Abstract
What are the genetic and neural components that support adaptive learning from positive and negative outcomes? Here, we show with genetic analyses that three independent dopaminergic mechanisms contribute to reward and avoidance learning in humans. A polymorphism in the DARPP-32 gene, associated with striatal dopamine function, predicted relatively better probabilistic reward learning. Conversely, the C957T polymorphism of the DRD2 gene, associated with striatal D2 receptor function, predicted the degree to which participants learned to avoid choices that had been probabilistically associated with negative outcomes. The Val/Met polymorphism of the COMT gene, associated with prefrontal cortical dopamine function, predicted participants' ability to rapidly adapt behavior on a trial-to-trial basis. These findings support a neurocomputational dissociation between striatal and prefrontal dopaminergic mechanisms in reinforcement learning. Computational maximum likelihood analyses reveal independent gene effects on three reinforcement learning parameters that can explain the observed dissociations.
Keywords: basal ganglia, prefrontal cortex, computational model
A wealth of evidence points to a key role for the neurotransmitter dopamine (DA) in reinforcement learning (1–4). Transient bursts of DA firing are observed after unexpected rewards (1–3). These signals are thought to enhance learning through DA effects on corticostriatal plasticity so that rewarding actions are more likely to be selected in the future. Similarly, DA firing “dips” below baseline when rewards are expected but not received (1–3). Nevertheless, whether DA plays a functional role in the opposite kind of learning, that is, in avoiding nonrewarding or aversive actions, is controversial (2, 5).
Neural network models of the basal ganglia show how DA bursts and dips, by facilitating synaptic plasticity in separate neuronal populations, can support “Go” learning to make good choices and “NoGo” learning to avoid those that are less adaptive in the long run (4, 6, 7). Supporting this account, monkey studies reveal separate neural populations in the striatum coding for actions that are probabilistically associated with positive and negative outcomes (8). In the models, DA bursts support positive (Go) reinforcement learning by modulating striatal plasticity through D1 receptors, whereas DA dips support avoidance (NoGo) learning through D2 receptors. Several convergent lines of research support this overall functionality. Physiologically, D1 and D2 pharmacological agonists/antagonists differentially modulate activity and gene expression in separate Go and NoGo striatal populations (9). Behaviorally, striatal D1 receptor blockade prevents the speeding of responses to obtain large rewards (impaired Go learning), whereas striatal D2 blockade slows responding to obtain small rewards (enhanced NoGo learning) (10). In humans, DA medications modulate behavioral Go and NoGo learning in opposite directions (4, 11, 12). Nevertheless, it has not been demonstrated whether naturally occurring individual differences in this learning arise from dopaminergic mechanisms. Below, we show that genetic polymorphisms associated with striatal D1 and D2 function are directly predictive of Go and NoGo learning.
In addition to the basal ganglia, other brain regions contribute to reinforcement learning and action selection. Recent computational models explore how the prefrontal cortex (PFC), and local DA signals therein, can complement functions of the striatum (13, 14). In these models, the striatal system continues to integrate the long-term probability of positive and negative outcomes through incremental changes in synaptic plasticity, as described above. This idea is consistent with a striatal role in slow habitual learning (15, 16). In contrast, prefrontal cortical regions contribute to learning on a shorter time scale by actively maintaining recent reinforcement experiences in a working memory-like state (13). These PFC representations are stabilized by frontal DA levels (17, 18) and are used to modify ongoing behavior by top-down influences on subcortical structures (19). According to this scheme, tonically elevated DA levels in PFC, particularly during negative events (20, 21), can be beneficial for a form of avoidance learning. That is, PFC–DA-dependent mechanisms support robust maintenance of recent reinforcement outcomes so as to rapidly adapt behavior on a trial-to-trial basis (13). Supporting this account, increased prefrontal DA levels in rats predicted enhanced ability to shift from one rule to another (22, 23).
Together, the combined striatal/prefrontal models predict three distinct DA-related contributions to reinforcement learning. Striatal DA efficacy, by modulating incremental changes in synaptic plasticity, should modulate Go and NoGo learning to choose and avoid actions that are probabilistically associated with positive and negative outcomes, through D1 and D2 receptors, respectively. In contrast, tonically elevated prefrontal DA levels should support trial-to-trial adjustments that depend on maintaining recent negative outcomes in working memory. Thus, this predicted dissociation suggests multiple independent forms of learning, each of which may be linked with individual differences in genetic variation. The aim of the present study was to test these hypotheses by using both behavioral and computational methods.
