Abstract
Posttranslational mechanisms are implicated in the development of epithelial cell polarity, but little is known about the patterns of gene expression and transcriptional regulation during this process. We characterized temporal patterns of gene expression during cell–cell adhesion-initiated polarization of cultured human Caco-2 cells, which develop structural and functional polarity resembling enterocytes in vivo. A distinctive switch in gene expression patterns occurred upon formation of cell–cell contacts. Comparison to gene expression patterns in normal human colon and colon tumors revealed that the pattern in proliferating, nonpolarized Caco-2 cells paralleled patterns seen in human colon cancer in vivo, including expression of genes involved in cell proliferation. The pattern switched in polarized Caco-2 cells to one more closely resembling that in normal colon tissue, indicating that regulation of transcription underlying Caco-2 cell polarization is similar to that during enterocyte differentiation in vivo. Surprisingly, the temporal program of gene expression in polarizing Caco-2 cells involved changes in signaling pathways (e.g., Wnt, Hh, BMP, FGF) in patterns similar to those during migration and differentiation of intestinal epithelial cells in vivo, despite the absence of morphogen gradients and interactions with stromal cells characteristic of enterocyte differentiation in situ. The full data set is available at http://microarray-pubs.stanford.edu/CACO2.
INTRODUCTION
In the intestine, enterocytes are generated from stem cells located in crypts between the villi (Nishimura et al., 2003), become postmitotic and differentiate as they migrate from the crypt up the villus, and finally undergo apoptosis and are sloughed off from the villus tip, a process that takes ∼5 d (Heath, 1996). Morphogens including Wnts and transforming growth factor β (TGFβ), and Ephrin and Notch guidance cues are expressed in gradients along the crypt-villus axis and are required for normal regulation of the cell cycle, cell exit from the stem cell niche, and cell migration and differentiation (Sancho et al., 2004).
Spatially regulated Wnt signaling is critical for maintaining the intestinal epithelial stem cell population and for appropriately regulated differentiation (Cadigan and Nusse, 1997; Polakis, 2000; Nelson and Nusse, 2004; Clevers, 2006). Several Wnt isoforms are secreted by cells surrounding the crypt resulting in a Wnt gradient that decreases from the crypt to villus (van de Wetering et al., 2002; Pinto et al., 2003; Gregorieff et al., 2005). Wnt signaling through Frizzled receptors inhibits β-catenin destruction (Polakis, 2000), leading to its stabilization, nuclear translocation and heterodimerization with proteins of the transcription factor (TCF)/lymphoid enhancer-binding factor (LEF) family to form a transcriptional activator of Wnt target genes (Behrens et al., 1996; Molenaar et al., 1996). Several of these target genes are involved in the maintenance of a self-renewing stem cell population (van de Wetering et al., 2002; Pinto and Clevers, 2005; Crosnier et al., 2006). In the absence of Wnt, the β-catenin destruction complex, composed of the adenomatous polyposis coli (APC) and Axin scaffolding proteins (Behrens et al., 1998; Hart et al., 1998; Ikeda et al., 1998; Fagotto et al., 1999) and the serine/threonine kinases Casein Kinase I (CKI) and Glycogen Synthase Kinase-3β (GSK3β; Ikeda et al., 2000; Gao et al., 2002), phosphorylates β-catenin, thereby targeting it for degradation by the proteosome (Aberle et al., 1997; Marikawa and Elinson, 1998; Kitagawa et al., 1999). Additional regulation of Wnt signaling occurs extracellularly through secreted inhibitors including the Secreted Frizzled-Related Protein (SFRP) family and Dickkopf, and in the nucleus by inhibitors including the transducin-like enhancer of split (TLE) Groucho homologues and activators (Clevers, 2006). Mutant, truncated forms of APC are unable to form a functional destruction complex, resulting in the accumulation of β-catenin and inappropriate expression of Wnt target genes, and are a common early event in development of familial and sporadic colon cancers (Fearon and Vogelstein, 1990; Groden et al., 1991; Powell et al., 1992; Fodde et al., 2001; Fodde, 2002).
Wnt can also activate a noncanonical, or β-catenin–independent signaling pathway (Veeman et al., 2003). This pathway is also known as the Wnt/Ca2+ pathway because some Wnt family proteins can trigger Ca2+ release, activating calcium-sensitive kinases such as protein kinase C (PKC) and calmodulin-dependent kinase II (CaMKII), and via CaMKII, TAK1 and NLK mitogen-activated protein (MAP) kinases (Ishitani et al., 2003). Phosphorylation of TCF by TAK1/NLK negatively regulates its activity (Ishitani et al., 1999; Meneghini et al., 1999). The noncanonical Wnt pathway may converge with the canonical Wnt cascade to modulate TCF activity. Studies in Caenorhabditis elegans have shown that POP-1 (the homolog of mammalian TCF) relocalizes from the nucleus to the cytoplasm when phosphorylated (Rocheleau et al., 1999). The ability of Wnt to activate the TAK1/NLK pathway (Smit et al., 2004) suggests that this MAP kinase pathway could provide a means of Wnt signal-dependent negative control of the canonical Wnt signaling pathway.
Other signaling pathways act in conjunction with the Wnt pathway to pattern and maintain the intestinal epithelium (Sancho et al., 2004). The Notch signaling pathway may play a role in inhibiting proliferation and promoting exit from the cell cycle (Fre et al., 2005; van Es et al., 2005). Sonic Hedgehog (Shh), acting through three different transmembrane proteins, Smoothened (SMO), Patched (PTCH), and Hedgehog-interacting protein (HIP), has been shown to regulate intestinal homeostasis (Fukuda and Yasugi, 2002; Katoh and Katoh, 2006a). Indian Hedgehog (IHH), a member of the Shh protein family, regulates differentiation of mature colon cells by antagonizing Wnt signaling (van den Brink et al., 2004). Bone morphogenic protein (BMP)/transforming growth factor β (TGFβ), which signals through a cognate family of serine-threonine kinase receptors leading to activation of Smad transcription factors, participates in regulating diverse cellular processes including proliferation, differentiation, and apoptosis (Massague and Gomis, 2006). Fibroblast growth factor (FGF) signaling modulates both cell cycle arrest and differentiation depending on cell type (Dailey et al., 2005). Epidermal growth factor (EGF) has a role in intestinal epithelial cell growth and differentiation (Wong and Wright, 1999). Finally, the ephrin ligand-receptor system is essential for the positioning of cells along the crypt-villus axis, a process that is coupled to intestinal cell proliferation and differentiation by the Wnt signaling pathway (Batlle et al., 2002).
Although progress has been made in understanding the roles of these signaling morphogens in the intestine, it remains unclear whether they have any direct roles in regulating the structural and functional polarization of enterocytes. Because of the complex architecture of the intestine it is difficult to study the development of cell polarity during enterocyte differentiation in situ. However, enterocyte polarization can be studied in vitro using Caco-2 cells, which are derived from a human colon adenocarcinoma but retain the ability to polarize and form a transporting epithelial monolayer in cell culture (Grasset et al., 1985; Wice et al., 1985). Caco-2 cells develop structural and functional polarity during 21 d in culture, and the resulting monolayer has many characteristics of polarized intestinal enterocytes in situ.
