters showed distinct transcriptional responses to LL-37 (Fig. 3d) in contrast with the indistinguishable Mg2+ responses described above (Fig. 3b). Thus, the increased PhoQ stimulation from exposure to LL-37 resulted in differential regulation of all four promoters.
The promoter with the highest fold increase in transcription was phoPQ (Fig. 3d), which suggests that the phoPQ promoter has the largest PhoP-P dissociation constant among the promoters tested in this study. Interestingly, this would provide maximal amplification of other PhoP-regulated promoters from phoPQ autoregulation before saturating PhoP production. However, this result is in striking contrast with the results from in vitro measurements that suggest the PhoP dissociation constant of the phoPQ promoter is lower than those of mgrB and hemL (see ref. 7 and SI Fig. 6). The in vitro measurements were made with unphosphorylated PhoP (see ref. 7 and SI Text). We were not able to produce sufficiently high levels of phosphorylated PhoP to perform the analogous in vitro measurements with PhoP-P. A previous study has shown that high-level expression of PhoP activates transcription of PhoP-regulated promoters independently of PhoQ or of phosphorylation (27). To determine whether overexpression of unphosphorylated PhoP leads to differential regulation of mgtA, mgrB, phoPQ, and hemL promoters, we overexpressed PhoP in ΔphoQ reporter strains. We found the differential regulation was similar to the in vivo results described above for phoQ+ strains and in marked contrast with the in vitro PhoP binding measurements (SI Fig. 8). These results suggest that the observed differences between the in vitro and in vivo data are due to additional factors that affect PhoP binding affinity at PhoP-regulated promoters in vivo that are absent in the in vitro measurements.
Our model predicts that a promoter containing mutations that weaken the affinity of PhoP-P for its PhoP-box should also exhibit differential regulation, relative to the wild-type promoter, at sufficiently high levels of PhoP-P. To test this, we measured the transcriptional response to LL-37 for the mgrB promoters with either T7A or A6C substitutions in the PhoP-box (Fig. 3a). Both mutated promoters showed fold increases in transcription from stimulation with LL-37 that were above that of the wild-type mgrB promoter. These results are also consistent with the observation that the T7A substitution was the weakest of the three promoters: the T7A substitution showed the most attenuated Mg2+ response curve (SI Fig. 7 Left) and displayed the largest fold increase in transcription (Fig. 3d).
Taken together, the above results suggest that the mgtA, phoPQ, mgrB, and hemL promoters are differentially regulated under conditions that strongly stimulate the PhoQ–PhoP system (e.g., the presence of LL-37).
Effective PhoP-P Binding Curves.
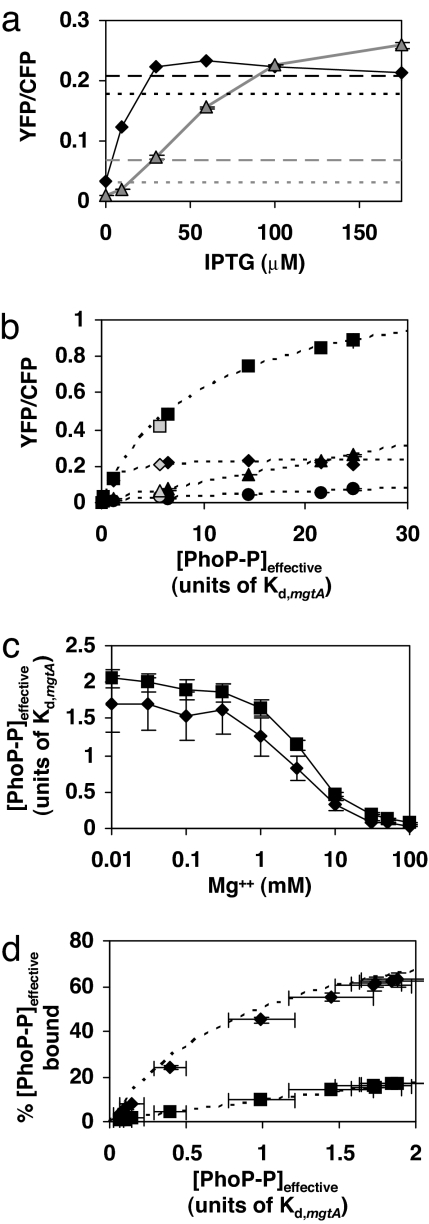
Within the context of our model, we can associate each transcription level of a PhoP-regulated promoter with a distinct value of [PhoP-P]. We call these values effective PhoP-P concentrations ([PhoP-P]effective) because they are model dependent and may not reflect the actual concentrations of PhoP-P within the cell. Because [PhoP-P]effective should be the same in every reporter strain for a given growth condition, it provides a means to relate transcription levels of the PhoP-regulated promoters to each other. To extract a complete curve of transcription as a function of [PhoP-P]effective for