Behavioral Results
We collected DNA from 69 healthy humans performing computerized learning tasks and analyzed three genes that have been clearly linked to striatal and prefrontal DA measures. We hypothesized that genetic differences in striatal DA efficacy would account for variability in probabilistic reward learning, and that genetic differences in striatal D2 receptor function would be associated with probabilistic avoidance learning. To test for striatal DA–reward learning associations, we analyzed a SNP in the gene coding for the DARPP-32 protein (24, 25). DARPP-32 potently modulates dopamine D1-dependent synaptic plasticity, is activated by D1 receptor stimulation, and is by far most abundant in the striatum (26–28). [Theoretically, a SNP in the D1 receptor itself could be examined, but D1 receptors are also prevalent in frontal cortex, whereas DARPP-32 protein is far more abundant in striatum (25, 26), making this SNP a better candidate to test specific striatal-D1 receptor predictions.] The particular genetic polymorphism that we examined (rs907094) was recently shown to strongly modulate striatal activity and function, with no direct effect of this gene on any other brain region (25). We hypothesized that this genetic marker of striatal DA would predict the extent to which participants learn to make decisions that had been probabilistically associated with positive outcomes. We also analyzed the C957T polymorphism within the DRD2 gene, which affects D2 mRNA translation and stability (29), and postsynaptic D2 receptor density in the striatum (30), without affecting presynaptic DA function (31). [This distinction may be critical: genetic modulation of presynaptic D2 function would presumably affect Go/reward learning during DA bursts (11), consistent with effects of another D2 polymorphism on reward-related brain activity (32).] We hypothesized that this genetic marker of postsynaptic striatal D2 function would predict the extent to which participants learn to avoid decisions that had been probabilistically associated with negative outcomes. Finally, we analyzed the functional Val158Met polymorphism within the COMT gene. This polymorphism is associated with individual difference in prefrontal DA, such that Met allele carriers have lower COMT enzyme activity and higher DA (23, 25, 33). We hypothesized that this genetic marker of prefrontal DA function would predict the extent to which participants maintain negative outcomes in working memory to quickly adjust their behavior on a trial-to-trial basis.
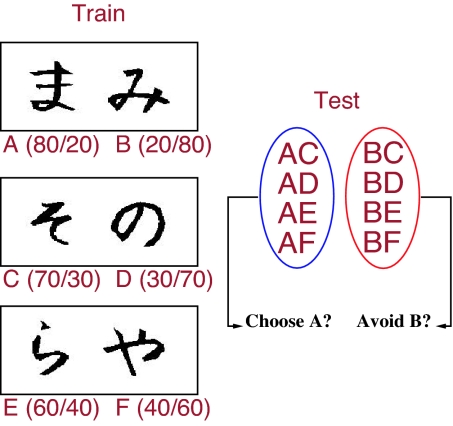
We administered a probabilistic selection task (4) that enables examination of all these behavioral measures within a single task. Three different stimulus pairs (AB, CD, and EF) are presented in random order, and participants have to learn to choose one of the two stimuli (Fig. 1). Feedback follows the choice to indicate whether it was correct or incorrect, but this feedback is probabilistic. In AB trials, a choice of stimulus A leads to correct (positive) feedback in 80% of AB trials, whereas a B choice leads to incorrect (negative) feedback in these trials (and vice versa for the remaining 20% of trials). CD and EF pairs are less reliable: stimulus C is correct in 70% of CD trials, E is correct in 60% of EF trials. Learning to choose A over B could be accomplished either by learning that A leads to positive feedback, or that B leads to negative feedback (or both). To evaluate the degree to which participants learned about the probabilistic outcomes of their decisions (both positive and negative), we subsequently tested them with novel combinations of stimulus pairs involving either an A or a B (each paired with a more neutral stimulus; no feedback was provided) (4). Positive-feedback learning is indicated by reliable choice of stimulus A in all test pairs in which it is present. Conversely, negative-feedback learning is indicated by reliable avoidance of stimulus B.
Fig. 1.
Probabilistic selection task. Example stimulus pairs, which minimize explicit verbal encoding by using Japanese Hiragana characters. Each pair is presented separately in different trials. Three different pairs are presented in random order; correct choices are determined probabilistically (percent positive/negative feedback shown in parentheses for each stimulus). A test (transfer) phase ensues in which stimuli A and B are repaired with all other more neutral stimuli; no feedback is provided in this test phase. Positive-reinforcement learning is assessed by choose-A accuracy; avoidance learning is assessed by avoid-B (4).
Prior studies revealed that Parkinson's patients, who have low striatal DA levels, were better at avoid-B than choose-A; DA medications reversed this bias as predicted by the models (4). In healthy participants, individual differences in positive and negative learning were accounted for by reinforcement-related brain activity thought to be linked to DA signals (34). We hypothesized that good choose-A performance would be associated with enhanced striatal DA/D1 efficacy, as reflected by the DARPP-32 gene. We further predicted that better avoid-B performance would be associated with striatal D2 receptor density, as indexed by the DRD2 gene (30).
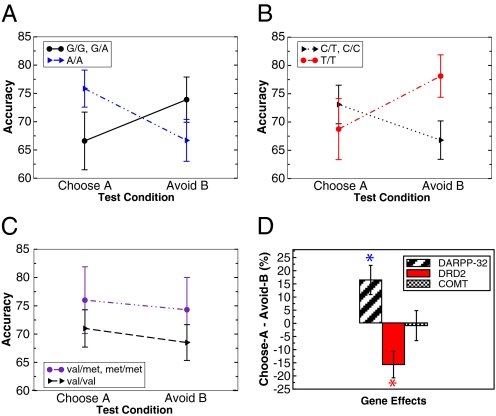
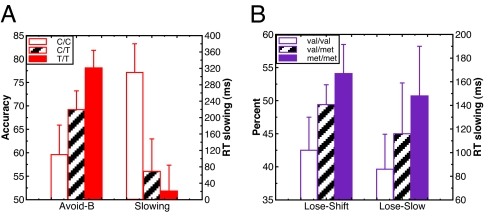
Results supported these predictions. Overall, participants were equally successful at choose-A and avoid-B pairs [F(1,68) = 0.9]. Nevertheless, there was an interaction between test pair condition and DARPP-32 genotype [F(1,67) = 4.1, P = 0.048], such that A/A homozygotes performed relatively better than G carriers at choose-A compared with avoid-B trials (Fig. 2A, Cohen's d = 0.5). The DRD2 gene also predicted relative choose-A versus avoid-B test performance, but in the opposite direction [F(1,67) = 7.6, P = 0.008, Cohen's d = 0.53]. In this case, T/T homozygotes, who have the highest D2 receptor availability (30), performed selectively better at avoid-B pairs (Fig. 2B; Cohen's d = 0.55). This effect was particularly evident by a gene–dose analysis (Fig. 4A), in which increasing numbers of T alleles were associated with better avoid-B performance [F(1,67) = 4.8, P = 0.03], with no effect on choose-A (P > 0.1). This DRD2/NoGo learning effect was further demonstrated at the level of reaction times [Fig. 4A and supporting information (SI) Table 1]. Both DARPP-32 and D2 genetic effects on Go and NoGo learning were found despite no effects on accuracy during the training phase (P > 0.25).