Although many studies have focused on posttranslational events involved in protein organization and distribution the global transcriptional program has not been studied (Le Gall et al., 1995; Yeaman et al., 1999; Rodriguez-Boulan et al., 2005). Although it is likely that reprogramming of gene expression programs is important in vivo as cells exit the crypt and start to differentiate, it is less clear whether these changes in gene expression play any role in polarization or to what extent they occur in cells grown in vitro in the absence of signaling from surrounding stromal cells and position-dependent morphogen gradients.
To address these questions, we used cDNA microarrays representing ∼24,500 unique human genes (based on Unigene clusters) to analyze genome-wide patterns of gene expression in Caco-2 cells as they developed from nonpolarized dispersed cells into a postmitotic polarized epithelial monolayer. Considering the lack of surrounding stroma and graded morphogens as well as the adenocarcinoma origin of Caco-2 cells, it might be expected that polarizing Caco-2 cells would have regulatory programs and gene expression patterns quite different from those of normal colon; for example, we might expect aberrant regulation of the Wnt pathway due to expression of a truncated APC in these cells (Ilyas et al., 1997; Chang et al., 2005). Therefore, we also compared the transcriptional program during Caco-2 cell polarization with those in normal and malignant human colon epithelial tissues. This analysis provides evidence of shared molecular targets involved in controlling epithelial cell proliferation and differentiation in the context of assembly of an epithelial layer in vitro and in vivo and in the aberrant growth of intestinal tumors.
MATERIALS AND METHODS
Cell Culture and Growth Conditions
The human intestinal epithelial cell line Caco-2 was obtained from Dr. Stanley Falkow (Stanford University; American Type Culture Collection, Manassas, VA; HTB-37). Cells were first grown in sparse culture on plastic 15-cm dishes (Nunc, Naperville, IL; Cat. no. 168381) to generate a population of “contact naïve” cells. These cells were combined and plated at confluency on rat tail collagen-coated Costar filter inserts (Sigma, St. Louis, MO; Cat. no. 3412) in DMEM (Sigma, Cat. no. D2902) with 10% fetal bovine serum. The day that cells were plated on filters is designated the zero time point. Cells were cultured for 26 d, during which time they develop structural and functional polarity. Media was changed every other day. Triplicate samples were harvested at 11 time points over the time course for microarray analysis and to analyze the development of cell polarity. The entire time course was performed twice.
RNA Isolation and Amplification
Caco-2 cells were lysed in buffer containing guanidine isothiocyanate (e.g., RLT buffer from QIAGEN, Chatsworth, CA) and scraped off the Costar filter inserts with a rubber policeman. Cells were homogenized using a rotor-stator homogenizer (∼45 s), and total RNA was isolated according to the manufacturer's protocol (QIAGEN; Cat. no. 75144). Total RNA from each cell sample was linearly amplified according to the Ambion MessageAmp procedure (Austin, TX; Cat. no. 1750). This amplification procedure was based on antisense RNA (aRNA) amplification first described by Van Gelder and Eberwine (Van Gelder et al., 1990). Total RNA was reversed-transcribed using a primer containing both oligo(dT) and a T7 RNA polymerase promoter sequence. After first-strand synthesis, DNA polymerase and RNase H were used to simultaneously degrade the RNA and synthesize second-strand cDNA. Double-stranded cDNA was next used as the template for in vitro transcription to produce linearly amplified aRNA. Finally, the amplified RNA was used as template for reverse transcription in the presence of CyDye-labeled dNTPs to generate labeled cDNA for microarray analysis. Supplementary Table S1 contains a summary of the samples subjected to array analysis.
Colon Tissue Samples for DNA Microarrays
Frozen tumor and normal colon mucosa were collected from colectomy specimens from Queen Mary Hospital, The University of Hong Kong. Tissue was frozen in liquid nitrogen within 30 min of resection. Nonneoplastic mucosa from colon was dissected free of muscle and histologically confirmed to be tumor free by frozen section. For cancer tissue, tumor purity of over 70% was confirmed by frozen section for each case before submission for RNA extraction. Information on the patient's age and sex, as well as the Duke's tumor stage is included in Supplementary Table S2. Total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA) from each tissue sample and processed for microarray hybridization. This study was approved by the Ethics Committee of The University of Hong Kong and the Internal Review Board of University of California, San Francisco.
cDNA Microarrays, Hybridization, and Data Filtering and Analysis
We used human cDNA microarrays containing 42,000 elements that represent ∼24,500 unique genes (based on Unigene clusters). The arrays were printed at the Stanford Functional Genomic Facility (www.microarray.org). Fluorescently labeled cDNA prepared from amplified RNA was hybridized to the array in a two-color comparative format, with Caco-2 cell samples labeled with Cy-5, and a reference pool of human mRNAs labeled with Cy-3 (Stratagene, La Jolla, CA; Cat. no. 740000). Array images were scanned using an Axon Scanner 4000B (Axon Instruments, Union City, CA), and data were analyzed using GenePix 3.0 (Axon Instruments). Data were normalized and retrieved as the log2 ratio of fluorescence intensities of the sample (Cy5) and the reference (Cy3). The ratio of intensities of the two fluors (Cy5/Cy3) at each microarray element provides a standardized measure of the abundance of the corresponding mRNA. Data were next filtered to exclude elements that did not have at least a twofold intensity over background ratio in the Cy3- or the Cy5-channel in at least 80% of the arrays. Detailed microarray protocols are available at http://brownlab.stanford.edu/protocols.html.
Genes that were differentially expressed upon in vitro epithelial cell polarization were analyzed by the two-class (unpaired) significance analysis of microarrays (SAM) algorithm (Tusher et al., 2001) to select a set of ∼4000 cDNA elements that were consistently differentially expressed between day 0 and day 14 in cultured Caco-2 cells, with a false discovery rate (FDR) <5%. This set of ∼4000 cDNA elements was next used to extract patterns of gene expression from the normal and tumor human colonic data sets described above.
Epithelial Cell Fractionation
Caco-2 cells grown as described above were homogenized as in Yeaman et al. (2004) and centrifuged at 2000 rpm for 5 min to pellet nuclei. The postnuclear supernatant was centrifuged at 100,000 × g for 45 min. The resulting membrane pellet was washed twice and resuspended. NP-40 was added to both nuclear and membrane fractions to a final concentration of 0.5%, followed by dounce homogenization.
Immunoblot and Immunoprecipitation
Equal protein from each subcellular fraction was separated by SDS-PAGE and transferred onto nitrocellulose. Membranes were probed with a β-catenin mAb (Transduction Laboratories, Lexington, KY) and E-cadherin polyclonal (Hinck et al., 1994a) so that membrane contamination within nuclear and cytoplasmic fractions could be corrected for. Experiments were done in triplicate. Immunoprecipitation (IP) of nuclear fractions was performed as in Barker et al. (1999) using monoclonal TCF4 antibody from Chemicon (Temecula, CA) and anti-GFP mAb (Molecular Probes, Eugene, OR) as a control. IPs were probed with β-catenin polyclonal antibody (Hinck et al., 1994b).
