Abstract
Emerging evidence over the past decade indicates a central role for transcription factors in the embryonic development of pancreatic islets and the consequent maintenance of normal glucose homeostasis. Pancreatic and duodenal homeobox 1 (Pdx1) is the best studied and perhaps most important of these factors. Whereas deletion or inactivating mutations of the Pdx1 gene causes whole pancreas agenesis in both mice and humans, even haploinsufficiency of the gene or alterations in its expression in mature islet cells causes substantial impairments in glucose tolerance and the development of a late-onset form of diabetes known as maturity onset diabetes of the young. The study of Pdx1 has revealed crucial phenotypic interrelationships of the varied cell types within the pancreas, particularly as these impinge upon cellular differentiation in the embryo and neogenesis and regeneration in the adult. In this review, we describe the actions of Pdx1 in the developing and mature pancreas and attempt to unify these actions with its known roles in modulating transcriptional complex formation and chromatin structure at the molecular genetic level.
Keywords: Pancreas, insulin, transcription, diabetes, islet
Introduction
The β cells of the pancreatic islets of Langerhans are solely responsible for the transcription, synthesis, and release of insulin in response to elevations in extracellular glucose concentration. Although β cells comprise 70–80% of islet mass, islets themselves comprise only about 1% of total pancreatic mass [1]; this latter feature explains, in part, the relatively poor functional reserve of β cells and the consequent high prevalence of diabetes mellitus in many countries. Diabetes mellitus is a metabolic disorder characterized by hyperglycemia and susceptibility to a host of micro- and macro-vascular complications, including cardiovascular disease, kidney disease, retinal disease, and nerve disease. The loss or impairment of β cell function appears to underlie virtually all forms of diabetes mellitus. Recently, much of the molecular and genetic research in the field of diabetes has focused upon ways to restore the number or function of β cells as a means of reversing the metabolic consequences of insulin deficiency. Both cell-intrinsic components (e.g. transcription factors) and cell-extrinsic components (e.g. signaling growth factors) have been implicated in the formation and maintenance of β cell mass. One cell intrinsic factor, pancreatic and duodenal homeobox 1 (Pdx1), has received attention over the years as perhaps the most crucial factor in the β cell.
Pdx1, previously referred to as STF1, IDX1, IPF1, IUF1, was identified as a β cell-specific transactivator of the insulin and somatostatin genes that appeared to be the mammalian ortholog of the endoderm-specific Xenopus laevis XlHbox8 homeobox protein [2–9]. As with other transcription factors, Pdx1 appears to function by binding to specific DNA sequences within the 5’ enhancer regions of target genes, and subsequently recruiting complexes of proteins that ultimately regulate the rate of transcription of that gene. The gene encoding Pdx1 (Pdx1) is a central member of a mammalian Parahox gene cluster on mouse chromosome 5. This cluster is so-named because it represents a group of developmentally important genes in mammals that is found outside the classical Hox (homeobox) cluster of genes [10]. The Parahox cluster is comprised of three genes, Gsh1, Pdx1, and Cdx2/3, all of which are expressed in specific pancreatic cell types [11]. In this regard, the importance of Pdx1 in the pancreas is emphasized by the near-absence of pancreas formation in Pdx1-null mice [12–14]. This dramatic phenotype also occurs in humans with homozygous mutations of the human ortholog of the Pdx1 gene (known as Ipf1) [15,16], thereby underscoring the relevance of this factor to human pancreas development. Pdx1 also appears to be crucial for the function of the mature β cell. Heterozygous missense and frameshift mutations of the Ipf1 gene in humans (while not impairing pancreas formation) result in defective insulin secretion and the development of a form of diabetes known as maturity onset diabetes of the young 4 (MODY4) [17–21]. Similarly, studies of animal models of insulin resistance suggest that the down regulation of pdx1 expression in the β cell may underlie the pathogenesis of β cell failure and type 2 diabetes [22–24]. Thus, Pdx1 plays both a broad role in pancreas development and a much more specific role in β cell function in the adult mammal. That Pdx1 can achieve these differential actions may arise from its ability to associate in protein complexes that have varying transcriptional outcomes. This review describes the role of Pdx1 in the differentiation of specific cell types in the developing pancreas, and how Pdx1 mechanistically achieves its transcriptional effects at target genes to maintain normal glucose homeostasis.
Role of Pdx1 in Pancreas Development
Pdx1 and early pancreas development
The mature pancreas is composed of endocrine (hormone-secreting), acinar (hydrolytic enzyme-secreting), and duct cells. Whereas the exocrine cells are scattered throughout the pancreas and form the bulk of pancreatic mass, the endocrine cells occur in circumscribed clusters known as islets of Langerhans. Notwithstanding the disparity in organization and function of the different cellular types, all cells within the pancreas appear to arise from Pdx1-expressing precursors during development [25,26]. During embryogenesis, the pancreas emanates initially as two cellular extensions of endodermal origin located dorsally and ventrally to the primitive gut tube at approximately embryonic day 9.0–9.5 (E9.0–9.5) in the mouse [1]. During this early period of pancreas development, often referred to as the “primary transition,” there is branching of the epithelial cells within the developing buds, a process that is regulated by signals from the notochord and the surrounding mesenchyme (see Fig. 1).
Figure 1. Pdx1 expression and gross pancreatic morphology during mouse embryonic development.

Shown are schematic representations of the developing mouse pancreas at different embryonic ages. At approximately embryonic day 8.5 (E8.5) signals from the notochord, including fibroblast growth factor 2 (FGF2) and activin, repress expression of hedgehog proteins (HH) in the region of the gut endoderm that is destined to develop into pancreatic buds (dark blue). Initial pancreatic bud outgrowth occurs between E9–9.5 (primary transition), and subsequent branching morphogenesis is dependent upon the expression of Pdx1 (see text). During the secondary transition (between E11–15), there is differentiation of islet cell types (e.g. α and β cells) and acinar cells that is dependent upon the action of Pdx1. After E15, the two buds fuse to form the single organ seen in adult mammals. In adults, Pdx1 expression persists at highest levels in islets (particularly β cells) and at low levels in duct cells and acinar cells.
The notochord factors fibroblast growth factor 2 (Fgf2) and activin (transforming growth factor (TGF)-β signaling molecule) repress endodermal sonic hedgehog (Shh) protein in the pancreas-specific endoderm, thus permitting expression of pancreatic genes such as Pdx1 [27,28]. Studies in chicken and mice have demonstrated that ectopic expression of Shh during early development results in a loss of Pdx1 expression (and other pancreatic β-cell markers), and in the transformation of pancreatic mesenchyme into duodenal mesenchyme [29] [30]. In addition, continued expression of hedgehog proteins (Shh and Indian hedgehog, Ihh) after initial pancreas formation (at about E12.5 in the mouse) result in reductions in pancreatic mass, including both acinar and endocrine tissues [31]. These findings point to an important role for inhibition of hedgehog signaling in early pancreas development. At least some of the phenotypes described with early ectopic hedgehog signaling can be explained by the consequent absence of Pdx1 expression in the pancreatic endoderm. In this regard, in studies of Pdx1−/− mice, the initial buds of the pancreas form but subsequent branching and morphogenesis of these buds is arrested [12]. By contrast, in the mature pancreas and related islet-derived cell lines, ongoing expression of hedgehog proteins appear to be important for the maintenance of both Pdx1 and insulin expression [32,33]. These findings suggest a complex interplay between hedgehog proteins and Pdx1 expression, and that regulation of Pdx1 gene expression appears to be differentially regulated in the developing pancreas vs. the mature pancreas.
Apart from its expression throughout the early pancreatic endoderm, Pdx1 is also expressed in regions of the developing stomach and duodenum. In addition to loss of pancreas formation, Pdx1−/− mice also exhibit loss of Brunner’s glands and deficiency of enteroendocrine differentiation in the stomach and duodenum [13,34,35]. Factors controlling the activation of the Pdx1 gene during early embryogenesis have been identified using both mouse genetic and biochemical approaches. A phylogenetically-conserved region in mammals at ~2 kilo-basepairs (kb) upstream of the transcriptional start site of the mouse Pdx1 gene known as Area I-II-III (or PH-1, -2, and -3 in humans) harbors binding sites for endodermal and pancreas-specific transcription factors (Foxa2, Ptf1a, HNF1α, MafA, HNF6, Pax6, and Pdx1 itself) [36–40]. Area III appears to control the pancreas-wide expression of Pdx1 during early bud formation [41], whereas Areas I and II appear to control islet-specific expression in later development and adulthood [42–44]. Homozygous deletion of the entire Area I-II-III region in mice results in agenesis of the ventral pancreatic bud and hypoplasia of the dorsal bud, but without abnormalities in the stomach or duodenum [45,46]. These findings suggest that the spatial regulation of Pdx1 gene expression in the early endoderm is regulated by discrete cis-elements within its promoter that are bound by trans-acting factors.
Beyond its importance in specifying the fate of progenitor cells within the early pancreatic buds, Pdx1 also plays a distinct role in the proliferation of differentiating cells. This proliferation function is mediated through a discrete amino acid sequence (FPWMK) within the amino terminal domain of Pdx1 that is believed to interact with the ubiquitously expressed TALE (three amino acid loop extension) homeodomain protein Pbx1 (pre-B cell leukemia homeobox 1) [47]. In transgenic complementation studies of Pdx1−/− mice, mutation of the FPWMK motif results in a severely hypoplastic pancreas with presence of all cell types (endocrine, exocrine, and duct) [48]. Consistent with this finding, Pbx1−/− mice also develop pancreatic hypoplasia, and defects in endocrine and exocrine cell formation [49]. The physical interaction between Pdx1 and Pbx1 may also play a role in the mature pancreas, as compound heterozygotes (Pdx1+/−; Pbx1+/−) develop frank diabetes, suggesting a defect in islet function [49].
Pdx1 and mid to late pancreas development
After formation of the initial dorsal and ventral pancreatic buds at E8.5 in mice, these structures undergo branching morphogenesis and proliferation as they evaginate into the surrounding mesenchyme. Between E13.5 and E15.5, endocrine and exocrine gene products dramatically magnify their levels of expression, coincident with the exponential increase in cellular mass; this period of time is referred to as the “secondary transition.” By E15.5, gut rotation causes the ventral and dorsal buds to fuse, forming a single pancreatic organ as seen in adults [50] (see Fig. 1). The role of Pdx1 in the development of the embryonic pancreas beyond the elaboration of the initial buds has been addressed in elegant studies of R. MacDonald and colleagues. Using a mouse model in which the expression of Pdx1 can be selectively repressed by administration of doxycycline, they studied the effect of Pdx1 deletion at various time points after the formation of the initial pancreatic buds [51]. Deletion of Pdx1 at E12.5 led to the formation of a pancreas nearly devoid of acinar cells and islets; the precursor pancreatic epithelium was observed to grow and branch out, producing a truncated ductal tree with immature duct-like cells. The lack of acinar cell formation coincided with the loss of Pft1a/p48, a transcription factor necessary for acinar cell development [52]. Deletion of Pdx1 at the start of the secondary transition (E13.5) results in the formation of incompletely differentiated acinar cells [52]. Thus, although expression of Pdx1 is necessary for the initial differentiation and growth of precursors within the developing pancreatic buds at E8.5–9.5, ongoing expression of Pdx1 during the secondary transition (after E12.5) appears to be necessary for the subsequent differentiation and expansion of acinar tissue and islets, whereas ducts appear to be only partially dependent upon Pdx1 after this stage. These findings are also consistent with lineage tracing studies, which demonstrate Pdx1 expression in duct precursors until about E12.5, but not thereafter [26]. However, it should be noted that these lineage tracing approaches relied upon transgenic expression of a reporter protein, a technique that may not fully and accurately recapitulate the expression pattern of the endogenous gene.