Fig. 2.
Go/NoGo generalizations. (A) DARPP32 gene. A/A homozygotes were relatively better than G carriers at choose-A relative to avoid-B performance. (B) DRD2 gene. T/T homozygotes were selectively better C carriers at avoiding the most negative stimulus B, with no DRD2 effect on choose-A. (C) COMT gene. No significant effects were observed. (D) Summary of within-subject positive–negative learning biases. COMT = Met carriers − Val/Val; DARPP-32 = A/A − G carriers; DRD2 = T/T − C carriers. Asterisks indicate significant (P < 0.05) effects of each gene on relative positive to negative learning. Error bars reflect standard error.
Fig. 4.
Gene–dose effects on two forms of avoidance learning. (A) Avoid-B performance, DRD2 gene–dose effect. Individuals with more T alleles performed better, and were relatively faster, at avoid-B test pairs. RTs are assessed on correct trials, and slowing is measured by subtracting choose-A from avoid-B RTs. (B) Trial-to-trial switching, COMT gene–dose effect. Individuals with more met alleles showed a greater propensity to switch after negative feedback (particularly in early training trials, shown here). These subjects also showed more slowing of reaction times in these trials (previous trial positive-feedback subtracted from previous trial negative-feedback RTs). Error bars reflect standard error.
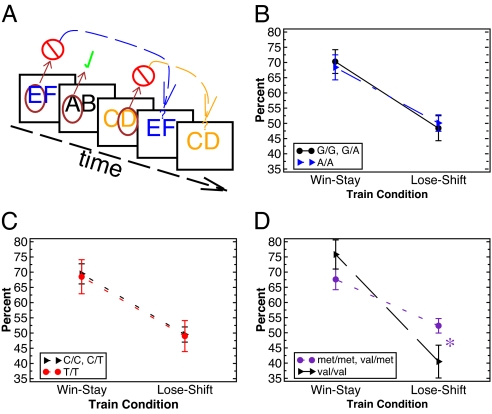
In contrast to these striatal DA-related genes, the COMT gene (number of Met alleles) was not predictive of either choose-A or avoid-B performance (Fig. 2C), or their relative reaction times (all P > 0.25). Motivated by the neurocomputational models described above, we predicted that the COMT gene should instead be associated with trial-to-trial adaptation after negative outcomes. To test this idea, we analyzed performance during the training phase of the task. We kept track of trials in which participants selected a particular stimulus (e.g., E in an EF trial), got negative feedback, and used this feedback to avoid selecting the same stimulus in the next trial in which it appeared (Fig. 3A). Note that this ability requires holding the negative outcome experience in mind over the course of multiple intervening trials (e.g., AB and CD) that could occur before the next EF occurrence. Thus our assessment of trial-to-trial negative feedback switching depends on robust maintenance of reinforcement values in working memory in the face of ongoing processing, a function widely thought to be modulated by PFC DA (17, 18).
Fig. 3.
Trial-to-trial adjustments. (A) Illustration of trial-to-trial switching assessment for an example stimulus train. Brown circles indicate the selected stimulus; check marks and cross-out symbols represent the feedback (positive or negative) received. In the example shown, lose–shift performance would involve a switch from selecting E in the first EF trial to F in the second EF trial and similarly for CD. Note that this shifting depends on maintaining the negative outcome value of a particular stimulus context (e.g., EF) in the face of intervening trials (AB and CD). Win–stay performance is assessed similarly (repeated choice of the A stimulus in the next AB trial; not shown). (B) DARPP32 effects on win–stay and lose–shift performance. (C) DRD2 effects. (D) COMT effects. Val/Val homozygotes were significantly less likely than others to switch their responses after negative feedback (lose–shift). Results are shown in early training trials (first 5 trials of each type, 15 trials total), when negative feedback is maximally informative.