Target Validation
Select gene profiles were validated using semiquantitative RT-PCR from RNA isolated as described above. All reactions were done using SuperScript III One-Step RT-PCR Kit with Platinum Taq (Invitrogen), following the manufacturer's specifications. Between 8–16 pg total RNA (depending on the target) was added to each reaction, and the annealing temperature was 55°C. Primer sequences were as follows: (5′ to 3′) flFZD4: GGC ATG GAG GTG TTT CTG AA (sense), GAC ATC GAT GCA GAC ATA CT (anti-sense); sFZD4: TTA TTT CAA GTG TTG TGC AAA GAG G (sense), GAA TGC TTC CAG GCA ATC TA (anti-sense); EGF: GTA GCC AGC TCT GCG TTC CTC TTA (sense), GAA TCT ACG GCC AAT CCA GTC CAC (anti-sense); and EGFR: ACT GCT GGG TGC GGA AGA GAA AGA (sense), CAG GGC ACG GTA GAA GTT GGA GTC (anti-sense).
RESULTS
Overview of Approach and Gene Expression Patterns during Development of Cell Polarity
To synchronize the development of cell polarity, Caco-2 cells were first grown at a sparse density to generate a population of “contact naïve” cells. Cells were then plated at confluent density on Costar filter inserts (Acton, MA) to initiate synchronous monolayer formation, and allowed to adhere for 4 h at which point the first samples were collected (designated 0 h). Cells were cultured for 26 d during which time RNA was collected and the development of structural and functional polarity was assessed (see accompanying article in this issue, Halbleib et al., 2007). Triplicate samples were harvested at 11 time points for analysis of gene expression, and the entire time course was performed twice. Because this experimental design allowed us to both evaluate trends in transcript expression over time and calculate error measurements for individual time points, additional validation was performed only on a few select targets.
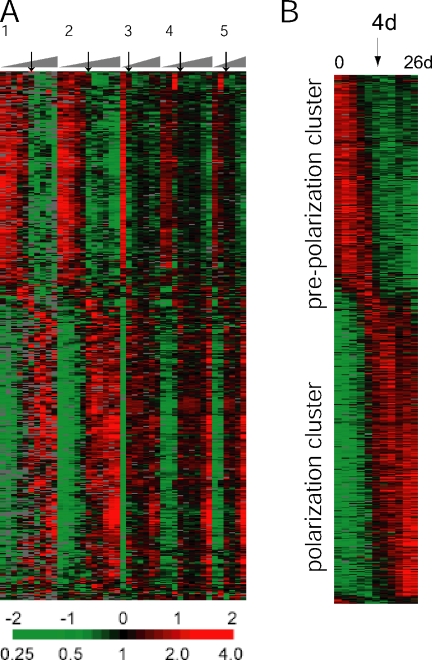
Total RNA was isolated from each cell sample, linearly amplified, and then processed for microarray hybridization. The transcriptional profile of five of these sample sets was analyzed with human cDNA microarrays as illustrated in Figure 1A. An overview of all cell samples for microarray analysis is shown in Supplementary Table S1. Each hybridization compared Cy5-labeled cDNA reverse-transcribed from Caco-2 amplified RNA with Cy3-labeled cDNA reverse-transcribed from a reference mRNA; this reference sample was a combined pool of mRNA isolated from 10 different human cell lines. The ratio of intensity of the two fluorophores (Cy5/Cy3) at each microarray element provided a standardized measure of the abundance of the corresponding mRNA. A total of 513 genes, whose transcript levels differed by at least threefold from the mean across all time points in at least 3 of the 43 samples, were further analyzed by hierarchical clustering. The abundance of each transcript measured in each Caco-2 sample, relative to its mean abundance across all samples, is represented in Figure 1 using a color key (red for expression levels above the mean for that gene and green for expression levels below the mean). The expression patterns of those genes, organized by hierarchical clustering, are displayed in Figure 1A. In Figure 1B, transcript levels of each of the 513 genes were averaged across the five replicate time-course experiments and sorted by the time at which a major increase in expression occurred (Bozdech et al., 2003). There were two distinct clusters of transcripts separated by an abrupt change in gene program at day 4. Genes expressed while the cells were still dividing and before stabilization of cell–cell contacts are shown in the “prepolarization cluster.” Genes that increased in expression after Caco-2 cells became postmitotic and began to polarize are found in the “polarization cluster.”
Figure 1.
Hierarchical cluster analysis of Caco-2 cell polarization in vitro. (A) Thumbnail overview of 513 genes, whose expression varies in at least three samples by a factor of at least three from the mean across all samples, illustrating the temporally staged transcriptional reprogramming during establishment of polarity in Caco-2 cells. Genes (rows) are organized by hierarchical clustering based on overall similarity in expression pattern, and samples (columns) are in temporal order from left to right. Expression levels of each gene relative to its mean expression level over the time course are displayed in a log2 scale. Relative expression levels above or below the mean over the time course are indicated by red or green, respectively. Five replicate time-course experiments are shown. Arrows indicate the 4-d time point in each individual time course. (B) Transcript levels of each of the 513 genes were averaged across the five replicate time-course experiments and sorted by the time at which a major increase in expression occurred, as described in work by DeRisi and coworkers (Bozdech et al., 2003). The abrupt transition in gene expression profiles at the 4-d time point is indicated with an arrow.
Comparison of the Gene Expression Patterns of Polarizing Caco-2 Cells in Culture and Those of Normal Colon and Colon Cancers
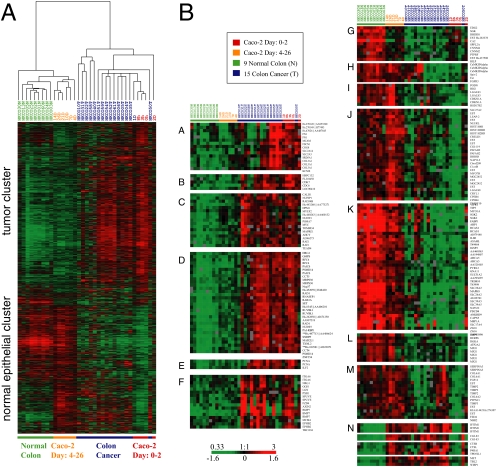
We compared expression patterns of Caco-2 genes with those in normal human colon and primary colon tumors (Figure 2A). A supervised analysis (SAM) was performed to identify genes that were significantly induced or repressed during Caco-2 cell polarization. A set of ∼4000 identified cDNA elements was then used to extract patterns of gene expression from normal human colon tissue and colon tumor data sets. Hierarchical clustering of the Caco-2 time-course samples with normal colon and colon cancer samples based on patterns of expression of these genes, separated the samples into two branches. The first branch comprised the normal colon samples that clustered with Caco-2 samples from 4 to 26 d after initiation of polarization, during which time the cells became postmitotic and developed structural and functional polarity. The second branch showed parallel patterns of expression in colon cancer samples and Caco-2 cells sampled from 0 to 2 d, when the cells were still nonpolarized and proliferating. Hierarchical clustering organized the genes (rows) into two main groups, termed the “tumor cluster” and the “normal epithelial cluster.” Selected examples of genes in these two clusters are illustrated in Figure 2B, cluster A–F and cluster G–M, respectively.
Figure 2.