After the secondary transition, Pdx1 protein is downregulated in acinar cells [53], suggesting that the maintenance of acinar cells in their differentiated state is not dependent upon Pdx1. In fact, continued expression of Pdx1 in acinar cells appears detrimental; studies of Heller, et al. demonstrated that ongoing expression of Pdx1 in acinar cells after the secondary transition (using an elastase-1 promoter-driven Pdx1 transgene in mice) results in dysmorphogenesis of the acinar compartment with a high rate of cellular turnover (both proliferation and apoptosis) and eventual fatty infiltration [54]. The importance of selective expression of Pdx1 during pancreas development is dramatically emphasized in studies by Miyastuka, et al., which showed that forced expression of Pdx1 throughout all cells of the developing pancreas from ~E10 onward results in pancreatic hypoplasia and an acinar compartment that has been replaced by duct-like structures; this phenotype appears to be a consequence of the ectopic activation by Pdx1 of the signal transducer and activator of transcription 3 (STAT3) [55]. On the one hand, these findings appear contradictory to the studies described above showing that Pdx1 is necessary for acinar, but not ductal, differentiation and growth. On the other hand, these findings emphasize that Pdx1 action is heavily nuanced by the cellular context; thus, in early pancreatic precursors, Pdx1 is necessary to drive proliferation and pancreatic outgrowth, and in subsequent time points, it is necessary for lineage determination, proliferation, and maintenance of selected cell types. The molecular mechanisms underlying this selectivity may be influenced by both the physical accessibility (“chromatin structure”) of potential downstream target genes and the nature of the transcriptional protein complexes formed by Pdx1. These mechanisms will be addressed later in this review.
Role of Pdx1 in the Adult Pancreas
Pdx1 in the maintenance of functional β cell mass
At the end of gestation (about e18.5 in the mouse), there is formation of distinct circumscribed structures of hormone-producing cells known as islets of Langerhans. In the late gestational and mature pancreas, Pdx1 expression is restricted primarily to insulin-producing β cells of the islet, with expression in some somatostatin-producing δ cells, and low-level expression in subpopulations of acinar and duct cells [53] (see Fig. 2). Pdx1 haploinsufficiency does not appear to physically or functionally impinge upon pancreas development in Pdx1+/− mice [12,13], however, such mice exhibit strikingly impaired glucose tolerance and glucose-stimulated insulin secretion with age [56,57]. Similarly impaired β cell function in the setting of Pdx1 deficiency has been observed in several other animal models (rat, fish, and sand rat [58–62]), as well as in humans with single allelic mutations in Ipf1 (where the development of impaired glucose responsiveness and frank diabetes occurs) [17–21]. The sand rat (Psammomys obesus) is a particularly an interesting case, as Pdx1 is not normally found in islets from these animals; however, gene transfer experiments show that repletion of Pdx1 in these islets greatly enhances insulin content and glucose-stimulated insulin transcription [62]. Complete or near-complete deletion of Pdx1 in late embryonic [63] or adult mice [64] also result in a later diabetic phenotype. Although subtle developmental defects cannot be ruled out, these observations suggest a dramatically different requirement for Pdx1 gene dosage in the developing pancreas vs. the mature β cell, the latter of which may require more tightly regulated transcription and/or translation. In this regard, in some mouse models (and quite possibly in obese humans) the progressive loss of Pdx1 protein levels with age might account for the increasing incidence of β cell dysfunction and impaired glucose tolerance [22,57,65,66].
Figure 2. Pdx1 protein expression in the adult pancreas.
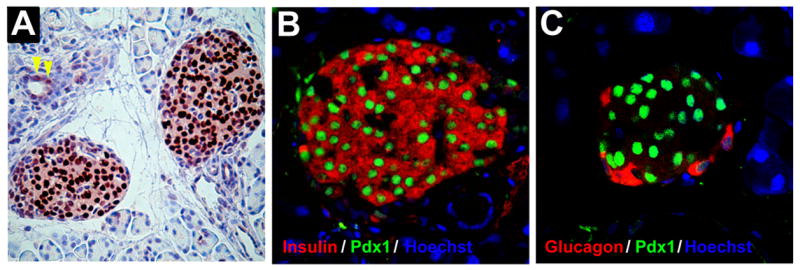
Pancreas from a 10 week-old mouse was fixed in paraformaldehyde and embedded in paraffin. Sections were then hematoxylin stained and subject to immunohistochemistry or immunofluorescence. A, horseradish peroxidase-based immunostaining for Pdx1. Pdx1 (dark brown color) demonstrates an islet-specific expression pattern with nuclear localization. Occasional duct cells are also observed to express Pdx1 (yellow arrowheads). B, dual immunofluorescence of an islet using antibodies against Pdx1 (green) and insulin (red). Pdx1 is seen to localize within the nuclei of insulin-producing β cells. C, dual immunofluorescence of an islet using antibodies against Pdx1 (green) and glucagon (red). Pdx1 is not expressed in glucagon-positive α cells. In panels B and C nuclei are counterstained with hoechst H33342 dye (blue).
The cause of β cell functional impairment in Pdx1 deficiency has been studied in multiple systems, including both cell line and mouse models. Based upon these studies, several mechanisms have been postulated: (a) increased β cell apoptosis via and Bcl-2), with resulting loss of downregulation of anti-apoptotic genes (BclXL functional cell mass [56,67], (b) loss of activity of key Pdx1 target genes whose products are involved in glucose-stimulated insulin transcription and secretion (including Glut2, glucokinase, MafA, Nkx6.1, insulin) [57,63,68–76], and (d) loss of new β cell formation/regeneration [64,77]; in this regard, studies suggest that the action of glucagon-like peptide 1 (GLP1) in enhancing β cell growth and formation in the adult may rely upon activation of Pdx1 in β cells and potential precursor cell types, such as those residing within ducts [74,78–82].
Role of Pdx1 in adaptive β cell hyperplasia and β cell regeneration
The effects of Pdx1 on β cell growth and survival noted above raise the tantilizing possibility that it may play a central role in the β cell hyperplasia and hyperinsulinemia observed in states of insulin resistance, including obesity and severe stress [22,66]. Amongst the most well-studied pathways that regulate β cell mass include the insulin receptor substrate 1 and 2 (IRS1 and IRS2) branch of insulin receptor (IR)/insulin-like growth factor 1 receptor (IGF1R) signaling [83,83,84]. Several studies suggest that Pdx1 may act downstream of the IR/IGF1R signaling to promote β cell proliferation [83,85–89]. Studies of Kulkarni, et al. reveal that haploinsufficiency of Pdx1 in two mouse models of insulin resistance—insulin receptor/IRS1 double heterozygous mice and liver insulin receptor knockout mice—virtually completely abrogated the adaptive islet hyperplastic response, resulting instead in β cell apoptosis and premature death of the animals [88]. Importantly, there is also loss of β cell proliferation in these models, suggesting that an important component of islet hyperplasia in insulin resistant states may be an enhanced replication rate of pre-existing β cells (rather than β cell neogenesis). β catenin signaling, a pathway mediating pancreas growth, may in part underlie proliferation of mature β cells. In this regard, it appears that a full complement of Pdx1 is necessary to promote the dissociation of β catenin from its membranous E cadherin anchor, thereby allowing the cytoplasmic and nuclear translocation of β catenin [88]. Precisely how Pdx1 action impinges upon β catenin release is unclear; nonetheless, this pathway may not represent an exclusive mechanism of Pdx1-mediated proliferation, as overexpression of active β catenin alone in late pancreas development appears to have minimal, if any, effects on β cell proliferation ([90]). These latter data suggest that the confluence of pathways (IR/IRS and β catenin signaling) in the setting of a normal Pdx1 complement is necessary for adaptive β cell proliferation.
There is some evidence that certain non-β cells, such as duct cells of the pancreas, may also contribute to adaptive β cell hyperplasia in a Pdx1-dependent fashion. For example, in Irs2−/− mice there is diminished β cell mass with impaired glucose tolerance [89,91]; in this model, superposition of haploinsufficiency of the Forkhead transcription factor Foxo1 causes activation of Pdx1 expression in duct cells, and the subsequent recovery of β cell mass [92]. In support of this observation, overexpression of Pdx1 alone in Irs2−/− animals promotes recovery of β cell mass and correction of glucose tolerance throughout life [93]. Whether duct cells per se give rise to β cells in this model, however, was not directly demonstrated in these studies and must await more rigorous lineage tracing studies. Activation of Pdx1 in duct cells of the pancreas has also been observed following partial pancreatectomy in mice [85] and in insulin resistant Zucker fatty (ZF) rats [83]. Importantly in this regard, Pdx1 expression is often observed in proliferating duct cells [77,94,95], and such cells have been suggested to serve as substrate for β cell neogenesis, particularly in cases of adaptive hyperplasia or pancreas regeneration [92,96–99]. A recent study suggests that Pdx1, in complex with TALE transcription factors, negatively regulates the duct-specific keratin 19 gene in mature duct cells [100]. This finding raises the possibility that enhanced levels of Pdx1 protein in duct cells may promote β cell transdifferentiation by direct repression of the duct cell phenotype (through suppression of keratin 19 and related genes) and simultaneous activation of β cell-specific genes [101–103]. Currently, there is little evidence that any other cell type contributes to β cell neogenesis. Although some studies had suggested the potential role of Pdx1 in promoting β cell transdifferentiation from acinar tissue [83,104–106], a recent elegant lineage tracing analysis suggests that such cells do not contribute to new β cells in models of regeneration in vivo [107].