Overall accuracy in the training phase did not differ across COMT genotypes [F(1,66) = 0.1]. Nevertheless, Val/Val homozygotes, who have the lowest PFC DA (33), were less likely than Met carriers to switch responses after negative outcomes on a trial-to-trial basis [lose–shift; F(1,66) = 5.2, P = 0.026, Cohen's d = 0.6]; there was no COMT effect on win-stay performance [F(1,66) = 1.5, not significant]. The condition by genotype interaction was significant [F(1,66) = 4.6, P = 0.037]. There was also a gene–dose effect such that increasing Met allele expression was associated with enhanced lose–shift performance [Fig. 4B; F(1,66) = 7.0, P = 0.01], again with no effect on win–stay [F(1,66 = 0.7]. These COMT differences were significant even in the very first 5 training trials of each type [15 trials total, F(1,66) = 4.6, P = 0.03; Fig. 3D] and reduced as training progressed, as indicated by a marginal genotype by training trial interaction [F(1,66) = 3.5, P = 0.067]. This latter effect was due to a decrease in lose–shift behavior with increasing trials as individual negative feedback experiences became less informative (SI Fig. 6). These COMT effects were also observed at the level of reaction times: Met carriers were more likely to slow responses after negative feedback [lose–slow; Fig. 4B; F(1,66) = 4.2, P = 0.04]. Finally, there was no effect of DRD2 or DARPP-32 genotypes on trial-to-trial switching or slowing (Fig. 3 B and C; P > 0.2).
Computational Model
We further investigated the above genetic effects using computational reinforcement “Q-learning” algorithms (35) (see SI Text for detailed methods). These simulations attempt to embody some of the core computational principles derived from the biological neural network implementations (6, 13) by using more analytically tractable equations and a minimal number of parameters. Moreover, unlike neural network models (which have many more parameters), these algorithms can quantitatively fit individual subjects' trial-to-trial data by generating parameters that are not directly observable in the behavioral data (36). Individual differences in these learning parameters have thus far been considered nuisance variables; here, we investigate whether these differences can be accounted for in a principled way by genetic mutations.
Because our neural models suggest separate mechanisms for positive- and negative-feedback learning, we fit behavioral data using separate learning-rate parameters αG and αL for gains and losses (see ref. 37 for related methods). Specifically, we compute a Q value for selecting each stimulus i during trial t, such that the value of the chosen stimulus is modified by reinforcement feedback:
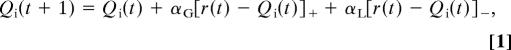 |
where r(t) = 1 for positive and 0 for negative feedback, and the learning rate applied is either αG or αL depending on whether the outcome is better or worse than expected, respectively. Choice behavior was then modeled by using a “softmax” logistic function (35), with inverse gain parameter β, such that the probability of choosing one stimulus over another (e.g., A over B) was computed as:
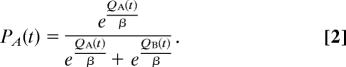 |
The same equation applies for other trial-types, replacing A and B with C, D, E, or F, as appropriate. Each participant‘s trial-by-trial choices were fit with three free parameters, αG, αL, and β, which were selected to maximize fit (likelihood) of each participant's trial-by-trial sequence of choices in the training phase of the task (SI Text, Eq. 7, and SI Tables 2–5).
We reasoned that the best fitting parameters to participants' training data would be forced to accommodate rapid adaptations in response to recent reinforcement experiences. If the COMT gene modulates the ability to rapidly adjust behavior as a function of working memory for recent losses, then this gene should predict individual differences in αL values. Indeed, there was a gene–dose effect of COMT, such that increasing Met allele expression was associated with higher αL values [F(1,66) = 4.6, P = 0.035; Fig. 5C and SI Fig. 7], and relatively higher αL than αG [F(1,66) = 4.1, P = 0.047]. There was no effect of the D2 or DARPP-32 genes on these measures (P > 0.2).
Fig. 5.
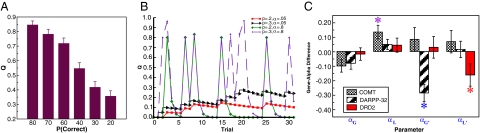
Q-learning simulations. (A) Mean Q values for each stimulus after the training phase using the gain-loss Q model, showing a significant descending trend with reinforcement probability. (B) Q values: probability and learning rate. Shown are simulations to illustrate how learning rates affect the ability to represent and compare probabilistic Q values across time. High learning rates (e.g., 0.8) result in trial-to-trial fluctuations and thereby capture recency (e.g., working memory) effects, whereas low learning rates (e.g., 0.05) enable a more stable estimate of values that can then be compared for subtle probabilistic discriminations. In this case, discriminating between two negative stimuli (20% and 30% reward probability) is accomplished reliably by a low learning rate. (C) Summary plot showing triple dissociation of three genetic polymorphisms on Q learning parameters. Difference scores are shown for each parameter and gene group (COMT: Met/Met − Val/Val, DARPP-32: A/A − G carriers; DRD2: T/T − C/C). Increasing COMT Met allele expression was associated with higher αL, increasing DARPP-32 A allele expression was associated with lower α′G, increasing DRD2 T allele expression was associated with lower α′L. Asterisks indicate P <.05; error bars reflect standard error. Raw parameters for each group (and associated gene–dose effects) are shown in SI Fig. 7.