Comparison between gene expression patterns underlying establishment of Caco-2 cell polarity in vitro to the gene expression signature identified in healthy human colon tissue and colon cancer. (A) Approximately 4000 array elements selected for differential expression by two-class significance analysis of microarrays (SAM) analysis were used to extract patterns of gene expression from normal human colon tissue and colon tumor data sets. Hierarchical cluster analysis of polarizing Caco-2 cells and the healthy colon and colon cancer data sets together clearly separates the samples (columns) into two branches, and genes (rows) into two distinct clusters, demonstrating that Caco-2 cells undergo a transcriptional transition from a gene expression program paralleling that of colon tumor samples (0–2 d in culture; right branch) to a program very similar to that observed in normal human colon tissue (4–26 d in culture; left branch). (B) High-resolution view of genes representing the “normal epithelial cluster” (cluster A–F) and the “tumor cluster” (cluster G–M). Overall, our analysis reveals remarkably similarities in the expression profiles of proliferating Caco-2 cells and colon tumor cells and in postmitotic, polarized Caco-2 cells and normal colon cells. However, some exceptions, which are illustrated in cluster N, were identified. Transcript levels determined by microarray analysis are shown relative to a reference pool of human mRNAs.
The tumor cluster contains sets of genes whose expression is relatively elevated in colonic tumors and early in the Caco-2 cell time course. These genes are involved in the cell cycle, including cell cycle control (CDC6, CDK7; cluster B), cell cycle checkpoints (RAD1 and MAD2L1; cluster D), and DNA synthesis (RCF4 and PCNA; cluster D–E). Also present in the tumor cluster were genes encoding specific extracellular matrix (ECM) components, including fibronectin (FN1), a member of the collagen protein family (COL3A1, cluster A), and an integrin family member (ITGA6, cluster F); signaling components and downstream effectors of the canonical Wnt pathway were also prominent (e.g., AXIN2, EPHB2; cluster F) consistent with activation of the Wnt signaling pathway (see below).
The normal epithelial cluster comprises genes expressed in normal colon and in postmitotic, polarizing Caco-2 cells. These include, first, components characteristic of structurally and functionally polarized enterocytes such as brush border proteins (MYO1A), cell–cell adhesion components (TJP3 and PVRL3), and membrane transporters that facilitate ion (SLC39A5), sulfate (SLC26A2), and sodium phosphate transport (SLC17A4; Figure 2B, cluster K). Second, genes involved in cell cycle inhibition (p21/CDKN1A, cluster I) and apoptosis control (TRAIL/TNFSF10, cluster J) were expressed at higher levels in healthy colon compared with colon cancer, and their expression also increased during Caco-2 cell polarization in vitro. Interestingly, the temporal expression profile of p21 was opposite to that of Myc in Caco-2 cells (Figure 2B, cluster C); p21 and Myc are expressed in a similarly reciprocal manner along the intestinal crypt-villus axis in vivo (Melhem et al., 1992; el-Deiry et al., 1995). Another example of reciprocal expression patterns are TRAIL ligand and its receptor (TNFSRSF11B/OPG; Almasan and Ashkenazi, 2003) and Survivin, an inhibitor of apoptosis (Lens et al., 2006; see below). Third, a cluster of transcripts encoding members of several protein kinase families including SGK2, MAPK8 and CamKII (cluster H and K) increased during polarization and in vivo differentiation, indicating roles in polarized enterocytes. Fourth, transcripts of a group of genes encoding ECM proteins (COL6A1 and COL6A2; cluster M) and ECM remodeling enzymes (TIMP2, TIMP3; cluster M) were expressed in higher amounts in normal colon and polarized Caco-2 cells than in tumor cells and proliferating Caco-2 cells. These data support the idea that the composition of the ECM plays an important role in formation of normal epithelia, and changes in cell organization in disease (Nelson and Bissell, 2006). Fifth, genes involved in immunological response and defense mechanisms including members of the interleukin family (IL10RB, IL6; cluster K) and a member of the defensin family (DEFB1), which encodes an antimicrobial peptide implicated in the resistance of epithelial surfaces to microbial colonization, were expressed at higher levels in polarized Caco-2 cells and normal colon tissue, consistent with the important functions of epithelial cells in intestinal immunology. Note also that members of the cyclin M family (CNNM2, CNNM4; cluster G) and some specific histone isoforms (HIST1H2BD and HIST1H1C; cluster J) were more highly expressed in polarized Caco-2 cells compared with proliferating cells. Although these genes code for proteins with currently unknown functions, this expression pattern suggests that they may have regulatory roles in differentiated enterocytes.
Overall, our analysis reveals remarkable similarities in the expression profiles of proliferating Caco-2 cells and colon tumor cells on the one hand, and polarized Caco-2 cells and normal colon cells on the other. However we noticed some exceptions of genes that were expressed at higher levels after polarization in Caco-2 cells, but were also more highly expressed in colon cancer than in normal colon (Figure 2B, cluster N). These included IFITM1, a member of the interferon-induced transmembrane protein family, genes encoding cathepsin family members (CTSB, CTSL), lactamase β 2 (CGI-83), and some genes involved in malignant transformation including a member of the Wnt1-inducible signaling pathway protein subfamily (WISP1) and the proto-oncogene MET which encodes hepatocyte growth factor receptor.
Cell Cycle-regulated Gene Expression Profile in Differentiating Caco-2 Cells and Normal and Tumor Colon Tissue
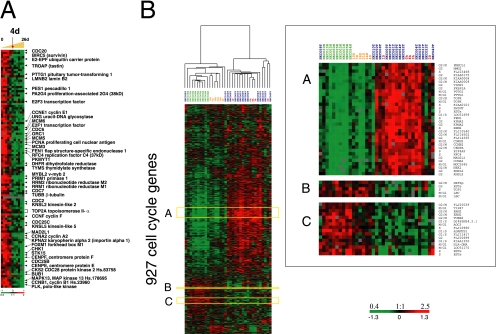
A prominent feature of the gene expression profile early in the time course, when Caco-2 cells are establishing extensive cell–cell contacts, is a set of genes characteristically expressed in proliferating cells. Clusters of genes coexpressed in concert with cell proliferation have been identified previously in genome-wide analyses of human tumors (Perou et al., 2000), and consist predominantly of genes periodically expressed during the cell cycle. The expression pattern of these genes as epithelial cells develop normally into a functional epithelium has not been directly studied. We examined this set of genes further by performing a supervised analysis of the time-course data in which a cluster of 112 genes previously associated with proliferation in breast tumors (Perou et al., 2000) were used to extract the corresponding proliferation pattern in Caco-2 cells (Figure 3). This analysis shows that proliferation genes and other cell cycle–regulated genes coordinately decrease in expression at day 4 (marked with a red arrow in Figure 3A), concomitant with the initiation of cell polarity (Halbleib et al., 2007).
Figure 3.
(A) Genes characteristically expressed during cell proliferation show a distinct temporal expression pattern during development of Caco-2 cell polarity in vitro. A set of 112 proliferation-related genes previously identified by Perou et al. (2000) in breast cancer samples are coordinately decreased in expression at the 4-d time point (marked with red arrow) in Caco-2 cells. A full list of cell cycle–regulated genes and their expression in polarizing Caco-2 cells and phase assignments can be viewed in Supplementary Figure S1. (B) Comparison of the coexpression of 927 cell cycle genes classified by their cyclic mitotic expression between differentiating Caco-2 cells and normal and colon human tissue. A set of cell cycle genes previously identified as “periodically expressed during human cell cycle” (Whitfield et al., 2002) was used to extract patterns of gene expression from the Caco-2 time course, normal and cancerous human colon. Hierarchical cluster analysis, in the gene and experiment dimension, of cell cycle genes expressed during in vitro epithelial cell polarization and human colon tissue (left) revealed the presence of two distinct gene clusters: the first representing early Caco-2 time points and colon tumor samples and the second representing late Caco-2 time points and normal colon tissue, also illustrated in the high-resolution inset (right). Transcript levels determined by microarray analysis are shown relative to a reference pool of human mRNAs.