a particular promoter, transcription measurements for sufficiently high levels of PhoQ stimulation such that the promoter approaches maximal binding to PhoP-P (saturated binding) are required. This will occur at PhoP-P concentrations well above the dissociation constant for the promoter (Fig. 2a). We were unable to observe saturation of any of the PhoP-regulated promoters studied here by varying either magnesium or LL-37 concentrations. Apparently, we were not able to reach sufficiently high levels of PhoP-P with either stimulus of PhoQ under our experimental conditions. Therefore, to reach higher levels of transcriptional activation, we used a constitutively active PhoP mutant, which we denote by PhoPca. When we expressed PhoPca from an inducible promoter, we found that transcription from the mgtA promoter leveled off at high levels of induction, whereas transcription from the mgrB, hemL, and phoPQ promoters continued to increase (Fig. 4a and data not shown). This suggests that at the highest levels of PhoPca reached in these experiments, the mgtA promoter is saturated.
Fig. 4.
Effective PhoP-P levels and resulting transcription and binding curves, inferred from the model of transcriptional activation described in Fig. 2. (a) Transcription of the mgtA (black diamonds) and phoPQ (gray triangles) promoters for various levels of PhoPca expression. (PhoPca is a constitutively active variant of PhoP.) Transcription levels corresponding to 100 μM [Mg2+] and stimulation with LL-37 are indicated with dotted and dashed lines, respectively. The upper (lower) pair of dotted and dashed lines is for the mgtA (phoPQ) promoter. (b) Transcription of mgrB (squares), mgtA (diamonds), hemL (circles), and phoPQ (triangles) as a function of [PhoP-P]effective. The filled symbols denote PhoPca expression data, and the dotted lines are the associated fits. Note that the PhoPca expression data for phoPQ fall on a line by construction. The open symbols denote transcription levels from stimulation with LL-37 in strains with wild-type phoP, which were not used in determining the fits. (c) [PhoP-P]effective as a function of [Mg2+] determined from the transcription data in Fig. 1b for mgtA (diamonds) and mgrB (squares). (d) Inferred [PhoP-P]effective bound at the mgtA (diamonds) and mgrB (squares) promoters for cells grown in the levels of magnesium shown in Fig. 1b. The [PhoP-P]effective values are the means of the corresponding values in c. The curves are derived from the fits in b. For a and b, each point is the mean of two independent cultures, and each bar, which is smaller than the data symbol in some cases, denotes the range. For c and d, bars denote the errors associated with the nonlinear fits. For details of the analysis, see SI Text.
By using this PhoPca-induction data and the model described in Fig. 2, we determined the curves of transcription as a function of [PhoP-P]effective for the mgtA, mgrB, and hemL promoters (Fig. 4b) (for details, see SI Text). Concentrations are expressed in units of Kd,mgtA, the dissociation constant of PhoP-P for the mgtA promoter. For the case of mgrB, the fit determined the dissociation constant for PhoP-P at the mgrB promoter to be ≈10Kd,mgtA. For the hemL promoter, we found the range of [PhoP-P]effective resulting from expression of PhoPca was too low to determine Kd,hemL. Although the curves in Fig. 4b were determined by using only PhoPca expression data, the points corresponding to LL-37 stimulation (open symbols) are in good agreement.