Mechanism of Pdx1 Action
Posttranslational modifications of Pdx1
As a member of the homeodomain class of proteins, Pdx1 is believed to exert its actions almost exclusively within the nucleus as a result of regulating gene transcription. Although some reports in the literature have described physiologic circumstances where the distribution of Pdx1 might be cytoplasmic, it is believed that cytoplasmic sequestration may represent more a mechanism to attenuate the nuclear action of Pdx1 under physiologic or pathologic conditions, rather than to promote a specific cytoplasmic function. For example, the negative effect of fatty acids on β cell function may be related to their eventual sequestration of Pdx1 in the cytoplasm [76], whereas the positive effect of glucose on insulin transcription may be a result of its enhancement of Pdx1nuclear translocation [108]. A basic amino acid sequence with the homeodomain of Pdx1, RRMKWKK, is believed to function as the nuclear targeting sequence [109]. Teleologically, regulation at the level of Pdx1 protein compartmentation (e.g. nuclear-cytoplasmic shuttling) or function would allow for acute alterations to target gene transcription, where control on the order of seconds to minutes might be crucial. At least 3 different reversible post-translational modifications, including phosphorylation [86,108,110–114], sumoylation [115], and glycosylation [116], have been proposed to explain nuclear-cytoplasmic shuttling and/or other functions of Pdx1. Consistent with these modifications, different molecular weight species of Pdx1 have been described in immunoblots [115]. Of the three modifications, phosphorylation is perhaps the best-studied. Elevated glucose levels have been suggested to promote Pdx1 phosphorylation through one or a combination of several pathways, including the stress-activated protein kinase (SAPK) and extracellular signal-regulated kinase (ERK) 1/2 pathways [86,108,112], phosphatidylinositol 3-kinase pathway [117,118], and/or the Per-Arnt-Sim (PAS) kinase [110]. Although the role of phosphorylation of specific residues within Pdx1 has not been rigorously analyzed, phosphorylation of Ser61 and Ser66 of Pdx1 via glycogen synthase kinase 3 appears to target Pdx1 for proteosomal degradation, thereby decreasing its half-life. This modification could partially explain diminished β cell function during endoplasmic reticulum stress and oxidative stress (both of which occur in the diabetic state), where GSK3 activity is enhanced [111,119]. Similarly, diminished β cell function following DNA damage might be explained by phosphorylation of Thr11 of Pdx1 by DNA-dependent kinases; this modification has also been proposed to lead to its more rapid intracellular degradation [114].
Target gene recognition by Pdx1
In the nucleus, the action of Pdx1 relies upon its binding to specific genes and subsequently regulating their rates of transcription. The homeodomain of Pdx1 recognizes A/T-rich DNA sequences (characteristically 5′-TAAT-3′) with affinity in the nanomolar range [120,121]. Interestingly, the range of DNA sequences bound by Pdx1 within the nucleus cannot be entirely predicted by its affinity for DNA in vitro; thus, some sequences that show preferential binding in vitro by electrophoretic mobility shift assays demonstrate little or no association within the nucleus by chromatin immunoprecipitation, and vice-versa [70,71]. At least two factors could account for this discrepancy. First, amino acid sequences outside of the homeodomain of Pdx1 itself may contribute to DNA binding either directly or though interactions with other proteins [120,121]. For example, the interaction of Pdx1 with the bHLH factor BETA2/NeuroD1 would allow for selective targeting to genes whose regulatory regions contain appropriately-spaced binding sites for both factors [122,123]. Second, the chromatin structure or conformation of DNA at specific genes within the nucleus could alter DNA site accessibility or binding affinity, respectively. In this regard, Pdx1 displays little or no ability to dynamically alter chromatin structure, and therefore appears to be highly susceptible to the nature chromatin accessibility [124]. Studies in our laboratory demonstrate that chromatin accessibility at the insulin gene in two different cultured pancreatic cell types (β cells vs. duct cells) is dramatically different, permitting the occupancy of Pdx1 in only one cell type (β cells) but not the other (duct cells) [124]. The limited accessibility in duct cells in that study could be accounted for by the presence of a nucleosome (DNA complexed with histone proteins) at a key Pdx1 binding site in the insulin promoter, and binding in the presence of this nucleosome can only be overcome by high concentrations of Pdx1 protein [124]. These data raise the attractive hypothesis that the higher levels of Pdx1 associated with transdifferentiating duct cells is a necessary trigger to initially overcome limited chromatin accessibility at β cell gene targets.
Even in the presence of accesssible or “open” chromatin, the local conformation (or “bending”) of DNA may subtly influence binding affinity and subsequent transcriptional rates. A recent crystallographic structure of the Pdx1 homeodomain by Longo, et al. demonstrates two distinctly different structures of Pdx1 when bound to DNA, each showing distinct side chain contacts depending upon the conformation of the DNA [125]. This “induced fit” model suggests that transcription factors bound at more distant sites (and possibly not even interacting with Pdx1) could alter the nature of the DNA conformation at the Pdx1 binding region. Thus, the nuclear composition of proteins (hence cellular context), may affect occupancy and consequent activity of Pdx1 at any given gene.
Mechanisms of transcriptional regulation by Pdx1
Based on studies using transient reporter gene assays in cell lines and electrophoretic mobility shift assays, a host of genes in the β cell initially emerged as potential candidates for direct regulation by Pdx1. These included the genes encoding insulin [68,70,71,126], Glut2 [127,128], glucokinase [129,130], islet amyloid polypeptide (IAPP) [63,75,131,132], Pax4 [133], Nkx6.1 [69,134], MafA [72], liver receptor homolog (Lrh) 1 [135], and Pdx1 itself [37]. In recent years, the direct regulation of many of these genes by Pdx1 has been confirmed using loss-of-function studies in islets and intact animals, and occupancy by Pdx1 of the endogenous genes in islets and β cell lines has been confirmed using the chromatin immunoprecipitation (ChIP) assay [70]. The insulin gene represents the best and most definitively characterized gene with respect to Pdx1 action. As such, much of our knowledge of the mechanism by which Pdx1 regulates target genes has emerged from study of the insulin enhancer. The general consensus from these studies has been that Pdx1 is an activator of gene transcription, with the activation function residing with the amino terminal domain of the protein, between amino acids 13–73 [136–138]. However, despite years of research, remarkably little is still known about the mechanisms by which Pdx1 directly influences gene transcription. For example, how does Pdx1 communicate across the gene with other transcription factors to achieve transcriptional synergy? How do proteins necessary for mRNA initiation and elongation interrelate with these transcription factors? How are all of these events achieved in the context of potentially repressive chromatin structure? Studies in our laboratory and others have elucidated that a more “dynamic” and intricate series of events ultimately contributes to transcriptional activation by Pdx1. These include (a) the formation of an activation complex involving other transcription factors and transcriptional coactivators, (b) the modulation of local chromatin structure to ensure a more “accessible” state of the gene, and (c) communication with components of the basal transcriptional machinery to alter final transcriptional responsiveness. Each of these events are briefly discussed below.
Formation of protein complexes
The participation of Pdx1 in a transcriptional complex is suggested by functional studies in cell lines, whereby addition of Pdx1 in conjunction with other β cell-specific transcription factors results in the synergistic activation of insulin gene-based reporter plasmids [139]. These interacting factors include bHLH proteins that bind to E-boxes (BETA2/NeuroD1 and E2A proteins) [122,123,140,141], the basic leucine zipper factor MafA that binds to the C-box [73,142], the homeodomain proteins Pbx1 and MEIS2 [47,48,143,144], and high mobility group (HMG) proteins that bind to the DNA backbone [123]. The free energy gained from these interactions may increase the likelihood that Pdx1 targets genes with binding sites for these factors [123,144]. Alternatively, these interactions may cause conformational changes in Pdx1 and/or DNA that enhance DNA binding affinity [125].
Apart from interaction with transcription factors, Pdx1 also appears to recruit transcriptional co-activators (proteins that bridge Pdx1 to the basal transcriptional machinery), including CBP/p300 [68,113,141,145,146], Set7/9 [147], Bridge-1 [148], PCIF1 [149], and histone deacetylases (HDACs) [150]. Once recruited, cofactors are believed to serve as a physical or functional “bridge” that allows Pdx1 to communicate with components of the basal transcriptional machinery. Although the majority of these cofactor interactions with Pdx1 are believed to enhance the activation function of Pdx1, there is the distinct possibility that certain cofactor interactions may allow Pdx1 to aid in the repression of some genes. In this regard, the well-defined repression of genes such as glucagon [63,64,70,134] and K19 [100] by Pdx1 may reflect a repression complex formed in association with corepressor molecules.
Modulation of chromatin structure
Although packaging of DNA in the context of chromatin is responsible for the efficient storage of genetic material within the nucleus, it also has the capacity to impede the accessibility of DNA to transcription factors and the transcriptional machinery [151]. In this regard, basic unit of chromatin is formed by the complex between DNA and histone proteins; the covalent modifications of the amino terminal tails of histone proteins (including acetylation and methylation of Lys residues) have been demonstrated to alter chromatin packaging, and hence directly affect rates of gene transcription [152]. Interestingly, although Pdx1 itself does not appear to have intrinsic abilility to either modify histones or directly alter chromatin structure, its depletion in β cell lines results in the loss of histone H3-Lys4 dimethylation (a mark of active, open chromatin) at the insulin gene. This result may be secondary to the recruitment of the histone methyltransferase Set7/9 by Pdx1 [147]. Similarly, the glucose-stimulated increase in histone H4 acetylation (another marker of active, open chromatin) at the insulin gene in both cell lines and islets by Pdx1 is consistent with its recruitment of the histone acetyltransferase p300 [145,153,154]. These results suggest that an important component of gene activation by the Pdx1 protein complex may be its direct alteration of chromatin to a more open state, thereby permitting enhanced accessibility to other transcription factors and the basal transcriptional machinery.
Communication with basal transcriptional machinery
The term “basal transcriptional machinery” is applied to a group of proteins responsible for the elongation of mRNAs and their subsequent processing. In eukaryotes, these include several general transcription factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH) and the RNA polymerase II holoenzyme [155]. Few studies in β cells have attempted to address a link between Pdx1 and the basal transcriptional machinery. Our laboratory has demonstrated that in the absence of Pdx1, the RNA polymerase II complex continues to be recruited to the insulin promoter, but transcriptional elongation by the complex is impaired [68]. Consistent with this observation, it appears that Pdx1 is necessary to ensure that the Ser5-phosphorylated isoform (the “initiation isoform”) is appropriately converted to the Ser2-phosphorylated isoform (the “elongation isoform”) [147]. These findings raise the possibility that the linkage between the Pdx1 transcriptional complex and the basal transcriptional machinery may involve an as yet unidentified kinase. Importantly, in yeast systems, these phosphorylation events (thought to be mediated by cyclin-dependent kinases) are inextricably linked to the presence of histone H3-Lys4 methylation [156]. Thus, the link between Pdx1 and the putative kinase may be indirect, and possibly secondary to the maintenance of the appropriate chromatin state by Pdx1.