We also performed a detailed analysis of the human cell cycle–regulated genes described by Whitfield et al. (2002); see also Supplementary Figure S1. In general, most of these cell cycle–regulated genes were expressed in Caco-2 cells up to day 4 in the time course, during which time the cells continue to proliferate, and then their expression levels greatly diminished as the cells became postmitotic and began to polarize. A few of these genes, particularly genes characteristically expressed in the M/G1 phase of the cell cycle, showed increasing expression over the time course and continued to be expressed in the postmitotic phase of Caco-2 cell differentiation. Because cell cycle control is required for cells to become postmitotic and initiate differentiation and loss of control is characteristic of malignant cell growth, we asked whether the expression pattern of these genes is shared between polarizing Caco-2 cells in vitro and either normal colon or colon tumor tissue. To answer this question, we used the set of cell cycle genes identified in Whitfield et al. (2002) to extract patterns of gene expression from the Caco-2 time course and normal and malignant human colon tissue data sets and performed a hierarchical cluster analysis of both the samples (columns) and the genes (rows), as is illustrated in Figure 3B. This analysis revealed the presence of two distinct gene clusters: one representing early Caco-2 time points and colon tumor samples (Figure 3B, cluster A) and the other representing late Caco-2 time points and normal colon tissue (Figure 3B, cluster B–C).
Because the cluster analysis of cell cycle genes clearly segregated the healthy colon from the colon tumor samples, we asked if these genes biased separation of the Caco-2 polarization time course and colon tissue samples into the normal epithelial cluster and tumor cluster shown in Figure 2A. Even when cell cycle genes are excluded from the cluster analysis, we found that there was still a good separation between colon cancer and proliferating Caco-2 cells (0–2 d in culture) on the one hand, and normal colon samples and postmitotic, differentiating Caco-2 cells (4–26 d in culture) on the other (Supplementary Figure S2).
Decreased Expression of Wnt Target Genes Accompanies Caco-2 Cell Polarization In Vitro
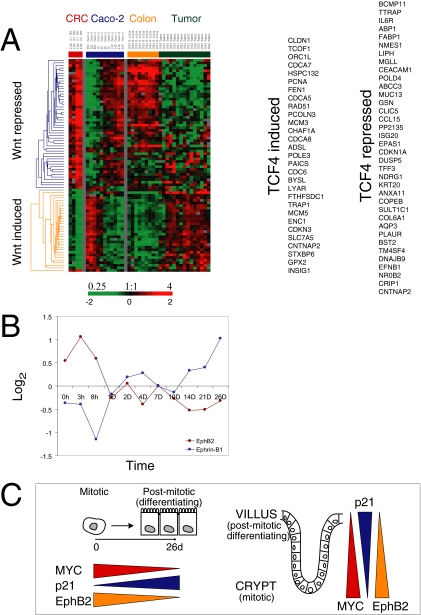
Wnt signaling has a critical role in controlling renewal of the intestinal epithelium in vivo, and uncontrolled β-catenin/TCF activity is linked to induction of colorectal cancer (Pinto et al., 2003). However, Caco-2 cells express a truncated APC protein unable to form a functional β-catenin degradation complex (Ilyas et al., 1997; Chang et al., 2005) potentially resulting in constitutive activation of Wnt signaling in these cells. We therefore investigated the activity of this pathway in Caco-2 cells and in normal colon and colon tumors by examining expression of a subset of Wnt target genes identified by van de Wetering et al. (2002) in colorectal cancer (Figure 4A). Genes that are either induced or repressed by TCF4 activity (the primary TCF expressed in the small intestine) in colorectal cancer are shown clustered with data from the Caco-2 time course and from normal colon and colon cancer.
Figure 4.
Global changes in expression of Wnt target genes during Caco-2 cell polarization in vitro. (A) A set of genes previously identified as regulated by TCF4 in colorectal cancer (CRC; A, left; van de Wetering et al., 2002) was used to extract patterns of gene expression from polarizing Caco-2 cells (A, middle), and normal human colon and colon tumor data sets (A, right). A reciprocal pattern was found for TCF4-induced and TCF4-repressed genes during early and late stages of in vitro epithelial polarization paralleling the trends between colon tumors and normal colon. The genes comprising each list are shown on the far right. (B) The similarity of expression patterns of TCF4 target genes between Caco-2 cells in vitro and enterocytes differentiating along the crypt-villus axis in vivo is illustrated by the expression of EphB2 and Ephrin-B1. EphB2 expression was highest in proliferating Caco-2 cells, whereas Ephrin-B1 transcripts increased as the cells polarized (B, left). (C) Overall expression of TCF4 induced or repressed genes during Caco-2 cell polarization in vitro mimics gene profiles in normal human colon where a progressively diminishing Wnt activity gradient occurs along the crypt-villus axis. Y-axis indicates fold change of transcript levels relative to a reference pool of human mRNAs on a log2 scale.
Genes identified as TCF4 induced were expressed in proliferating, nonpolarized Caco-2 cells, but their transcript levels decreased as cells became postmitotic and developed polarity (Figure 4A, middle). Conversely, genes identified as TCF4 repressed were expressed at significantly higher levels in postmitotic polarizing Caco-2 cells compared with their levels in proliferating, nonpolarized cells. Similar trends were found, as expected, in colon tissue (Figure 4A, right); TCF4-induced genes were expressed at higher levels in tumor tissue, whereas TCF4-repressed genes were expressed at higher levels in normal colon tissue.
TCF4-induced regulators and mediators of proliferation included MYC, members of the CDC family (CDC6, CDC25A, CDCA5, CDCA7, and CDCA8), MCM family members (MCM3, MCM5, and MCM6), ORC1L, PCNA, and the cell cycle inhibitor CDKN3 (Figure 4A). Other genes in this cluster, which may play roles in regulating renewal of intestinal epithelial stem cells, declined over the time course of Caco-2 polarization. These include the Wnt inhibitor DKK1, a BMP family member (BMP7) that is known to reverse epithelial-mesenchymal transition (Zeisberg et al., 2003) and survivin/BIRC5, a bifunctional regulator of cell proliferation and cell death (Lens et al., 2006). Additional Wnt target genes, including ITF2 (Kolligs et al., 2002), CD44 (a stem cell marker; Wielenga et al., 1999), CX43 (van der Heyden et al., 1998), and SOX9 (Blache et al., 2004), were expressed in proliferating Caco-2 cells and their levels decreased as cells polarized. Among the TCF4-repressed genes whose expression increased during Caco-2 polarization were p21 (CDKN1A), an important regulator of the cell cycle and cell growth, and many genes characteristic of differentiated intestinal epithelial cells including alkaline phosphatase (ALPI), VIPR1, TFF3, MUC13, GPA33, and cingulin (CGN; Figure 4A; see also Halbleib et al., 2007).
Transcripts encoding the EphB2 receptor were expressed in proliferating Caco-2 cells, but their levels decreased as cells polarized. Conversely, transcripts encoding the ligand Ephrin-B1 increased during Caco-2 polarization (Figure 4B). These results parallel previous reports of inverse levels of ephrins and EphB along the intestinal crypt-villus axis (Batlle et al., 2002), in which highest levels of receptors (EphB2, EphB3) were found in the crypt, and levels of Ephrin-B1 increased in a reciprocal gradient along the crypt-villus axis.