From this analysis, we also inferred [PhoP-P]effective for cells grown in various concentrations of magnesium (Fig. 4c). These were separately determined from the YFP/CFP data for the mgtA and mgrB promoters (Fig. 1b). The two resulting curves are in close agreement (Fig. 4c). For the lowest magnesium level (10 μM [Mg2+]), [PhoP-P]effective is well below the dissociation constant for mgrB, which is consistent with our conclusions above concerning the differential regulation of mgrB and mgtA promoters. This differential regulation is further illustrated in Fig. 4d, which shows the extent of PhoP-P binding to the two promoters (within the context of our model) as a function of magnesium. The dashed curves, which were determined from PhoPca expression data (Fig. 4b), are in close agreement. These results suggest that the plateau in the Mg2+ response curves at low magnesium (Fig. 1) is due to a corresponding plateau in the levels of PhoP-P as a function of [Mg2+] (Fig. 4c). Interestingly, we have found that PhoP-regulated transcription is further activated in a PhoQ-dependent manner when cells are exposed to growth-limiting levels of Mg2+, which are well below 10 μM (data not shown). There is considerable evidence that magnesium stimulation acts by direct interaction with PhoQ (10, 23–25, 28). The above results suggest this activation is not characterized by magnesium binding with a single dissociation constant. This is plausible given the multiple magnesium ions and multiple acidic residues in the PhoQ periplasmic domain that appear to be involved in this interaction (25).
For the above analysis, we have assumed for simplicity that each promoter binds a single PhoP-P molecule. However, the results can be easily extended to the case of binding by multiple PhoP-P molecules. For example, if PhoP-P binds promoters as a dimer, then the curves in Fig. 4 b–d remain unchanged provided the points corresponding to specific values of [PhoP-P]effective are now interpreted as values of [PhoP-P]2effective. For the general case of cooperative binding characterized by a Hill constant h, [PhoP-P]effective is replaced with [PhoP-P]heffective.
Concluding Remarks.
We found that the relative responses of three PhoP-regulated promoters were identical over a remarkably large range of magnesium concentrations (Fig. 1d). Consistent with the predictions of a simplified model of transcriptional activation, differential regulation of these genes emerged at higher levels of PhoQ stimulus by treatment with the antimicrobial peptide LL-37 (Fig. 3d). Interestingly, even at the highest levels of stimulation, the promoters for mgrB, hemL, and phoPQ were apparently far from the maximal activity associated with saturated binding by PhoP-P (Fig. 4b). It is possible that maximal activity could be reached with different environmental conditions, such as the presence of other antimicrobial compounds or growth in much lower levels of magnesium. However, it is also possible that some PhoP-regulated promoters will remain far from saturation under all growth conditions. In this case, these promoters would be maximally responsive to changes in PhoQ activity for the range of PhoQ stimulation encountered by E. coli under physiological conditions.