Conclusions
The prevalence of diabetes mellitus is increasing to alarming proportions. Current estimates from the Centers for Disease Control predict that 1 in 3 Americans born in the year 2000 will develop diabetes, and similar trends are predicted around the world [157]. Underlying all forms of diabetes is dysfunction at the level of the β cell. Given the broad role of Pdx1 in the differentiation of cell types with the pancreas and in the maintenance of β cell function and mass (see summary in Table I), preclinical studies have recently focused on the therapeutic potential for Pdx1 and related factors to reverse Type 1 diabetes. For example, viral-mediated expression of Pdx1 in liver cells appears to stimulate a host of pancreas-related genes (including insulin), leading to reversal of chemically-induced diabetes in mice [158–161]. From a delivery perspective, transduction of Pdx1 using viruses may not be requisite. Pdx1 also has been shown to harbor a protein transduction domain, thereby enabling delivery of the protein directly into potential precursor cells to induce the pancreatic gene program [162,163]. Thus, leveraging of the protein transduction domain may open therapeutic avenues via delivery of Pdx1 protein in vivo to stimulate potential β cell precursors. The results of these therapeutic intervention studies emphasize the central role of Pdx1 in the pancreatic transcriptional hierarchy (see Fig. 3) and suggest a relatively simplistic approach to cellular “reprogramming” for the generation of new β cells. Just as importantly, however, the disparate effects of Pdx1 in the context of different cell types emphasize our limited understanding of the mechanisms and pathways encompassing Pdx1 action. For example, what upstream signaling pathways are necessary for initiation and maintenance of Pdx1 gene expression? What factors distinguish why one cell type responds to Pdx1 by activating acinar genes, another responds by activating β cell genes, and still others activate neither acinar nor β cell genes? How does the level of Pdx1 protein in a given cell type nuance the activity of a given gene or, ultimately, the global gene expression pattern in that cell? The answers to these and related mechanistic questions may ultimately hold the clues to the identification and/or engineering of a renewable supply of β cells for individuals with diabetes.
TABLE I.
Expression Pattern and Function of Pdx1 in the Developing and Mature Mouse Pancreas
| Developmental Period | Pdx1 Expression Pattern | Functional Role of Pdx1 |
|---|---|---|
| Early Development (E8.5 – E12) | All cells of the early pancreatic endoderm, and portions of the stomach and duodenum | Necessary for:
|
| Mid to Late Pancreas Development (E12.5 – E18) | Endocrine and acinar cells | Necessary for acinar and islet cell formation |
| Adult Pancreas | Primarily β (insulin) and δ(somatostatin) endocrine cells of the islets of Langerhans; occasional duct cells and acinar cells. | Necessary for:
|
Figure 3. Upstream regulators and direct downstream targets of Pdx1.
The figure emphasizes the central role of Pdx within the pancreas. Cell-extrinsic factors emanating from the notochord (FGF2 and activin) as well as cell-intrinsic factors (transcription factors Foxa2 and HNF6) collaborate to activate Pdx1 expression in the primitive gut endoderm. During pancreas development, Pdx1 is believed to directly activate or repress a subset of genes that lead to differentiation of islet and acinar cells. In the mature pancreas, Pdx1 directly activates or represses genes that confer the β cell phenotype. The “+” sign indicates that the gene is activated by Pdx1, and the “−” sign indicates that the gene is repressed by Pdx1.
Acknowledgments
We wish to acknowledge the following organizations for their support of the research in the Mirmira laboratory: National Institutes of Health (grants R01 DK60581, T32 GM007055, and P30 DK063609), American Diabetes Association, Juvenile Diabetes Research Foundation, the Goldman Philanthropic Partnerships, and the Meade Family.
ABBREVIATIONS
- bHLH
basic helix-loop-helix
- Pbx1
pre-B cell leukemia homeobox 1
- Pdx1
pancreatic and duodenal homeobox 1
- Shh
sonic hedgehog
- TALE
three amino acid loop extension
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kim SK, Hebrok M. Intercellular signals regulating pancreas development and function. Genes Dev. 2001;15:111–127. doi: 10.1101/gad.859401. [DOI] [PubMed] [Google Scholar]
- 2.Scott V, Clark AR, Hutton JC, Docherty K. Two proteins act as the iuf1 insulin gene enhancer binding factor. FEBS Lett. 1991;290:27–30. doi: 10.1016/0014-5793(91)81217-v. [DOI] [PubMed] [Google Scholar]
- 3.Ohlsson H, Edlund T. Sequence-specific interactions of nuclear factors with the insulin gene enhancer. Cell. 1986;45:35–44. doi: 10.1016/0092-8674(86)90535-0. [DOI] [PubMed] [Google Scholar]
- 4.Ohlsson H, Karlsson O, Edlund T. A beta-cell-specific protein binds to the two major regulatory sequences of the insulin gene enhancer. Proc Natl Acad Sci U S A. 1988;85:4228–4231. doi: 10.1073/pnas.85.12.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohlsson H, Thor S, Edlund T. Novel insulin promoter- and enhancer-binding proteins that discriminate between pancreatic alpha- and beta-cells. Mol Endocrinol. 1991;5:897–904. doi: 10.1210/mend-5-7-897. [DOI] [PubMed] [Google Scholar]
- 6.Ohlsson H, Karlsson K, Edlund T. Ipf1, a homeodomain-containing transactivator of the insulin gene. Embo J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peshavaria M, Gamer L, Henderson E, Teitelman G, Wright CV, Stein R. Xihbox 8, an endoderm-specific xenopus homeodomain protein, is closely related to a mammalian insulin gene transcription factor. Mol Endocrinol. 1994;8:806–816. doi: 10.1210/mend.8.6.7935494. [DOI] [PubMed] [Google Scholar]
- 8.Leonard J, Peers B, Johnson T, Ferreri K, Lee S, Montminy MR. Characterization of somatostatin transactivating factor-1, a novel homeobox factor that stimulates somatostatin expression in pancreatic islet cells. Mol Endocrinol. 1993;7:1275–1283. doi: 10.1210/mend.7.10.7505393. [DOI] [PubMed] [Google Scholar]
- 9.Miller CP, McGehee REJ, Habener JF. Idx-1: A new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. EMBO J. 1994;13:1145–1156. doi: 10.1002/j.1460-2075.1994.tb06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brooke NM, Garcia-Fernandez J, Holland PW. The parahox gene cluster is an evolutionary sister of the hox gene cluster. Nature. 1998;392:920–922. doi: 10.1038/31933. [DOI] [PubMed] [Google Scholar]
- 11.Rosanas-Urgell A, Marfany G, Garcia-Fernandez J. Pdx1-related homeodomain transcription factors are distinctly expressed in mouse adult pancreatic islets. Mol Cell Endocrinol. 2005;237:59–66. doi: 10.1016/j.mce.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- 13.Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. Pdx-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- 14.Ahlgren U, Jonsson J, Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in ipf1/pdx1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- 15.Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, Habener JF. Pancreatic agenesis attributable to a single nucleotide deletion in the human ipf1 gene coding sequence. Nat Genet. 1997;15:106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 16.Schwitzgebel VM, Mamin A, Brun T, Ritz-Laser B, Zaiko M, Maret A, Jornayvaz FR, Theintz GE, Michielin O, Melloul D, Philippe J. Agenesis of human pancreas due to decreased half-life of insulin promoter factor 1. J Clin Endocrinol Metab. 2003;88:4398–4406. doi: 10.1210/jc.2003-030046. [DOI] [PubMed] [Google Scholar]
- 17.Stoffers DA, Ferrer J, Clarke WL, Habener JF. Early-onset type-ii diabetes mellitus (mody4) linked to ipf1. Nat Genet. 1997;17:138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 18.Stoffers DA, Stanojevic V, Habener JF. Insulin promoter factor-1 gene mutation linked to early-onset type 2 diabetes mellitus directs expression of a dominant negative isoprotein. J Clin Invest. 1998;102:232–241. doi: 10.1172/JCI2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hani EH, Stoffers DA, Chevre JC, Durand E, Stanojevic V, Dina C, Habener JF, Froguel P. Defective mutations in the insulin promoter factor-1 (ipf-1) gene in late-onset type 2 diabetes mellitus. J Clin Invest. 1999;104:R41–8. doi: 10.1172/JCI7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clocquet AR, Egan JM, Stoffers DA, Muller DC, Wideman L, Chin GA, Clarke WL, Hanks JB, Habener JF, Elahi D. Impaired insulin secretion and increased insulin sensitivity in familial maturity-onset diabetes of the young 4 (insulin promoter factor 1 gene) Diabetes. 2000;49:1856–1864. doi: 10.2337/diabetes.49.11.1856. [DOI] [PubMed] [Google Scholar]
- 21.Macfarlane WM, Frayling TM, Ellard S, Evans JC, Allen LI, Bulman MP, Ayres S, Shepherd M, Clark P, Millward A, Demaine A, Wilkin T, Docherty K, Hattersley AT. Missense mutations in the insulin promoter factor-1 gene predispose to type 2 diabetes. J Clin Invest. 1999;104:R33–9. doi: 10.1172/JCI7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weir GC, Sharma A, Zangen DH, Bonner-Weir S. Transcription factor abnormalities as a cause of beta cell dysfunction in diabetes: A hypothesis. Acta Diabetol. 1997;34:177–184. doi: 10.1007/s005920050071. [DOI] [PubMed] [Google Scholar]
- 23.Laybutt DR, Sharma A, Sgroi DC, Gaudet J, Bonner-Weir S, Weir GC. Genetic regulation of metabolic pathways in beta-cells disrupted by hyperglycemia. J Biol Chem. 2002;277:10912–10921. doi: 10.1074/jbc.M111751200. [DOI] [PubMed] [Google Scholar]