Prior work suggests that basal ganglia (BG) and PFC systems learn in parallel and that the two systems can both cooperate and compete for behavioral control (13, 14). Whereas the PFC guides rapid behavioral adjustments during training, we hypothesized that the BG system may be more adept at discriminating between subtly different probabilistic reinforcement values at test (participants were told to use “gut-level intuition” for these test choices). If this is the case, the learning rates found by fitting participants' trial-to-trial training choices may reflect rapid adaptation to changing outcomes, but would conceal the learning rates of the BG system (which would observably affect behavior at test). To address this issue, we ran a parallel Q-learning simulation, in an attempt to uncover the learning rates of a second Q′ system that learns during training but exerts control over behavior during the test phase. The Q′ value updating equation is similar to that above:
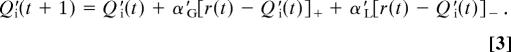 |
Rather than trying to fit trial-by-trial sequential data, we instead optimized α′G and α′L values to select those that produce final Q′ values (i.e., at the end of training), which correspond best to participants' subsequent test choices. Thus, the goal was to estimate the learning rates of the system that had integrated reinforcement probabilities during training and could then discriminate between them at test. Furthermore, because there was no feedback during the test phase, there were no trial-to-trial adjustments that would otherwise be captured by these simulations (see SI Text for additional justification and alternative models). To compute the probability of choosing a given stimulus in the test phase, we modify Eq. 2 to reflect a softmax comparison of final accumulated Q′ values, and assume that these Q′ values do not change across the test phase (given the lack of feedback). For example, if the subject chose stimulus A in test pair AC, we compute the probability that the model would make that same choice, given final Q′ values:
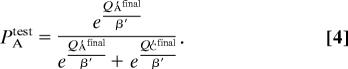 |
To find each subject's best fitting α′G and α′L parameters, we applied Eq. 3 repeatedly for different parameter combinations and selected those producing final Q′ values that maximize fit to their test phase choices. We then examined whether the parameters of this Q′ system could predict choose-A and avoid-B test choices. Indeed, we found that increasing choose-A performance was associated with smaller α′G values [F(1,66) = 8.3, P = 0.005], whereas increasing avoid-B performance was associated with smaller α′L [F(1,66) = 4.4, P = 0.04]. There was no relationship between α′G and avoid-B performance or α′L and choose-A (P > 0.15).
The negative relationship among α′G, α′L, and probabilistic test performance suggests that the ability to discriminate between subtly different probabilistic values requires a low learning rate, as opposed to the above-described effects of αL, where increasing values were associated with rapid adaptations. This fundamental tradeoff between working memory recency and probabilistic integration is shown by Q learning simulations (Fig. 5B). Larger learning rates lead to high sensitivity to recent outcomes, whereas lower learning rates support integration over multiple trials (37) (see also ref. 38 for a similar tradeoff in the episodic memory domain). Thus, the system in control over behavior at test is better off having had a lower learning rate during training, so that it is not overly influenced by just the most recent reinforcement experiences.
Having shown that lower α′G and α′L are associated with better test phase generalization, we hypothesized that these learning rate differences would be accounted for by the striatal genes. Notably, both DARPP-32 and D2 genes modulated these α′ parameters. For DARPP-32, A/A homozygotes had substantially lower α′G parameters than the other groups [F(1,67) = 12.1, P < 0.001; Fig. 5C]; there was no effect of this gene on α′L [F(1,67) = 0.04]. This effect was selective to slow reward integration and not trial-by-trial adaptation, as revealed by a significant interaction between DARPP-32 genotype and α′G vs. αG [F(1,67) = 3.9, P = 0.05]. For DRD2, there was a gene–dose effect of increasing T allele expression, leading to relatively smaller α′L than αL values [F(1,67) = 5.7, P = 0.02; Fig. 5D and SI Fig. 7]. Again, this D2 gene effect was selective; it was not observed for α′G (by itself or relative to αG), and there were no COMT effects on any of these measures (all P > 0.15). Finally, there was no effect of any gene on β or β′ (P > 0.1). (All of the reported effects held regardless of whether we fit a new β′ parameter for each subject in the Q′ simulations, or whether we fixed it to be the same as β found in the training simulations. Thus the computational findings are robust.) In sum, smaller α′G values in DARPP-32 A/A homozygotes, and smaller α′L values in D2 T/T homozygotes, demonstrate that these genes modulate slow integration of positive and negative outcomes.
Discussion
These collective findings provide evidence for three distinct dopaminergic mechanisms in reinforcement learning. Each of the gene/behavior effects were accounted for by independent genetic modulation of reinforcement learning parameters in a computational model. First, the DARPP-32 gene predicted the ability to choose stimuli that had been probabilistically associated with positive feedback [Go learning (4, 6)]. Computational analyses show that this gene modulates the learning rate that enables discrimination between probabilistic reward values (Fig. 5C and SI Fig. 8).
In contrast, enhanced DRD2 genetic function predicted the ability to avoid the probabilistically most negative stimuli, accompanied by associated changes in α′L values. This finding confirms a specific neurocomputational prediction that postsynaptic striatal D2 receptors are critical for integrating and learning from low DA levels during negative outcomes (4, 6, 7). Neural evidence for this claim comes from studies showing that postsynaptic D2 receptor blockade (simulating the lack of DA during dips) enhances corticostriatal long-term potentiation (39), associated with NoGo learning in the models (6). Nevertheless, this claim is controversial. Several theorists argue that although DA bursts support positive-reinforcement learning, the firing rate changes associated with DA dips do not have sufficient dynamic range to be functionally effective (2, 40). The present findings suggest that striatal D2 receptors are indeed effective in avoidance learning, potentially because of their enhanced sensitivity to low DA levels and their potent role in synaptic plasticity (11, 41, 42). This function seems to depend highly on DRD2 genotype and associated striatal D2 receptor density.