In summary, we found that a large set of Wnt target genes were coordinately regulated during polarization of Caco-2 cells, a cell line of adenocarcinoma origin and lacking full-length APC, in a pattern that closely paralleled their expression along the intestinal crypt-villus axis in vivo (Figure 4C).
Regulation of Wnt Target Gene Expression during Caco-2 Cell Differentiation In Vitro
The unexpected, concerted decrease in transcripts of Wnt target genes in Caco-2 cells, which express a truncated and functionally defective APC, suggested the action of a mechanism that did not involve β-catenin degradation for regulating the Wnt pathway in response to epithelial polarization cues. We therefore explored where in the Wnt pathway (Figure 5A) this unexpected regulation might operate.
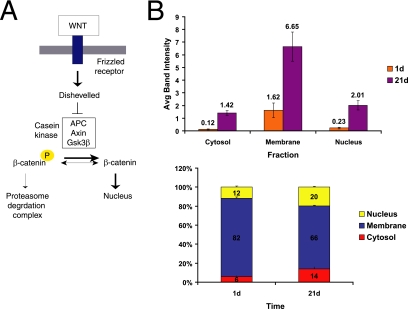
Figure 5.
Subcellular distribution of β-catenin during Caco-2 cell polarization. (A) Simplified schematic of classical Wnt signaling. Wnt signaling through Frizzled receptors inhibits destruction of the transcription cofactor β-catenin by the APC/Axin/GSK3β complex, leading to β-catenin stabilization, nuclear translocation, and complex formation with the TCF/LEF family of transcriptional cofactors that activate Wnt target genes. (B) Levels of β-catenin were compared in membrane, cytoplasmic, and nuclear fractions in nonpolarized (1d) and polarized (21d) Caco-2 cells as both raw band intensity (top graph) and percentage of the entire β-catenin pool (bottom graph). Nuclear β-catenin increased more than eightfold in polarized cells, compared with nonpolarized cells. This is consistent with the absence of a functional APC destruction complex in Caco-2 cells, leading to accumulation of nuclear β-catenin. n = 3. Error bars, SE.
We first investigated whether the mechanism acted upstream or downstream of β-catenin stabilization and nuclear localization. To analyze the subcellular localization of β-catenin, we fractionated nonpolarized (1 d) and polarized (21 d) Caco-2 cells into membrane, cytoplasmic, and nuclear fractions and compared the levels of β-catenin in each fraction. We observed an increase in overall β-catenin levels during Caco-2 polarization, including a more than eightfold increase in the level of nuclear β-catenin in polarized cells compared with nonpolarized cells (Figure 5B). This indicates that as Caco-2 cells polarize β-catenin is not being degraded efficiently via APC. Coimmunoprecipitation with TCF4 revealed a corresponding increase in the amount of β-catenin associated with TCF4 in the nuclear fraction of polarized cells (data not shown). These results show that the decrease in Wnt target gene expression during Caco-2 cell polarization cannot be explained by decreased ligand-mediated receptor activation, β-catenin degradation, inhibition of β-catenin localization to the nucleus, or inhibition of the TCF4–β-catenin interaction. Thus neither secreted inhibitors, the β-catenin destruction complex, TLE/Groucho proteins nor interference with the formation of TCF—β-catenin complexes were responsible for the decrease in Wnt target gene expression. This suggests that the decrease in Wnt pathway target gene transcripts during Caco-2 polarization was due to an inhibitor acting downstream of TCF–β-catenin complex formation.
Although the critical negative regulation of Wnt pathway activity during Caco-2 polarization appears to act downstream of TCF–β-catenin complex formation, the larger gene expression program in these cells suggests a multilayered regulation of the Wnt signaling pathway. Such regulation may be important for the exquisite spatial and temporal control of proliferation, migration, polarization, and ultimately differentiation required to build the precise three-dimensional architecture of a functional villus. Therefore we examined other components of the Wnt signaling system to determine how their expression varied during polarization (Figure 6A) and between normal colon and colon tumors (Supplementary Figure S3). Interestingly, we did not observe a notable alteration in the expression of Wnt genes themselves as Caco-2 cells polarized, consistent with the surrounding stroma as the source of ligand in vivo.
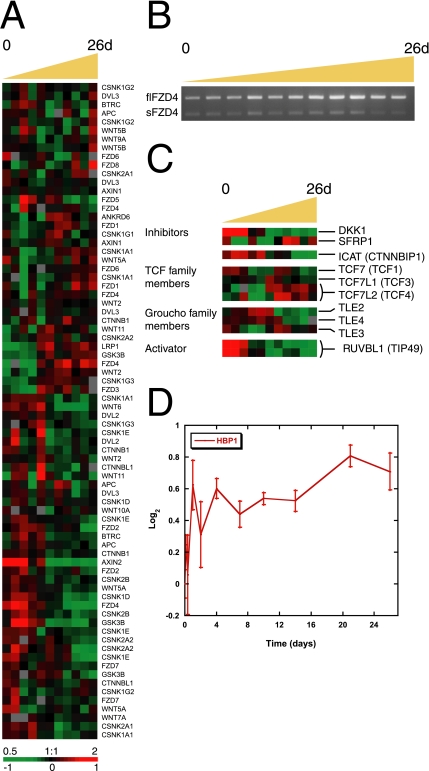
Figure 6.
Gene-expression patterns of Wnt pathway components during Caco-2 cell polarization in vitro. (A) Transcriptional levels of components of the Wnt pathway including the β-catenin destruction complex changed over time, although not as a cohort. (B) Isoform-specific RT-PCR showed that full-length FZD4 (flFZD4) mRNA increased during polarization, whereas that of the soluble form (sFZD4) had a more modest transcriptional regulation. (C) Proteins with an inhibitory effect on Wnt signaling, including DKK1, SFRP, and ICAT, or activators of the pathway such as TCF isoforms and RUVBL1 (TIP49), were temporally regulated during Caco-2 cell polarization. Members of the Groucho (TLE) family of Wnt suppressors were expressed from the beginning to the middle of the time course. (D) Expression of HBP1, which inhibits activation of Wnt target genes by preventing the interaction of the β-catenin-TCF4 complex with DNA, increased during Caco-2 cell polarization. Y-axis indicates fold change of transcript levels relative to a reference pool of human mRNAs on a log2 scale. Error bars, SE.
Components of the β-Catenin Destruction Complex.
A major control point in the Wnt pathway is the degradation of β-catenin by the APC destruction complex. Transcript levels of several regulators of β-catenin stability (GSK3β, Axin1, Axin2, APC, Casein kinase-1, Deversin) changed over time, although not as a cohort (Figure 6A). GSK3β was most highly expressed in proliferating Caco-2 cells before polarization. AXIN and AXIN2 were expressed in different temporal profiles during Caco-2 cell polarization, consistent with their different distributions in normal and neoplastic colon (Anderson et al., 2002); AXIN2 is exclusively expressed in crypt cells, and is induced by Wnt signaling. AXIN1, Casein kinase-1 (gamma subunits G1 and G3 [CSNK1G1, G3]), APC, and Deversin (ANKRD6) were all more highly expressed at the time when Caco-2 cells established cell–cell contacts and developed polarity.
TCF Family Members.