- 24.Laybutt DR, Glandt M, Xu G, Ahn YB, Trivedi N, Bonner-Weir S, Weir GC. Critical reduction in beta-cell mass results in two distinct outcomes over time. Adaptation with impaired glucose tolerance or decompensated diabetes. J Biol Chem. 2003;278:2997–3005. doi: 10.1074/jbc.M210581200. [DOI] [PubMed] [Google Scholar]
- 25.Gannon M, Herrera PL, Wright CV. Mosaic cre-mediated recombination in pancreas using the pdx-1 enhancer/promoter. Genesis. 2000;26:143–144. doi: 10.1002/(sici)1526-968x(200002)26:2<143::aid-gene13>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 26.Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: Ngn3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 27.Hebrok M, Kim SK, Melton DA. Notochord repression of endodermal sonic hedgehog permits pancreas development. Genes Dev. 1998;12:1705–1713. doi: 10.1101/gad.12.11.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hebrok M, Kim SK, St Jacques B, McMahon AP, Melton DA. Regulation of pancreas development by hedgehog signaling. Development. 2000;127:4905–4913. doi: 10.1242/dev.127.22.4905. [DOI] [PubMed] [Google Scholar]
- 29.Apelqvist A, Ahlgren U, Edlund H. Sonic hedgehog directs specialised mesoderm differentiation in the intestine and pancreas. Curr Biol. 1997;7:801–804. doi: 10.1016/s0960-9822(06)00340-x. [DOI] [PubMed] [Google Scholar]
- 30.Hebrok M. Hedgehog signaling in pancreas development. Mech Dev. 2003;120:45–57. doi: 10.1016/s0925-4773(02)00331-3. [DOI] [PubMed] [Google Scholar]
- 31.Kawahira H, Scheel DW, Smith SB, German MS, Hebrok M. Hedgehog signaling regulates expansion of pancreatic epithelial cells. Dev Biol. 2005;280:111–121. doi: 10.1016/j.ydbio.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Thomas MK, Lee JH, Rastalsky N, Habener JF. Hedgehog signaling regulation of homeodomain protein islet duodenum homeobox-1 expression in pancreatic beta-cells. Endocrinology. 2001;142:1033–1040. doi: 10.1210/endo.142.3.8007. [DOI] [PubMed] [Google Scholar]
- 33.Thomas MK, Rastalsky N, Lee JH, Habener JF. Hedgehog signaling regulation of insulin production by pancreatic beta-cells. Diabetes. 2000;49:2039–2047. doi: 10.2337/diabetes.49.12.2039. [DOI] [PubMed] [Google Scholar]
- 34.Jepeal LI, Fujitani Y, Boylan MO, Wilson CN, Wright CV, Wolfe MM. Cell-specific expression of glucose-dependent-insulinotropic polypeptide is regulated by the transcription factor pdx-1. Endocrinology. 2005;146:383–391. doi: 10.1210/en.2004-0223. [DOI] [PubMed] [Google Scholar]
- 35.Larsson LI, Madsen OD, Serup P, Jonsson J, Edlund H. Pancreatic-duodenal homeobox 1 -role in gastric endocrine patterning. Mech Dev. 1996;60:175–184. doi: 10.1016/s0925-4773(96)00609-0. [DOI] [PubMed] [Google Scholar]
- 36.Gerrish K, Gannon M, Shih D, Henderson E, Stoffel M, Wright CV, Stein R. Pancreatic beta cell-specific transcription of the pdx-1 gene. The role of conserved upstream control regions and their hepatic nuclear factor 3beta sites. J Biol Chem. 2000;275:3485–3492. doi: 10.1074/jbc.275.5.3485. [DOI] [PubMed] [Google Scholar]
- 37.Gerrish KE, Cissell MA, Stein R. The role of hepatic nuclear factor 1a and pdx-1 in transcriptional regulation of the pdx-1 gene. J Biol Chem. 2001;276:47775–47784. doi: 10.1074/jbc.M109244200. [DOI] [PubMed] [Google Scholar]
- 38.Samaras SE, Cissell MA, Gerrish K, Wright CV, Gannon M, Stein R. Conserved sequences in a tissue-specific regulatory region of the pdx-1 gene mediate transcription in pancreatic beta cells: Role for hepatocyte nuclear factor 3 beta and pax6. Mol Cell Biol. 2002;22:4702–4713. doi: 10.1128/MCB.22.13.4702-4713.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerrish K, Van Velkinburgh JC, Stein R. Conserved transcriptional regulatory domains of the pdx-1 gene. Mol Endocrinol. 2004;18:533–548. doi: 10.1210/me.2003-0371. [DOI] [PubMed] [Google Scholar]
- 40.Marshak S, Benshushan E, Shoshkes M, Havin L, Cerasi E, Melloul D. Functional conservation of regulatory elements in the pdx-1 gene: Pdx-1 and hepatocyte nuclear factor 3beta transcription factors mediate beta-cell-specific expression. Mol Cell Biol. 2000;20:7583–7590. doi: 10.1128/mcb.20.20.7583-7590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiebe PO, Kormish JD, Roper VT, Fujitani Y, Alston NI, Zaret KS, Wright CV, Stein RW, Gannon M. Ptf1A binds to and activates area iii, a highly conserved region of the pdx1 promoter that mediates early pancreas-wide pdx1 expression. Mol Cell Biol. 2007 doi: 10.1128/MCB.01978-06. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H, Fujitani Y, Wright CV, Gannon M. Efficient recombination in pancreatic islets by a tamoxifen-inducible cre-recombinase. Genesis. 2005;42:210–217. doi: 10.1002/gene.20137. [DOI] [PubMed] [Google Scholar]
- 43.Gannon M, Gamer LW, Wright CV. Regulatory regions driving developmental and tissue-specific expression of the essential pancreatic gene pdx1. Dev Biol. 2001;238:185–201. doi: 10.1006/dbio.2001.0359. [DOI] [PubMed] [Google Scholar]
- 44.Van Velkinburgh JC, Samaras SE, Gerrish K, Artner I, Stein R. Interactions between areas I and ii direct pdx-1 expression specifically to islet cell types of the mature and developing pancreas. J Biol Chem. 2005;280:38438–38444. doi: 10.1074/jbc.M508594200. [DOI] [PubMed] [Google Scholar]
- 45.Fujitani Y, Fujitani S, Boyer DF, Gannon M, Kawaguchi Y, Ray M, Shiota M, Stein RW, Magnuson MA, Wright CV. Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for pdx1 in foregut organ differentiation and pancreas formation. Genes Dev. 2006;20:253–266. doi: 10.1101/gad.1360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boyer DF, Fujitani Y, Gannon M, Powers AC, Stein RW, Wright CV. Complementation rescue of pdx1 null phenotype demonstrates distinct roles of proximal and distal cis-regulatory sequences in pancreatic and duodenal expression. Dev Biol. 2006;298:616–631. doi: 10.1016/j.ydbio.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 47.Peers B, Sharma S, Johnson T, Kamps M, Montminy M. The pancreatic islet factor stf-1 binds cooperatively with pbx to a regulatory element in the somatostatin promoter: Importance of the fpwmk motif and of the homeodomain. Mol Cell Biol. 1995;15:7091–7097. doi: 10.1128/mcb.15.12.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dutta S, Gannon M, Peers B, Wright C, Bonner-Weir S, Montminy M. Pdx:pbx complexes are required for normal proliferation of pancreatic cells during development. Proc Natl Acad Sci U S A. 2001;98:1065–1070. doi: 10.1073/pnas.031561298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim SK, Selleri L, Lee JS, Zhang AY, Gu X, Jacobs Y, Cleary ML. Pbx1 inactivation disrupts pancreas development and in ipf1-deficient mice promotes diabetes mellitus. Nat Genet. 2002;30:430–435. doi: 10.1038/ng860. [DOI] [PubMed] [Google Scholar]
- 50.Slack JM. Developmental biology of the pancreas. Development. 1995;121:1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- 51.Holland AM, Hale MA, Kagami H, Hammer RE, MacDonald RJ. Experimental control of pancreatic development and maintenance. Proc Natl Acad Sci U S A. 2002;99:12236–12241. doi: 10.1073/pnas.192255099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hale MA, Kagami H, Shi L, Holland AM, Elsasser HP, Hammer RE, MacDonald RJ. The homeodomain protein pdx1 is required at mid-pancreatic development for the formation of the exocrine pancreas. Dev Biol. 2005;286:225–237. doi: 10.1016/j.ydbio.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 53.Guz Y, Montminy MR, Stein R, Leonard J, Gamer LW, Wright CV, Teitelman G. Expression of murine stf-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development. 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- 54.Heller RS, Stoffers DA, Bock T, Svenstrup K, Jensen J, Horn T, Miller CP, Habener JF, Madsen OD, Serup P. Improved glucose tolerance and acinar dysmorphogenesis by targeted expression of transcription factor pdx-1 to the exocrine pancreas. Diabetes. 2001;50:1553–1561. doi: 10.2337/diabetes.50.7.1553. [DOI] [PubMed] [Google Scholar]
- 55.Miyatsuka T, Kaneto H, Shiraiwa T, Matsuoka TA, Yamamoto K, Kato K, Nakamura Y, Akira S, Takeda K, Kajimoto Y, Yamasaki Y, Sandgren EP, Kawaguchi Y, Wright CV, Fujitani Y. Persistent expression of pdx-1 in the pancreas causes acinar-to-ductal metaplasia through stat3 activation. Genes Dev. 2006;20:1435–1440. doi: 10.1101/gad.1412806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson JD, Ahmed NT, Luciani DS, Han Z, Tran H, Fujita J, Misler S, Edlund H, Polonsky KS. Increased islet apoptosis in pdx1+/− mice. J Clin Invest. 2003;111:1147–1160. doi: 10.1172/JCI16537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brissova M, Shiota M, Nicholson WE, Gannon M, Knobel SM, Piston DW, Wright CV, Powers AC. Reduction in pancreatic transcription factor pdx-1 impairs glucose-stimulated insulin secretion. J Biol Chem. 2002;277:11225–11232. doi: 10.1074/jbc.M111272200. [DOI] [PubMed] [Google Scholar]
- 58.Seufert J, Weir GC, Habener JF. Differential expression of the insulin gene transcriptional repressor ccaat/enhancer-binding protein beta and transactivator islet duodenum homeobox-1 in rat pancreatic beta cells during the development of diabetes mellitus. J Clin Invest. 1998;101:2528–2539. doi: 10.1172/JCI2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yee NS, Yusuff S, Pack M. Zebrafish pdx1 morphant displays defects in pancreas development and digestive organ chirality, and potentially identifies a multipotent pancreas progenitor cell. Genesis. 2001;30:137–140. doi: 10.1002/gene.1049. [DOI] [PubMed] [Google Scholar]