Overall, the finding that lower α′G and α′L values were associated with better test phase performance supports the notion that slow feedback integration is necessary for generalization of probabilistic positive and negative reinforcement values. We posit that this slow integration is performed by the “habit-learning” BG system (15), consistent with the observed striatal genetic effects. However, these findings raise the question of how enhanced DARPP-32 or D2 function might lower learning rates, mechanistically. We offer two possible resolutions to this conundrum. First, detailed biophysical models suggest that DARPP-32 contributes to synaptic plasticity precisely by integrating DA bursts over extended periods of time (28). Participants with enhanced DARPP-32 function (because of genetic or other factors) may therefore be better at performing such integration, which would be fit by lower learning rates (37). Second, the computational approach used here is highly abstract (to minimize the number of parameters needed to fit behavioral data); the derived learning rates cannot capture those of individual synapses and, instead, reflect adaptation of the entire behavioral system. Thus, although we argue that one is better off relying on BG (rather than PFC) when choosing among long-term probabilistic reward values, the extent to which individuals actually do this can itself vary. We speculate that those with enhanced DARPP-32 or D2 genetic factors may rely relatively more on BG, and less on PFC, in the test phase of our task, leading to lowered effective learning rates. In contrast, those with poorer striatal function may be overly influenced by only their most recent reinforcement experiences (at the end of the training phase), a strategy that would be fit by higher learning rates. This interpretation is in accord with suggestions that the degree to which striatal and prefrontal systems contribute to behavior is governed by the relative certainty of each system's predictions (14). Those with enhanced striatal function would have greater certainty in the predictions of the BG system, leading to greater BG than PFC recruitment.
This line of reasoning is further supported by our COMT analysis. Increasing COMT Met allele expression was associated with greater trial-to-trial adjustments after a single instance of negative feedback; this behavior was fit by higher αL values in the computational analysis. Notably, COMT Met allele expression is associated with elevated DA levels in PFC (33), with little to no COMT effect in striatum (25, 43). These prefrontal DA differences may be particularly evident after negative events, which have been associated with temporally extended DA elevations in PFC (and usually not striatum) (20, 21). Thus, we suggest that the COMT gene modulates the elevation of frontal DA levels after negative events. In turn, these DA levels stabilize frontal working memory representations (17), including those encoding recent reinforcement values, enabling adaptive behavior in the face of changing reinforcement contingencies (13). Animal studies support this notion, showing that PFC DA elevations, including those induced by COMT manipulation, enhance behavioral shifting (23, 22). Moreover, schizophrenic patients, who consistently show impaired prefrontal function linked to COMT (33), are impaired at rapid lose–shift performance in the same task used here, while showing intact probabilistic NoGo learning (44).
At first glance, our interpretation conflicts with recent physiological data showing that striatal learning rates are actually faster than those in PFC (45). Those data seem to challenge the widely held notion that the striatum supports gradual habit-based learning (16). Our neural models provide a possible resolution to this issue, whereby initial BG/DA learning is required for later habits to be “ingrained” in the more slowly learning corticcortical projections (6, 13). The critical difference is that here, we view the high learning rate PFC system to reflect not fast synaptic modification but, rather, the ability to actively maintain reinforcement values in working memory, which can then be immediately applied to affect behavior. This distinction has been explored in computational models, in which the synaptic learning of prefrontal abstract representations is very slow but that, once established, can be actively maintained and rapidly applied to modify behavior by top-down control (46). It is the latter function that we attribute to the fast PFC system here.
Finally, although prior studies have linked each of the genetic polymorphisms with the neural function of interest, we cannot absolutely discount the possibility that our observed effects are mediated by different neural mechanisms than those posited here. We simply note that the polymorphisms examined were specifically selected based on a priori theoretical and converging empirical work. Future research is required to confirm that the genetic effects are accompanied by brain-related changes in the context of the same behaviors here. Furthermore, although our moderately large sample size (n = 69) enabled examination of independent genetic effects on model parameters and behavioral measures of interest, it is in principle possible that these genetic effects also interact with each other. Preliminary analysis revealed no such gene–gene interactions, but larger sample sizes may be required to reliably exclude this possibility.
In addition to corroborating a neurocomputational account of learning in basal ganglia and frontal cortex, our findings also lend insight into the genetic basis for learning differences. We limited our analysis to just three polymorphisms that have been clearly associated with the dopaminergic measures of interest in striatum and PFC, motivated by theoretical modeling predictions. We found relatively large effects of normal genetic variation in DA function on our cognitive measures. These findings provide evidence that when cognitive tasks and candidate genes are chosen based on formal theories of brain function, substantial genetic components can be revealed (47). Moreover, understanding how dopaminergic variation affects learning and decision making processes may have substantial implications for patient populations such as Parkinson's disease, attention-deficit hyperactivity disorder, and schizophrenia. Thus, further research into the functions of DA in the basal ganglia and PFC should provide important insights into improving human cognition and learning.
Methods
Sample.
Our sample was 69 healthy participants (30 females, 39 males), between the ages of 18 and 35 (median = 21). The vast majority of participants were white, with three participants categorizing themselves as “more than one race.” We were unable to obtain COMT genotypes for one subject. The breakdown of COMT genotypes was 17:32:19 (Val/Val:Val/Met:Met/Met). The breakdown of C957T genotypes was 13:38:18 (C/C:C/T:T/T). The breakdown of DARPP32 genotypes was 4:25:40 (G/G:G/A:A/A). Because of the low number of participants in the G/G group, we compared A/A (n = 40) to G/A and G/G carriers combined (n = 29). The number of T/Met alleles in the DRD2/COMT SNPs were not correlated [r (68) = 0.06, P = 0.7), and the groupings for DARPP-32 (A/A vs. others) did not correlate with either of the other SNPs (P > 0.15). Thus, the three genetic effects reported above were independent from one another.