β-catenin induces target gene expression by acting as a heterodimer with members of the LEF/TCF family of transcription factors; thus, decreased levels of LEF/TCF could diminish expression of Wnt targets. However, levels of the principal intestinal isoform, TCF4 (gene name TCF7L2), increased as cells became postmitotic and developed polarity (Figure 6C). This result is in accordance with accumulated levels of the β-catenin/TCF4 complex observed during Caco-2 cell polarization (Figure 5B). TCF1 (gene name TCF7) had two peaks of expression during Caco-2 cell polarization, first in proliferating cells (day 0) and again at later time points (7–10 d). Data from Roose et al. (1999) indicate that TCF1 is a target gene of β-catenin/TCF4 activity in epithelial cells and that the most abundant isoforms, which lack a β-catenin binding domain, may cooperate with APC as a feedback repressor of Wnt signaling (Roose et al., 1999). A third member of the TCF family of transcription factors (TCF3 or TCF7L1) had a peak level of expression at day 4, which is the time at which Caco-2 cells became postmitotic and initiated polarization. This intricate modulation of TCFs may play a role in achieving precise control of Wnt signaling in vivo.
Wnt Antagonists.
The secreted frizzled-related protein family (SFRP) and the Dickkopf (DKK) family comprise secreted inhibitors of Wnt signaling (see Introduction). Representatives of both families of proteins were expressed in Caco-2 cells (Figure 6C). DKK1 is a Wnt target gene and as expected is expressed in higher amounts in proliferating Caco-2 cells when expression of Wnt-induced genes is highest, whereas the highest levels of SFRP1 transcripts were detected in postmitotic differentiated Caco-2 cells. The distinct temporal expression patterns of these two secreted inhibitors provide further evidence for multilayered control of Wnt signaling in this system and suggest that understanding the differences in the function and regulatory logic of these proteins might yield insights into Wnt pathway regulation.
Frizzled-4 Isoforms.
A reciprocal expression pattern was detected by two array elements representing two different Frizzled-4 (FZD4) isoforms. The soluble FZD4 splice variant (FZD4S, R26141) was most highly expressed at early time points during Caco-2 polarization, whereas the full-length FZD4 receptor isoform (FZD4FL, AA677200) was more highly expressed later in the time course in polarized Caco-2 cells (Supplementary Figure S4A). These two FZD4 isoforms were also differentially expressed in normal colon and colon tumor samples. FZD4S transcripts increased in colon tumors compared with normal tissue and FZD4FL had greater expression levels in normal colon (Supplementary Figure S4B). The soluble FZD4 isoform encodes a polypeptide with the N-terminal portion of the FZD4 extracellular domain and has been identified as a context-dependent regulator of Wnt activity (Sagara et al., 2001). We performed isoform-specific RT-PCR to confirm mRNA expression of the two splice variants. We found a large increase in the level of the full-length FZD4 transcript as Caco-2 cells polarized, whereas levels of the transcript encoding soluble FZD4 were unexpectedly only slightly higher during the middle time points (Figure 6B). This apparent conflict with the array data may be due to partial overlap of the IMAGE clone corresponding to FZD4S (R26141) with the sequence of FZD4FL mRNA or to the presence of additional, uncharacterized FZD4 isoforms. The precise role of Frizzled-4 alternative splicing in Wnt signaling during Caco-2 polarization is unknown.
Inhibitors and Activators of Transcriptional Regulation by β-Catenin.
Several factors have been identified that directly interfere with β-catenin function in the nucleus. The TLE family of Drosophila Groucho homologues bind TCFs and prevent their interaction with β-catenin (Cavallo et al., 1998; Roose et al., 1998; Brantjes et al., 2001). During Caco-2 polarization, TLE3 transcripts were expressed most highly between 7 and 10 d, immediately after the onset of polarization, and although TLE2 and TLE4 mRNA expression decreased over the time course, their levels remained high through day 4, perhaps allowing these proteins to play a role in down-regulating Wnt signaling (Figure 6C). ICAT is another β-catenin–interacting protein that inhibits β-catenin binding to TCF4 and as a result represses Wnt signaling (Tago et al., 2000); overexpression of ICAT in tumor cells, even in the presence of elevated β-catenin levels, inhibits cell proliferation by inducing G2 arrest (Sekiya et al., 2002). However, we found higher levels of ICAT (CTNNBIP1) transcripts in proliferating Caco-2 cells than in polarized, postmitotic cells (Figure 6C), and a correspondingly high level of ICAT was found in colon tumor tissue compared with normal colon cells. These data are inconsistent with a role for ICAT in inhibiting Wnt signaling during Caco-2 cell polarization. However, the level of HBP1, which inhibits β-catenin transcription by displacing the entire β-catenin/TCF4 complex from DNA (Sampson et al., 2001), increased slightly as Caco-2 cells polarized and therefore may act in this system to inhibit Wnt target gene expression (Figure 6D).
Noncanonical Wnt Pathway.
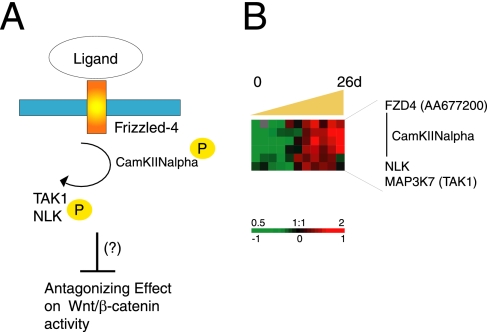
It has been suggested that the noncanonical Wnt pathway converges with the canonical Wnt cascade to down-regulate TCF activity (Topol et al., 2003; Mikels and Nusse, 2006; Figure 7A). We found increasing levels of transcripts encoding components of the noncanonical Wnt/Ca2+ pathway in polarizing Caco-2 cells. As illustrated in Figure 7B, CamKII, TAK1/MAP3K7, and NLK transcripts had similar expression profiles that increased over time with highest levels from day 4 to day 26 when Caco-2 cells became postmitotic and polarized (Figure 7B). CamKII expression levels were significantly higher in normal colon tissue compared with tumor tissue (Figure 2B, cluster H). It has previously been shown that the FZD4 receptor can activate CaMKII and PKC (Sheldahl et al., 1999; Kuhl et al., 2000; Robitaille et al., 2002). Expression of full-length FZD4 (AA677200) also increased as polarization proceeded. The reciprocal relationship between expression profiles of components of the noncanonical Wnt pathway and target genes of canonical Wnt signaling supports a possible role for the noncanonical Wnt pathway in regulating intestinal cell proliferation and differentiation, by antagonizing canonical Wnt signaling as intestinal epithelial cells differentiate.
Figure 7.
Regulation of the noncanonical Wnt pathway. (A) Schematic of noncanonical Wnt pathway proteins and their possible effect on canonical Wnt signaling. (B) Transcriptional levels of noncanonical Wnt pathway components increased during Caco-2 cell polarization in vitro. Transcript levels determined by microarray analysis are shown relative to a reference pool of human mRNAs.
Taken together, our results indicate that in a stroma-free Caco-2 cell monolayer modulation of Wnt signaling during cell polarization and differentiation are most likely due to intracellular factors acting downstream of β-catenin/TCF4 in the nucleus. However, they also reveal an intricate, multilayered regulation of extracellular and intracellular components and modulators of the Wnt pathway. In view of the striking parallels between the gene expression programs accompanying Caco-2 polarization in culture and enterocyte differentiation in vivo, this intricate program may reflect the multilayered regulation that has evolved to provide precise and robust spatial and temporal control of this critical pathway in vivo.