- 60.Huang H, Liu N, Lin S. Pdx-1 knockdown reduces insulin promoter activity in zebrafish. Genesis. 2001;30:134–136. doi: 10.1002/gene.1048. [DOI] [PubMed] [Google Scholar]
- 61.Milewski WM, Duguay SJ, Chan SJ, Steiner DF. Conservation of pdx-1 structure, function, and expression in zebrafish. Endocrinology. 1998;139:1440–1449. doi: 10.1210/endo.139.3.5768. [DOI] [PubMed] [Google Scholar]
- 62.Leibowitz G, Ferber S, Apelqvist A, Edlund H, Gross DJ, Cerasi E, Melloul D, Kaiser N. Ipf1/pdx1 deficiency and beta-cell dysfunction in psammomys obesus, an animal with type 2 diabetes. Diabetes. 2001;50:1799–1806. doi: 10.2337/diabetes.50.8.1799. [DOI] [PubMed] [Google Scholar]
- 63.Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. Beta-cell-specific inactivation of the mouse ipf1/pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Holland AM, Gonez LJ, Naselli G, Macdonald RJ, Harrison LC. Conditional expression demonstrates the role of the homeodomain transcription factor pdx1 in maintenance and regeneration of beta-cells in the adult pancreas. Diabetes. 2005;54:2586–2595. doi: 10.2337/diabetes.54.9.2586. [DOI] [PubMed] [Google Scholar]
- 65.Thomas MK, Devon ON, Lee JH, Peter A, Schlosser DA, Tenser MS, Habener JF. Development of diabetes mellitus in aging transgenic mice following suppression of pancreatic homeoprotein idx-1. J Clin Invest. 2001;108:319–329. doi: 10.1172/JCI12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weir GC, Bonner-Weir S. Five stages of evolving beta-cell dysfunction during progression to diabetes. Diabetes. 2004;53(Suppl 3):S16–21. doi: 10.2337/diabetes.53.suppl_3.s16. [DOI] [PubMed] [Google Scholar]
- 67.Johnson JD, Bernal-Mizrachi E, Alejandro EU, Han Z, Kalynyak TB, Li H, Beith JL, Gross J, Warnock GL, Townsend RR, Permutt MA, Polonsky KS. Insulin protects islets from apoptosis via pdx1 and specific changes in the human islet proteome. Proc Natl Acad Sci U S A. 2006;103:19575–19580. doi: 10.1073/pnas.0604208103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iype T, Francis J, Garmey JC, Schisler JC, Nesher R, Weir GC, Becker TC, Newgard CB, Griffen SC, Mirmira RG. Mechanism of insulin gene regulation by the pancreatic transcription factor pdx-1: Application of pre-mrna analysis and chromatin immunoprecipitation to assess formation of functional transcriptional complexes. J Biol Chem. 2005;280:16798–16807. doi: 10.1074/jbc.M414381200. [DOI] [PubMed] [Google Scholar]
- 69.Iype T, Taylor DG, Ziesmann SM, Garmey JC, Watada H, Mirmira RG. The transcriptional repressor nkx6.1 also functions as a deoxyribonucleic acid context-dependent transcriptional activator during pancreatic beta-cell differentiation: Evidence for feedback activation of the nkx6.1 gene by nkx6.1. Mol Endocrinol. 2004;18:1363–1375. doi: 10.1210/me.2004-0006. [DOI] [PubMed] [Google Scholar]
- 70.Chakrabarti SK, James JC, Mirmira RG. Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor, pdx1. Importance of chromatin structure in directing promoter binding. J Biol Chem. 2002;277:13286–13293. doi: 10.1074/jbc.M111857200. [DOI] [PubMed] [Google Scholar]
- 71.Cissell MA, Zhao L, Sussel L, Henderson E, Stein R. Transcription factor occupancy of the insulin gene in vivo. Evidence for direct regulation by nkx2.2. J Biol Chem. 2003;278:751–756. doi: 10.1074/jbc.M205905200. [DOI] [PubMed] [Google Scholar]
- 72.Raum JC, Gerrish K, Artner I, Henderson E, Guo M, Sussel L, Schisler JC, Newgard CB, Stein R. Foxa2, nkx2.2, and pdx-1 regulate islet beta-cell-specific mafa expression through conserved sequences located between base pairs -8118 and -7750 upstream from the transcription start site. Mol Cell Biol. 2006;26:5735–5743. doi: 10.1128/MCB.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao L, Guo M, Matsuoka TA, Hagman DK, Parazzoli SD, Poitout V, Stein R. The islet beta cell-enriched mafa activator is a key regulator of insulin gene transcription. J Biol Chem. 2005;280:11887–11894. doi: 10.1074/jbc.M409475200. [DOI] [PubMed] [Google Scholar]
- 74.Wang H, Iezzi M, Theander S, Antinozzi PA, Gauthier BR, Halban PA, Wollheim CB. Suppression of pdx-1 perturbs proinsulin processing, insulin secretion and glp-1 signalling in ins-1 cells. Diabetologia. 2005;48:720–731. doi: 10.1007/s00125-005-1692-8. [DOI] [PubMed] [Google Scholar]
- 75.Wang H, Maechler P, Ritz-Laser B, Hagenfeldt KA, Ishihara H, Philippe J, Wollheim CB. Pdx1 level defines pancreatic gene expression pattern and cell lineage differentiation. J Biol Chem. 2001;276:25279–25286. doi: 10.1074/jbc.M101233200. [DOI] [PubMed] [Google Scholar]
- 76.Hagman DK, Hays LB, Parazzoli SD, Poitout V. Palmitate inhibits insulin gene expression by altering pdx-1 nuclear localization and reducing mafa expression in isolated rat islets of langerhans. J Biol Chem. 2005;280:32413–32418. doi: 10.1074/jbc.M506000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharma A, Zangen DH, Reitz P, Taneja M, Lissauer ME, Miller CP, Weir GC, Habener JF, Bonner-Weir S. The homeodomain protein idx-1 increases after an early burst of proliferation during pancreatic regeneration. Diabetes. 1999;48:507–513. doi: 10.2337/diabetes.48.3.507. [DOI] [PubMed] [Google Scholar]
- 78.Zhou J, Pineyro MA, Wang X, Doyle ME, Egan JM. Exendin-4 differentiation of a human pancreatic duct cell line into endocrine cells: Involvement of pdx-1 and hnf3beta transcription factors. J Cell Physiol. 2002;192:304–314. doi: 10.1002/jcp.10143. [DOI] [PubMed] [Google Scholar]
- 79.Koizumi M, Doi R, Fujimoto K, Ito D, Toyoda E, Mori T, Kami K, Kawaguchi Y, Gittes GK, Imamura M. Pancreatic epithelial cells can be converted into insulin-producing cells by glp-1 in conjunction with virus-mediated gene transfer of pdx-1. Surgery. 2005;138:125–133. doi: 10.1016/j.surg.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 80.Bai L, Meredith G, Tuch BE. Glucagon-like peptide-1 enhances production of insulin in insulin-producing cells derived from mouse embryonic stem cells. J Endocrinol. 2005;186:343–352. doi: 10.1677/joe.1.06078. [DOI] [PubMed] [Google Scholar]
- 81.Holz GG, Chepurny OG. Diabetes outfoxed by glp-1? Sci STKE. 2005;2005:pe2. doi: 10.1126/stke.2682005pe2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yue F, Cui L, Johkura K, Ogiwara N, Sasaki K. Glucagon-like peptide-1 differentiation of primate embryonic stem cells into insulin-producing cells. Tissue Eng. 2006;12:2105–2116. doi: 10.1089/ten.2006.12.2105. [DOI] [PubMed] [Google Scholar]
- 83.Jetton TL, Lausier J, LaRock K, Trotman WE, Larmie B, Habibovic A, Peshavaria M, Leahy JL. Mechanisms of compensatory beta-cell growth in insulin-resistant rats: Roles of akt kinase. Diabetes. 2005;54:2294–2304. doi: 10.2337/diabetes.54.8.2294. [DOI] [PubMed] [Google Scholar]
- 84.Ueki K, Okada T, Hu J, Liew CW, Assmann A, Dahlgren GM, Peters JL, Shackman JG, Zhang M, Artner I, Satin LS, Stein R, Holzenberger M, Kennedy RT, Kahn CR, Kulkarni RN. Total insulin and igf-I resistance in pancreatic beta cells causes overt diabetes. Nat Genet. 2006;38:583–588. doi: 10.1038/ng1787. [DOI] [PubMed] [Google Scholar]
- 85.Peshavaria M, Larmie BL, Lausier J, Satish B, Habibovic A, Roskens V, Larock K, Everill B, Leahy JL, Jetton TL. Regulation of pancreatic beta-cell regeneration in the normoglycemic 60% partial-pancreatectomy mouse. Diabetes. 2006;55:3289–3298. doi: 10.2337/db06-0017. [DOI] [PubMed] [Google Scholar]
- 86.Elrick LJ, Docherty K. Phosphorylation-dependent nucleocytoplasmic shuttling of pancreatic duodenal homeobox-1. Diabetes. 2001;50:2244–2252. doi: 10.2337/diabetes.50.10.2244. [DOI] [PubMed] [Google Scholar]
- 87.Da Silva Xavier G, Qian Q, Cullen PJ, Rutter GA. Distinct roles for insulin and insulin-like growth factor-1 receptors in pancreatic beta-cell glucose sensing revealed by RNA silencing. Biochem J. 2004;377:149–158. doi: 10.1042/BJ20031260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kulkarni RN, Jhala US, Winnay JN, Krajewski S, Montminy M, Kahn CR. Pdx-1 haploinsufficiency limits the compensatory islet hyperplasia that occurs in response to insulin resistance. J Clin Invest. 2004;114:828–836. doi: 10.1172/JCI21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Withers DJ, Burks DJ, Towery HH, Altamuro SL, Flint CL, White MF. Irs-2 coordinates igf-1 receptor-mediated beta-cell development and peripheral insulin signalling. Nat Genet. 1999;23:32–40. doi: 10.1038/12631. [DOI] [PubMed] [Google Scholar]
- 90.Heiser PW, Lau J, Taketo MM, Herrera PL, Hebrok M. Stabilization of beta-catenin impacts pancreas growth. Development. 2006;133:2023–2032. doi: 10.1242/dev.02366. [DOI] [PubMed] [Google Scholar]
- 91.Withers DJ, Gutierrez JS, Towery H, Burks DJ, Ren JM, Previs S, Zhang Y, Bernal D, Pons S, Shulman GI, Bonner-Weir S, White MF. Disruption of irs-2 causes type 2 diabetes in mice. Nature. 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 92.Kitamura T, Nakae J, Kitamura Y, Kido Y, Biggs WH, 3rd, Wright CV, White MF, Arden KC, Accili D. The forkhead transcription factor foxo1 links insulin signaling to pdx1 regulation of pancreatic beta cell growth. J Clin Invest. 2002;110:1839–1847. doi: 10.1172/JCI200216857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kushner JA, Ye J, Schubert M, Burks DJ, Dow MA, Flint CL, Dutta S, Wright CV, Montminy MR, White MF. Pdx1 restores beta cell function in irs2 knockout mice. J Clin Invest. 2002;109:1193–1201. doi: 10.1172/JCI14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ko SH, Suh SH, Kim BJ, Ahn YB, Song KH, Yoo SJ, Son HS, Cha BY, Lee KW, Son HY, Kang SK, Bonner-Weir S, Weir GC, Yoon KH, Park CG. Expression of the intermediate filament vimentin in proliferating duct cells as a marker of pancreatic precursor cells. Pancreas. 2004;28:121–128. doi: 10.1097/00006676-200403000-00002. [DOI] [PubMed] [Google Scholar]