Procedures.
Procedures were approved by the University of Colorado Human Research Committee. Detailed procedures for the PS task have been described (4, 11) and are reiterated in SI Text.
Data Analysis.
We performed general linear model regressions to test comparisons of interest, using the number of Met/T alleles as a continuous variable; for details, please see SI Text.
Genotyping.
DNA was collected by use of buccal swabs. Subjects swabbed their cheeks with three cotton swabs, followed by a rinse of the mouth with water, after which all contents were placed in a 50-ml sterile conical tube and stored at 4°C until extraction. Genomic DNA was extracted from buccal cells by using a modification of published procedures (48). Before SNP analyses, the concentration of genomic DNA was adjusted to 20 ng/μl. SNP analyses were performed by using an ABI PRISM 7500 instrument (Applied Biosystems, Foster City, CA) using TaqMan chemistry.
Supplementary Material
Acknowledgments
We thank Nathaniel Daw and Mike Cohen for helpful comments and Casey DeBuse, Rex Villanueva, and Breanna Miller for help with data collection and DNA extraction. This work was supported by National Institute on Drug Abuse Grant DA022630 and National Institute on Alcohol Abuse and Alcoholism Grant AA012238.
Abbreviations
- DA
dopamine
- PFC
prefrontal cortex.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706111104/DC1.
References
- 1.Schultz W. Neuron. 2002;36:241–263. doi: 10.1016/s0896-6273(02)00967-4. [DOI] [PubMed] [Google Scholar]
- 2.Bayer HM, Glimcher PW. Neuron. 2005;47:129–141. doi: 10.1016/j.neuron.2005.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satoh T, Nakai S, Sato T, Kimura M. J Neurosci. 2003;23:9913–9923. doi: 10.1523/JNEUROSCI.23-30-09913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank MJ, Seeberger LC, O'Reilly RC. Science. 2004;306:1940–1943. doi: 10.1126/science.1102941. [DOI] [PubMed] [Google Scholar]
- 5.Ungless MA, Magill PJ, Bolam JP. Science. 2004;303:583–586. doi: 10.1126/science.1093360. [DOI] [PubMed] [Google Scholar]
- 6.Frank MJ. J Cognit Neurosci. 2005;17:51–72. doi: 10.1162/0898929052880093. [DOI] [PubMed] [Google Scholar]
- 7.Brown JW, Bullock D, Grossberg S. Neural Networks. 2004;17:471–510. doi: 10.1016/j.neunet.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Samejima K, Ueda Y, Doya K, Kimura Science. 2005;310:1337–1340. doi: 10.1126/science.1115270. [DOI] [PubMed] [Google Scholar]
- 9.Robertson GS, Vincent SR, Fibiger HC. Neuroscience. 1992;49:285–296. doi: 10.1016/0306-4522(92)90096-k. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura K, Hikosaka O. J Neurosci. 2006;26:5360–5369. doi: 10.1523/JNEUROSCI.4853-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank MJ, O'Reilly RC. Behav Neurosci. 2006;120:497–517. doi: 10.1037/0735-7044.120.3.497. [DOI] [PubMed] [Google Scholar]
- 12.Cools R, Altamirano L, D'Esposito M. Neuropsychologia. 2006;44:1663–1673. doi: 10.1016/j.neuropsychologia.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 13.Frank MJ, Claus ED. Psychol Rev. 2006;113:300–326. doi: 10.1037/0033-295X.113.2.300. [DOI] [PubMed] [Google Scholar]
- 14.Daw ND, Niv Y, Dayan P. Nat Neurosci. 2005;8:1704–1711. doi: 10.1038/nn1560. [DOI] [PubMed] [Google Scholar]
- 15.Jog MS, Kubota Y, Connolly CI, Hillegaart V, Graybiel AM. Science. 1999;286:1745–1749. doi: 10.1126/science.286.5445.1745. [DOI] [PubMed] [Google Scholar]
- 16.Yin HH, Knowlton BJ, Balleine BW. Eur J Neurosci. 2004;19:181–189. doi: 10.1111/j.1460-9568.2004.03095.x. [DOI] [PubMed] [Google Scholar]
- 17.Durstewitz D, Seamans JK, Sejnowski TJ. J Neurophysiol. 2000;83:1733–1750. doi: 10.1152/jn.2000.83.3.1733. [DOI] [PubMed] [Google Scholar]
- 18.Cohen JD, Braver TS, Brown JW. Curr Opin Neurobiol. 2002;12:223–229. doi: 10.1016/s0959-4388(02)00314-8. [DOI] [PubMed] [Google Scholar]
- 19.Miller EK, Cohen JD. Annu Rev Neurosci. 2001;24:167–202. doi: 10.1146/annurev.neuro.24.1.167. [DOI] [PubMed] [Google Scholar]
- 20.Di Chiara G, Loddo P, Tanda G. Biol Psychiatry. 1999;46:1624–1633. doi: 10.1016/s0006-3223(99)00236-x. [DOI] [PubMed] [Google Scholar]
- 21.Jackson ME, Moghaddam B. J Neurochem. 2004;88:1327–1334. doi: 10.1046/j.1471-4159.2003.02205.x. [DOI] [PubMed] [Google Scholar]