Our results also highlight how stimulus–response curves, when measured with high precision, can be used to infer the parameters characterizing cell regulatory circuits in situ. This approach is complementary to in vitro biochemical measurements, which may be difficult to carry out in some cases and which may not always accurately reflect the conditions inside the cell, e.g., because of biochemical and biophysical differences between the in vitro and in vivo environments (29). Indeed, our results suggest that current in vitro measurements of PhoP binding to promoters do not accurately reflect the relative binding affinities in vivo. Importantly, we find that this cannot be simply explained by the absence of PhoP phosphorylation in the in vitro measurements, because we find that high-level expression of PhoP in vivo in the absence of phosphorylation shows the same ordering of promoter activation (SI Fig. 8). Our analysis, which is based on the steady-state behavior of the system, has some features in common with studies of activation kinetics of genetic circuits (30, 31). These approaches depend on assumptions concerning the physical, chemical, and biological mechanisms controlling gene expression in vivo. Therefore, the interpretation of our measurements must be viewed within this context. In particular, we assume that interactions of PhoP-P with regulatory sites at promoters can be described by a simple model of equilibrium binding, and that the extent of transcriptional activation is proportional to the fraction of bound PhoP-P at the promoter. These assumptions, which are simplifications of more accurate models of transcriptional regulation (e.g., ref. 32), are sufficient to account for the experimental measurements described here. Regardless of whether [PhoP-P]effective has a simple relationship to actual intracellular levels of PhoP-P, this parameter provides a unifying framework for analyzing and comparing PhoP-regulated promoters. This is evident from the excellent agreement between the transcription data for PhoQ stimulation via low [Mg2+] or LL-37 and the transcription curves determined independently from PhoPca data (Fig. 4 b and d). Further studies with a larger range of stimuli, combined with precise kinetics measurements, will lead to a more refined model of the PhoQ–PhoP system and related regulatory circuits from in situ measurements.
Materials and Methods
Strains and Growth Conditions.
All E. coli strains used in this study were derived from the K-12 strain MG1655 (33). A table of strains and plasmids with the details of their construction is given in SI Text.
Before each experiment, cells were grown overnight at 37°C in minimal A medium (34) containing 1 mM MgSO4 and supplemented with 0.1% casamino acids and 0.2% glucose. Plasmids were maintained by using 50 μg/ml ampicillin. The pTrc-derived promoters were induced with isopropyl-β-D-thiogalactoside. For the data in Figs. 1 and 3 b and c, cultures were diluted 1:104 into prewarmed media with the level of MgSO4 indicated in the figure and grown at 37°C for 3.5 h. We verified steady-state conditions were reached by measuring cellular fluorescence at several time points (data not shown). In experiments involving LL-37 and PhoPca (Figs. 3d and 4, respectively), a dilution of 1:1,000 was used and cells were grown for 4.5 h. LL-37 was added after 3.5 h.
At appropriate times, cultures were cooled quickly in an ice-slurry, and streptomycin was added to 250 μg/ml to inhibit further translation. When necessary, cultures were centrifuged at 4°C and resuspended in ≈10 μl to concentrate the cells. Cellular fluorescence was measured by fluorescence microscopy as described previously (22). For further details, see SI Text.
Supplementary Material
Acknowledgments
We thank Andy Binns, Linda Kenney, Eleonora Garcia Vescovi, Carey Waldburger, Marjan van der Woude, Amy Vollmer, Jun Zhu, and members of the Binns, Zhu, and M.G. laboratories for helpful advice and discussions. This work was supported by National Science Foundation Grant MCB0212925 (to M.G.) and an American Heart Association predoctoral fellowship (to T.M.).
Abbreviations
- PhoP-P
phosphorylated PhoP
- YFP
yellow fluorescent protein
- CFP
cyan fluorescent protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700025104/DC1.