- 95.Taguchi M, Yamaguchi T, Otsuki M. Induction of pdx-1-positive cells in the main duct during regeneration after acute necrotizing pancreatitis in rats. J Pathol. 2002;197:638–646. doi: 10.1002/path.1134. [DOI] [PubMed] [Google Scholar]
- 96.Bonner-Weir S, Taneja M, Weir GC, Tatarkiewicz K, Song KH, Sharma A, O'Neil JJ. In vitro cultivation of human islets from expanded ductal tissue. Proc Natl Acad Sci U S A. 2000;97:7999–8004. doi: 10.1073/pnas.97.14.7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bonner-Weir S, Toschi E, Inada A, Reitz P, Fonseca SY, Aye T, Sharma A. The pancreatic ductal epithelium serves as a potential pool of progenitor cells. Pediatr Diabetes. 2004;5(Suppl 2):16–22. doi: 10.1111/j.1399-543X.2004.00075.x. [DOI] [PubMed] [Google Scholar]
- 98.Inada A, Nienaber C, Sharma A, Bonner-Weir S. Lineage tracing shows pancreatic ductal cells as islet progenitors in postnatal mice. American Diabetes Association, 66th Annual Scientific Sessions; Washington, D.C.. 2006. [Google Scholar]
- 99.Kritzik MR, Jones E, Chen Z, Krakowski M, Krahl T, Good A, Wright C, Fox H, Sarvetnick N. Pdx-1 and msx-2 expression in the regenerating and developing pancreas. J Endocrinol. 1999;163:523–530. doi: 10.1677/joe.0.1630523. [DOI] [PubMed] [Google Scholar]
- 100.Deramaudt TB, Sachdeva MM, Wescott MP, Chen Y, Stoffers DA, Rustgi AK. The pdx1 homeodomain transcription factor negatively regulates the pancreatic ductal cell-specific keratin 19 promoter. J Biol Chem. 2006;281:38385–38395. doi: 10.1074/jbc.M605891200. [DOI] [PubMed] [Google Scholar]
- 101.Heimberg H, Bouwens L, Heremans Y, Van De Casteele M, Lefebvre V, Pipeleers D. Adult human pancreatic duct and islet cells exhibit similarities in expression and differences in phosphorylation and complex formation of the homeodomain protein ipf-1. Diabetes. 2000;49:571–579. doi: 10.2337/diabetes.49.4.571. [DOI] [PubMed] [Google Scholar]
- 102.Heremans Y, Van De Casteele M, in't Veld P, Gradwohl G, Serup P, Madsen O, Pipeleers D, Heimberg H. Recapitulation of embryonic neuroendocrine differentiation in adult human pancreatic duct cells expressing neurogenin 3. J Cell Biol. 2002;159:303–312. doi: 10.1083/jcb.200203074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gasa R, Mrejen C, Leachman N, Otten M, Barnes M, Wang J, Chakrabarti S, Mirmira R, German M. Proendocrine genes coordinate the pancreatic islet differentiation program in vitro. Proc Natl Acad Sci U S A. 2004;101:13245–13250. doi: 10.1073/pnas.0405301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rooman I, Heremans Y, Heimberg H, Bouwens L. Modulation of rat pancreatic acinoductal transdifferentiation and expression of pdx-1 in vitro. Diabetologia. 2000;43:907–914. doi: 10.1007/s001250051468. [DOI] [PubMed] [Google Scholar]
- 105.Mashima H, Ohnishi H, Wakabayashi K, Mine T, Miyagawa J, Hanafusa T, Seno M, Yamada H, Kojima I. Betacellulin and activin a coordinately convert amylase-secreting pancreatic ar42j cells into insulin-secreting cells. J Clin Invest. 1996;97:1647–1654. doi: 10.1172/JCI118591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhou J, Wang X, Pineyro MA, Egan JM. Glucagon-like peptide 1 and exendin-4 convert pancreatic ar42j cells into glucagon- and insulin-producing cells. Diabetes. 1999;48:2358–2366. doi: 10.2337/diabetes.48.12.2358. [DOI] [PubMed] [Google Scholar]
- 107.Desai BM, Oliver-Krasinski J, De Leon DD, Farzad C, Hong N, Leach SD, Stoffers DA. Preexisting pancreatic acinar cells contribute to acinar cell, but not islet beta cell, regeneration. J Clin Invest. 2007;117:971–977. doi: 10.1172/JCI29988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Macfarlane WM, McKinnon CM, Felton-Edkins ZA, Cragg H, James RF, Docherty K. Glucose stimulates translocation of the homeodomain transcription factor pdx1 from the cytoplasm to the nucleus in pancreatic beta-cells. J Biol Chem. 1999;274:1011–1016. doi: 10.1074/jbc.274.2.1011. [DOI] [PubMed] [Google Scholar]
- 109.Moede T, Leibiger B, Pour HG, Berggren P, Leibiger IB. Identification of a nuclear localization signal, rrmkwkk, in the homeodomain transcription factor pdx-1. FEBS Lett. 1999;461:229–234. doi: 10.1016/s0014-5793(99)01446-5. [DOI] [PubMed] [Google Scholar]
- 110.An R, da Silva Xavier G, Hao HX, Semplici F, Rutter J, Rutter GA. Regulation by per-arnt-sim (pas) kinase of pancreatic duodenal homeobox-1 nuclear import in pancreatic beta-cells. Biochem Soc Trans. 2006;34:791–793. doi: 10.1042/BST0340791. [DOI] [PubMed] [Google Scholar]
- 111.Boucher MJ, Selander L, Carlsson L, Edlund H. Phosphorylation marks ipf1/pdx1 protein for degradation by glycogen synthase kinase 3-dependent mechanisms. J Biol Chem. 2006;281:6395–6403. doi: 10.1074/jbc.M511597200. [DOI] [PubMed] [Google Scholar]
- 112.Khoo S, Griffen SC, Xia Y, Baer RJ, German MS, Cobb MH. Regulation of insulin gene transcription by erk1 and erk2 in pancreatic beta cells. J Biol Chem. 2003;278:32969–32977. doi: 10.1074/jbc.M301198200. [DOI] [PubMed] [Google Scholar]
- 113.Mosley AL, Corbett JA, Ozcan S. Glucose regulation of insulin gene expression requires the recruitment of p300 by the beta-cell-specific transcription factor pdx-1. Mol Endocrinol. 2004;18:2279–2290. doi: 10.1210/me.2003-0463. [DOI] [PubMed] [Google Scholar]
- 114.Lebrun P, Montminy MR, Van Obberghen E. Regulation of the pancreatic duodenal homeobox-1 protein by DNA-dependent protein kinase. J Biol Chem. 2005;280:38203–38210. doi: 10.1074/jbc.M504842200. [DOI] [PubMed] [Google Scholar]
- 115.Kishi A, Nakamura T, Nishio Y, Maegawa H, Kashiwagi A. Sumoylation of pdx1 is associated with its nuclear localization and insulin gene activation. Am J Physiol Endocrinol Metab. 2003;284:E830–40. doi: 10.1152/ajpendo.00390.2002. [DOI] [PubMed] [Google Scholar]
- 116.Gao Y, Miyazaki J, Hart GW. The transcription factor pdx-1 is post-translationally modified by o-linked n-acetylglucosamine and this modification is correlated with its DNA binding activity and insulin secretion in min6 beta-cells. Arch Biochem Biophys. 2003;415:155–163. doi: 10.1016/s0003-9861(03)00234-0. [DOI] [PubMed] [Google Scholar]
- 117.Rafiq I, da Silva Xavier G, Hooper S, Rutter GA. Glucose-stimulated preproinsulin gene expression and nuclear trans-location of pancreatic duodenum homeobox-1 require activation of phosphatidylinositol 3-kinase but not p38 mapk/sapk2. J Biol Chem. 2000;275:15977–15984. doi: 10.1074/jbc.275.21.15977. [DOI] [PubMed] [Google Scholar]
- 118.Wu H, MacFarlane WM, Tadayyon M, Arch JR, James RF, Docherty K. Insulin stimulates pancreatic-duodenal homoeobox factor-1 (pdx1) DNA-binding activity and insulin promoter activity in pancreatic beta cells. Biochem J. 1999;344(Pt 3):813–818. [PMC free article] [PubMed] [Google Scholar]
- 119.Robertson LA, Kim AJ, Werstuck GH. Mechanisms linking diabetes mellitus to the development of atherosclerosis: A role for endoplasmic reticulum stress and glycogen synthase kinase-3. Can J Physiol Pharmacol. 2006;84:39–48. doi: 10.1139/Y05-142. [DOI] [PubMed] [Google Scholar]
- 120.Liberzon A, Ridner G, Walker MD. Role of intrinsic DNA binding specificity in defining target genes of the mammalian transcription factor pdx1. Nucleic Acids Res. 2004;32:54–64. doi: 10.1093/nar/gkh156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Taylor DG, Babu D, Mirmira RG. The c-terminal domain of the beta cell homeodomain factor nkx6.1 enhances sequence-selective DNA binding at the insulin promoter. Biochemistry. 2005;44:11269–11278. doi: 10.1021/bi050821m. [DOI] [PubMed] [Google Scholar]
- 122.Glick E, Leshkowitz D, Walker MD. Transcription factor beta2 acts cooperatively with e2a and pdx1 to activate the insulin gene promoter. J Biol Chem. 2000;275:2199–2204. doi: 10.1074/jbc.275.3.2199. [DOI] [PubMed] [Google Scholar]
- 123.Ohneda K, Mirmira RG, Wang J, Johnson JD, German MS. The homeodomain of pdx-1 mediates multiple protein-protein interactions in the formation of a transcriptional activation complex on the insulin promoter. Mol Cell Biol. 2000;20:900–911. doi: 10.1128/mcb.20.3.900-911.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Francis J, Babu DA, Deering TG, Chakrabarti SK, Garmey JC, Evans-Molina C, Taylor DG, Mirmira RG. Role of chromatin accessibility in the occupancy and transcription of the insulin gene by the pancreatic and duodenal homeobox factor 1. Mol Endocrinol. 2006;20:3133–3145. doi: 10.1210/me.2006-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Longo A, Guanga GP, Rose RB. Structural basis for induced fit mechanisms in DNA recognition by the pdx1 homeodomain. Biochemistry. 2007;46:2948–2957. doi: 10.1021/bi060969l. [DOI] [PubMed] [Google Scholar]
- 126.Le Lay J, Matsuoka TA, Henderson E, Stein R. Identification of a novel pdx-1 binding site in the human insulin gene enhancer. J Biol Chem. 2004;279:22228–22235. doi: 10.1074/jbc.M312673200. [DOI] [PubMed] [Google Scholar]
- 127.Lottmann H, Vanselow J, Hessabi B, Walther R. The tet-on system in transgenic mice: Inhibition of the mouse pdx-1 gene activity by antisense RNA expression in pancreatic beta-cells. J Mol Med. 2001;79:321–328. doi: 10.1007/s001090100229. [DOI] [PubMed] [Google Scholar]