- 22.Stefani MR, Moghaddam B. J Neurosci. 2006;26:8810–8818. doi: 10.1523/JNEUROSCI.1656-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tunbridge EM, Bannerman DM, Sharp T, Harrison PJ. J Neurosci. 2004;24:5331–5335. doi: 10.1523/JNEUROSCI.1124-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walaas SI, Aswad D, Greendard P. Nature. 1983;301:69–71. doi: 10.1038/301069a0. [DOI] [PubMed] [Google Scholar]
- 25.Meyer-Lindenberg A, Straub RE, Lipska BK, Verchinski BA, Goldberg T, Callicott JH, Egan MF, Huffaker SS, Mattay VS, Kolachana B, et al. J Clin Invest. 2007;117:672–682. doi: 10.1172/JCI30413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouimet CC, Miller PE, Hemmings HCJ, Walaass SI, Greengard P. J Neurosci. 1984;4:111–124. doi: 10.1523/JNEUROSCI.04-01-00111.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calabresi P, Gubellini P, Centonze D, Picconi B, Bernardi G, Chergui K, Svenningsson P, Fienberg AA, Greengard P. J Neurosci. 2000;20:8443–8451. doi: 10.1523/JNEUROSCI.20-22-08443.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindskog M, Kim M, Wikström MA, Blackwell KT, Kotaleski JH. PLoS Comput Biol. 2006;2:1045–1060. doi: 10.1371/journal.pcbi.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, Gejman PV. Hum Mol Genet. 2003;12:205–216. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- 30.Hirvonen M, Laakso A, Nagren K, Rinne J, Pohjalainen T, Hietala J. Mol Psychiatry. 2005;10:889. doi: 10.1038/sj.mp.4001561. [DOI] [PubMed] [Google Scholar]
- 31.Laakso A, Pohjalainen T, Bergman J, Kajander J, Haaparanta M, Solin O, Syvalahti E, Hietala J. Pharmacogenet Genomics. 2005;15:387–391. doi: 10.1097/01213011-200506000-00003. [DOI] [PubMed] [Google Scholar]
- 32.Cohen MX, Young J, Baek J-M, Kessler C, Ranganath C. Brain Res Cogn Brain Res. 2005;25:851–861. doi: 10.1016/j.cogbrainres.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 33.Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, Goldman D, Weinberger D. Proc Natl Acad Sci USA. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frank MJ, Woroch BS, Curran T. Neuron. 2005;47:495–501. doi: 10.1016/j.neuron.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 35.Sutton RS, Barto AG. Reinforcement Learning: An Introduction. Cambridge, MA: MIT Press; 1998. [Google Scholar]
- 36.Daw ND, O'Doherty JP, Dayan P, Seymour B, Dolan RJ. Nature. 2006;441:876–879. doi: 10.1038/nature04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yechiam E, Busemeyer JR, Stout JC, Bechara A. Psychol Sci. 2005;16:973–978. doi: 10.1111/j.1467-9280.2005.01646.x. [DOI] [PubMed] [Google Scholar]
- 38.McClelland JL, McNaughton BL, O'Reilly RC. Psychol Rev. 1995;102:419–457. doi: 10.1037/0033-295X.102.3.419. [DOI] [PubMed] [Google Scholar]
- 39.Centonze D, Usiello A, Costa C, Picconi B, Erbs E, Bernardi G, Borrelli E, Calabresi P. J Neurosci. 2004;24:8214–8222. doi: 10.1523/JNEUROSCI.1274-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daw ND, Kakade S, Dayan P. Neural Networks. 2002;15:603–616. doi: 10.1016/s0893-6080(02)00052-7. [DOI] [PubMed] [Google Scholar]
- 41.Creese I, Sibley DR, Hamblin MW, Leff SE. Annu Rev Neurosci. 1983;6:43–71. doi: 10.1146/annurev.ne.06.030183.000355. [DOI] [PubMed] [Google Scholar]
- 42.Kreitzer AC, Malenka RC. Nature. 2007;445:643–647. doi: 10.1038/nature05506. [DOI] [PubMed] [Google Scholar]
- 43.Gogos JA, Morgan M, Luine V, Santha M, Ogawa S, Pfaff D, Karayiorgou M. Proc Natl Acad Sci USA. 1998;95:9991–9996. doi: 10.1073/pnas.95.17.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waltz JA, Frank MJ, Robinson BM, Gold JM. Biol Psychiatry. 2007;62:756–764. doi: 10.1016/j.biopsych.2006.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pasupathy A, Miller EK. Nature. 2005;433:873–876. doi: 10.1038/nature03287. [DOI] [PubMed] [Google Scholar]
- 46.Rougier NP, Noelle D, Braver TS, Cohen JD, O'Reilly RC. Proc Natl Acad Sci USA. 2005;102:7338–7343. doi: 10.1073/pnas.0502455102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parasuraman R, Greenwood PM, Kumar R, Fossella J. Psychol Sci. 2005;16:200–207. doi: 10.1111/j.0956-7976.2005.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker A, Najarian D, White D, Jaffe J, Kanetsky P, Rebbeck T. Environ Health Perspect. 1999;107:517–520. doi: 10.1289/ehp.99107517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.