References
- 1.Kutsukake K, Ohya Y, Iino T. J Bacteriol. 1990;172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalir S, McClure J, Pabbaraju K, Southward C, Ronen M, Leibler S, Surette MG, Alon U. Science. 2001;292:2080–2083. doi: 10.1126/science.1058758. [DOI] [PubMed] [Google Scholar]
- 3.Cotter PA, Jones AM. Trends Microbiol. 2003;11:367–373. doi: 10.1016/s0966-842x(03)00156-2. [DOI] [PubMed] [Google Scholar]
- 4.Fujita M, Gonzalez-Pastor JE, Losick R. J Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waters CM, Bassler BL. Genes Dev. 2006;20:2754–2767. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groisman EA. J Bacteriol. 2001;183:1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minagawa S, Ogasawara H, Kato A, Yamamoto K, Eguchi Y, Oshima T, Mori H, Ishihama A, Utsumi R. J Bacteriol. 2003;185:3696–3702. doi: 10.1128/JB.185.13.3696-3702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zwir I, Shin D, Kato A, Nishino K, Latifi T, Solomon F, Hare JM, Huang H, Groisman EA. Proc Natl Acad Sci USA. 2005;102:2862–2867. doi: 10.1073/pnas.0408238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monsieurs P, De Keersmaecker S, Navarre WW, Bader MW, De Smet F, McClelland M, Fang FC, De Moor B, Vanderleyden J, Marchal K. J Mol Evol. 2005;60:462–474. doi: 10.1007/s00239-004-0212-7. [DOI] [PubMed] [Google Scholar]
- 10.Garcia Vescovi E, Soncini FC, Groisman EA. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 11.Soncini FC, Garcia Vescovi E, Solomon F, Groisman EA. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, Fang FC, Miller SI. Mol Microbiol. 2003;50:219–230. doi: 10.1046/j.1365-2958.2003.03675.x. [DOI] [PubMed] [Google Scholar]
- 13.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 14.Soncini FC, Vescovi EG, Groisman EA. J Bacteriol. 1995;177:4364–4371. doi: 10.1128/jb.177.15.4364-4371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst RK, Guina T, Miller SI. Microbes Infect. 2001;3:1327–1334. doi: 10.1016/s1286-4579(01)01494-0. [DOI] [PubMed] [Google Scholar]
- 16.Brodsky IE, Gunn JS. Mol Interv. 2005;5:335–337. doi: 10.1124/mi.5.6.4. [DOI] [PubMed] [Google Scholar]
- 17.Shin D, Groisman EA. J Biol Chem. 2005;280:4089–4094. doi: 10.1074/jbc.M412741200. [DOI] [PubMed] [Google Scholar]
- 18.Kato A, Tanabe H, Utsumi R. J Bacteriol. 1999;181:5516–5520. doi: 10.1128/jb.181.17.5516-5520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 20.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 21.Batchelor E, Silhavy TJ, Goulian M. J Bacteriol. 2004;186:7618–7625. doi: 10.1128/JB.186.22.7618-7625.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyashiro T, Goulian M. Methods Enzymol. 2007;423:458–475. doi: 10.1016/S0076-6879(07)23022-8. [DOI] [PubMed] [Google Scholar]
- 23.Castelli ME, Garcia Vescovi E, Soncini FC. J Biol Chem. 2000;275:22948–22954. doi: 10.1074/jbc.M909335199. [DOI] [PubMed] [Google Scholar]
- 24.Chamnongpol S, Cromie M, Groisman EA. J Mol Biol. 2003;325:795–807. doi: 10.1016/s0022-2836(02)01268-8. [DOI] [PubMed] [Google Scholar]
- 25.Cho US, Bader MW, Amaya MF, Daley ME, Klevit RE, Miller SI, Xu W. J Mol Biol. 2006;356:1193–1206. doi: 10.1016/j.jmb.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 26.Cromie MJ, Shi Y, Latifi T, Groisman EA. Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 27.Lejona S, Castelli ME, Cabeza ML, Kenney LJ, Garcia Vescovi E, Soncini FC. J Bacteriol. 2004;186:2476–2480. doi: 10.1128/JB.186.8.2476-2480.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montagne M, Martel A, Le Moual H. J Bacteriol. 2001;183:1787–1791. doi: 10.1128/JB.183.5.1787-1791.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Kuhlman T, Phillips R. Curr Opin Genet Dev. 2005;15:125–135. doi: 10.1016/j.gde.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ronen M, Rosenberg R, Shraiman BI, Alon U. Proc Natl Acad Sci USA. 2002;99:10555–10560. doi: 10.1073/pnas.152046799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalir S, Alon U. Cell. 2004;117:713–720. doi: 10.1016/j.cell.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 32.Bintu L, Buchler NE, Garcia HG, Gerland U, Hwa T, Kondev J, Phillips R. Curr Opin Genet Dev. 2005;15:116–124. doi: 10.1016/j.gde.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 34.Miller JH. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia Coli and Related Bacteria. Plainview, NY: Cold Spring Harbor Lab Press; 1992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.