- 128.Waeber G, Thompson N, Nicod P, Bonny C. Transcriptional activation of the glut2 gene by the ipf-1/stf-1/idx-1 homeobox factor. Mol Endocrinol. 1996;10:1327–1334. doi: 10.1210/mend.10.11.8923459. [DOI] [PubMed] [Google Scholar]
- 129.Watada H, Kajimoto Y, Miyagawa J, Hanafusa T, Hamaguchi K, Matsuoka T, Yamamoto K, Matsuzawa Y, Kawamori R, Yamasaki Y. Pdx-1 induces insulin and glucokinase gene expressions in alphatc1 clone 6 cells in the presence of betacellulin. Diabetes. 1996;45:1826–1831. doi: 10.2337/diab.45.12.1826. [DOI] [PubMed] [Google Scholar]
- 130.Watada H, Kajimoto Y, Umayahara Y, Matsuoka T, Kaneto H, Fujitani Y, Kamada T, Kawamori R, Yamasaki Y. The human glucokinase gene beta-cell-type promoter: An essential role of insulin promoter factor 1/pdx-1 in its activation in hit-t15 cells. Diabetes. 1996;45:1478–1488. doi: 10.2337/diab.45.11.1478. [DOI] [PubMed] [Google Scholar]
- 131.Macfarlane WM, Campbell SC, Elrick LJ, Oates V, Bermano G, Lindley KJ, Aynsley-Green A, Dunne MJ, James RF, Docherty K. Glucose regulates islet amyloid polypeptide gene transcription in a pdx1- and calcium-dependent manner. J Biol Chem. 2000;275:15330–15335. doi: 10.1074/jbc.M908045199. [DOI] [PubMed] [Google Scholar]
- 132.Watada H, Kajimoto Y, Kaneto H, Matsuoka T, Fujitani Y, Miyazaki J, Yamasaki Y. Involvement of the homeodomain-containing transcription factor pdx-1 in islet amyloid polypeptide gene transcription. Biochem Biophys Res Commun. 1996;229:746–751. doi: 10.1006/bbrc.1996.1875. [DOI] [PubMed] [Google Scholar]
- 133.Smith SB, Watada H, Scheel DW, Mrejen C, German MS. Autoregulation and maturity onset diabetes of the young transcription factors control the human pax4 promoter. J Biol Chem. 2000;275:36910–36919. doi: 10.1074/jbc.M005202200. [DOI] [PubMed] [Google Scholar]
- 134.Schisler JC, Jensen PB, Taylor DG, Becker TC, Knop FK, Takekawa S, German M, Weir GC, Lu D, Mirmira RG, Newgard CB. The nkx6.1 homeodomain transcription factor suppresses glucagon expression and regulates glucose-stimulated insulin secretion in islet beta cells. Proc Natl Acad Sci U S A. 2005;102:7297–7302. doi: 10.1073/pnas.0502168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Annicotte JS, Fayard E, Swift GH, Selander L, Edlund H, Tanaka T, Kodama T, Schoonjans K, Auwerx J. Pancreatic-duodenal homeobox 1 regulates expression of liver receptor homolog 1 during pancreas development. Mol Cell Biol. 2003;23:6713–6724. doi: 10.1128/MCB.23.19.6713-6724.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Peshavaria M, Henderson E, Sharma A, Wright CV, Stein R. Functional characterization of the transactivation properties of the pdx-1 homeodomain protein. Mol Cell Biol. 1997;17:3987–3996. doi: 10.1128/mcb.17.7.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Petersen HV, Peshavaria M, Pedersen AA, Philippe J, Stein R, Madsen OD, Serup P. Glucose stimulates the activation domain potential of the pdx-1 homeodomain transcription factor. FEBS Lett. 1998;431:362–366. doi: 10.1016/s0014-5793(98)00776-5. [DOI] [PubMed] [Google Scholar]
- 138.Lu M, Miller C, Habener JF. Functional regions of the homeodomain protein idx-1 required for transactivation of the rat somatostatin gene. Endocrinology. 1996;137:2959–2967. doi: 10.1210/endo.137.7.8770920. [DOI] [PubMed] [Google Scholar]
- 139.Chakrabarti SK, Mirmira RG. Transcription factors direct the development and function of pancreatic beta cells. Trends Endocrinol Metab. 2003;14:78–84. doi: 10.1016/s1043-2760(02)00039-5. [DOI] [PubMed] [Google Scholar]
- 140.Peers B, Leonard J, Sharma S, Teitelman G, Montminy MR. Insulin expression in pancreatic islet cells relies on cooperative interactions between the helix loop helix factor e47 and the homeobox factor stf-1. Mol Endocrinol. 1994;8:1798–1806. doi: 10.1210/mend.8.12.7708065. [DOI] [PubMed] [Google Scholar]
- 141.Qiu Y, Guo M, Huang S, Stein R. Insulin gene transcription is mediated by interactions between the p300 coactivator and pdx-1, beta2, and e47. Mol Cell Biol. 2002;22:412–420. doi: 10.1128/MCB.22.2.412-420.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Docherty HM, Hay CW, Ferguson LA, Barrow J, Durward E, Docherty K. Relative contribution of pdx-1, mafa and e47/beta2 to the regulation of the human insulin promoter. Biochem J. 2005;389:813–820. doi: 10.1042/BJ20041891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Swift GH, Liu Y, Rose SD, Bischof LJ, Steelman S, Buchberg AM, Wright CV, MacDonald RJ. An endocrine-exocrine switch in the activity of the pancreatic homeodomain protein pdx1 through formation of a trimeric complex with pbx1b and mrg1 (meis2) Mol Cell Biol. 1998;18:5109–5120. doi: 10.1128/mcb.18.9.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu Y, MacDonald RJ, Swift GH. DNA binding and transcriptional activation by a pdx1.pbx1b.meis2b trimer and cooperation with a pancreas-specific basic helix-loop-helix complex. J Biol Chem. 2001;276:17985–17993. doi: 10.1074/jbc.M100678200. [DOI] [PubMed] [Google Scholar]
- 145.Mosley AL, Ozcan S. Glucose regulates insulin gene transcription by hyperacetylation of histone h4. J Biol Chem. 2003;278:19660–19666. doi: 10.1074/jbc.M212375200. [DOI] [PubMed] [Google Scholar]
- 146.Stanojevic V, Habener JF, Thomas MK. Pancreas duodenum homeobox-1 transcriptional activation requires interactions with p300. Endocrinology. 2004;145:2918–2928. doi: 10.1210/en.2003-1188. [DOI] [PubMed] [Google Scholar]
- 147.Francis J, Chakrabarti SK, Garmey JC, Mirmira RG. Pdx-1 links histone H3-Lys-4 methylation to RNA polymerase II elongation during activation of insulin transcription. J Biol Chem. 2005;280:36244–36253. doi: 10.1074/jbc.M505741200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stanojevic V, Yao KM, Thomas MK. The coactivator bridge-1 increases transcriptional activation by pancreas duodenum homeobox-1 (pdx-1) Mol Cell Endocrinol. 2005;237:67–74. doi: 10.1016/j.mce.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 149.Liu A, Desai BM, Stoffers DA. Identification of pcif1, a poz domain protein that inhibits pdx-1 (mody4) transcriptional activity. Mol Cell Biol. 2004;24:4372–4383. doi: 10.1128/MCB.24.10.4372-4383.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mosley AL, Ozcan S. The pancreatic duodenal homeobox-1 protein (pdx-1) interacts with histone deacetylases hdac-1 and hdac-2 on low levels of glucose. J Biol Chem. 2004;279:54241–54247. doi: 10.1074/jbc.M410379200. [DOI] [PubMed] [Google Scholar]
- 151.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 152.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 153.Iguchi H, Ikeda Y, Okamura M, Tanaka T, Urashima Y, Ohguchi H, Takayasu S, Kojima N, Iwasaki S, Ohashi R, Jiang S, Hasegawa G, Ioka RX, Magoori K, Sumi K, Maejima T, Uchida A, Naito M, Osborne TF, Yanagisawa M, Yamamoto TT, Kodama T, Sakai J. Sox6 attenuates glucose-stimulated insulin secretion by repressing pdx1 transcriptional activity and is down-regulated in hyperinsulinemic obese mice. J Biol Chem. 2005;280:37669–37680. doi: 10.1074/jbc.M505392200. [DOI] [PubMed] [Google Scholar]
- 154.Evans-Molina C, Garmey JC, Ketchum RJ, Brayman K, Deng S, Mirmira RG. Glucose regulation of insulin gene transcription and pre-mrna processing in human islets. Diabetes. 2007;56:827–835. doi: 10.2337/db06-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kadonaga JT. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 156.Hampsey M, Reinberg D. Tails of intrigue: Phosphorylation of RNA polymerase II mediates histone methylation. Cell. 2003;113:429–432. doi: 10.1016/s0092-8674(03)00360-x. [DOI] [PubMed] [Google Scholar]
- 157.Narayan KM, Boyle JP, Thompson TJ, Sorensen SW, Williamson DF. Lifetime risk for diabetes mellitus in the united states. JAMA. 2003;290:1884–1890. doi: 10.1001/jama.290.14.1884. [DOI] [PubMed] [Google Scholar]
- 158.Ferber S, Halkin A, Cohen H, Ber I, Einav Y, Goldberg I, Barshack I, Seijffers R, Kopolovic J, Kaiser N, Karasik A. Pancreatic and duodenal homeobox gene 1 induces expression of insulin genes in liver and ameliorates streptozotocin-induced hyperglycemia. Nat Med. 2000;6:568–572. doi: 10.1038/75050. [DOI] [PubMed] [Google Scholar]
- 159.Kojima H, Fujimiya M, Matsumura K, Younan P, Imaeda H, Maeda M, Chan L. Neurod-betacellulin gene therapy induces islet neogenesis in the liver and reverses diabetes in mice. Nat Med. 2003;9:596–603. doi: 10.1038/nm867. [DOI] [PubMed] [Google Scholar]
- 160.Cao LZ, Tang DQ, Horb ME, Li SW, Yang LJ. High glucose is necessary for complete maturation of pdx1-vp16-expressing hepatic cells into functional insulin-producing cells. Diabetes. 2004;53:3168–3178. doi: 10.2337/diabetes.53.12.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kaneto H, Nakatani Y, Miyatsuka T, Matsuoka TA, Matsuhisa M, Hori M, Yamasaki Y. Pdx-1/vp16 fusion protein, together with neurod or ngn3, markedly induces insulin gene transcription and ameliorates glucose tolerance. Diabetes. 2005;54:1009–1022. doi: 10.2337/diabetes.54.4.1009. [DOI] [PubMed] [Google Scholar]
- 162.Noguchi H, Kaneto H, Weir GC, Bonner-Weir S. Pdx-1 protein containing its own antennapedia-like protein transduction domain can transduce pancreatic duct and islet cells. Diabetes. 2003;52:1732–1737. doi: 10.2337/diabetes.52.7.1732. [DOI] [PubMed] [Google Scholar]
- 163.Noguchi H, Matsushita M, Matsumoto S, Lu YF, Matsui H, Bonner-Weir S. Mechanism of pdx-1 protein transduction. Biochem Biophys Res Commun. 2005;332:68–74. doi: 10.1016/j.bbrc.2005.04.092. [DOI] [PubMed] [Google Scholar]