Components of Key Signaling Pathways Involved in Intestinal Epithelium Renewal In Vivo Are Expressed during Caco-2 Cell Polarization In Vitro
In addition to the Wnt, other signaling pathways including Hedgehog, Notch, and the TGFβ/BMP have been implicated in intestinal epithelium differentiation in vivo (Sancho et al., 2004). Signaling between cells in the surrounding connective tissue and epithelial cells is thought to be critical for normal enterocyte differentiation. Because Caco-2 cells in vitro are not surrounded by stromal cells it might be expected that they do not express components of different signaling pathways or cytokines that function in juxtacrine signaling between epithelial and stromal cells in vivo. To test this prediction and to advance an understanding of the interplay between different signaling pathways during epithelial cell polarization, we analyzed the expression patterns of components of different signaling pathways and cytokines upon Caco-2 cell polarization in vitro. Figure 8 presents an overview of transcripts representing signaling proteins and pathways differentially regulated over time in Caco-2 cells.
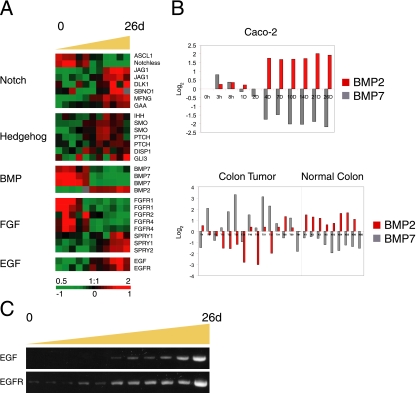
Figure 8.
Components of signaling pathways involved in intestinal epithelium formation in vivo are regulated during Caco-2 cell polarization in vitro. (A) Proteins in the Notch, Hedgehog. BMP, FGF, and EGF pathways were temporally expressed during Caco-2 cell polarization. A reciprocal expression pattern was identified for FGF receptors and FGF antagonists Sprouty1 and Sprouty 2. (B) Members of the BMP family (BMP2 and BMP7) showed an inverse expression pattern in polarizing Caco-2 cells, and a similar trend was identified in normal and colon cancer tissue. BMP2 expression was similar in polarized Caco-2 cells and healthy colon tissue, whereas BMP7 was more highly expressed in proliferating Caco-2 cells and cancerous colon tissue. Y-axis indicates fold change of transcript levels relative to a reference pool of human mRNAs on a Log2 scale. (C) Expression patterns of EGF and EGFR were confirmed by RT-PCR.
Components, modifiers and target genes of the Notch pathway were temporally regulated during Caco-2 cell polarization (Figure 8A). Proliferating Caco-2 cells expressed Notchless, a suppressor of Notch activity (Royet et al., 1998), and slightly higher amounts of the Achaete-Scute homologue gene (ASCL1/hASH1/mASH1) compared with polarized cells; ASCL1 inhibits Notch signaling and is itself transcriptionally repressed by the Notch target gene HES1 (Chen et al., 1997; Sriuranpong et al., 2002). In contrast, transcripts encoding Notch ligands (Jagged 1 and Delta-like 1), a member of the Fringe gene family (MFNG) that encodes a secreted protein known to act in the Notch pathway, and a Notch target gene (GAA) all increased with Caco-2 cell polarization. Similarly, levels of Notchless and ASCL were highest and the level of MFNG lowest in colon cancer compared with normal colon tissue (data not shown). These results are consistent with studies identifying Notch as a tumor suppressor and the signaling pathway as antimitogenic in several systems, primarily through up-regulation of p21 (Rangarajan et al., 2001; Qi et al., 2003; Noseda et al., 2004; Niimi et al., 2007), and indicate that this pathway may play a similar role in suppressing proliferation and promoting differentiation during Caco-2 polarization and perhaps during enterocyte differentiation in vivo.
The expression profiles of Hedgehog (Hh) pathway components in polarizing Caco-2 cells are shown in Figure 8A. As Caco-2 cells polarized, we found increasing levels of transcripts encoding the Indian Hedgehog (IHH) ligand; two Hh receptors, Smoothened (SMO) and Patched (PTCH); and the GLI3 transcription factor, which drives expression of Hh target genes. These results are consistent with reduced Hh signaling promoting epithelial proliferation in adult colon (van den Brink et al., 2004) and neonatal small intestine (Madison et al., 2005), although there is also evidence linking tumorigenesis to Hh pathway activation (Taipale and Beachy, 2001). Note that the expression profiles of Hedgehog components and Wnt-induced genes were reciprocal, indicating that these two pathways are differentially regulated and perhaps act in different phases of intestinal epithelial cell differentiation in vivo. Indeed, TCF4 is expressed in proliferating cells of the crypt and IHH is expressed in mature colonic epithelial cells (van den Brink et al., 2004), and the Wnt inhibitor SFRP1 is induced by Hedgehog signaling (Katoh and Katoh, 2006b).
We examined the expression profiles of members of the TGFβ/BMP family. BMP7 transcripts were expressed at high levels in proliferating Caco-2 cells and then decreased, whereas BMP2 levels increased over time as Caco-2 cells became postmitotic and polarized (Figure 8, A and B). We did not detect significant changes in BMP4 expression levels (data not shown). BMP7 is also expressed at high levels in tumor colon samples compared with normal tissue and, conversely, BMP2 is expressed at higher levels in normal tissue (Hardwick et al., 2004; Figure 8B). The expression pattern of BMP2 is consistent with the finding that BMP2 and BMP4 are coexpressed with Hh genes during mouse development (Bitgood and McMahon, 1995).
Little is known about the FGF signaling pathway in adult human intestine or expression of FGF pathway components in epithelial cells along the intestinal crypt-villus axis. We found that several members of the FGF receptor family (FGFR1, -2, and -4) were expressed at higher levels in proliferating Caco-2 cells and decreased as cells became postmitotic and polarized (Figure 8A). In contrast, two members of the Sprouty protein family (SPRY1 and SPRY2), which are implicated in negative-feedback regulation of FGF signaling (Mason et al., 2006), peaked in expression as Caco-2 cells became polarized, indicating that the responsiveness of enterocytes to FGF and perhaps other signals may decrease as the cells differentiate and polarize. Interestingly, secreted FGF ligands did not appear to be transcriptionally regulated as Caco-2 cells polarized, perhaps suggesting that these signals are normally provided by the surrounding stroma during enterocyte differentiation in vivo. Transcripts of both the EGF and its receptor increased during Caco-2 cell polarization (Figure 8, A and C).
In summary, we found that several signaling pathways, in addition to Wnt, are temporally regulated during Caco-2 cell polarization in vitro despite the lack of stromal cells and morphogen gradients. The temporal pattern of expression of these pathways is similar to the temporal and spatial pattern in which they are expressed during migration and differentiation of enterocytes along the crypt-villus axis in vivo. Moreover, the differences in expression of components of these pathways between polarized and unpolarized Caco2 cells parallel differences between normal colon and colon cancer.
DISCUSSION
The development of epithelial cell polarity is fundamental to the formation of many organs and tissues, and abnormalities in this process occur in a variety of disease states. The study of mechanisms involved in cell polarization has focused on tissues such as the intestine, in which enterocytes differentiate as they leave the stem cell niche located in the crypt, exit the cell cycle, and polarize as they migrate along the villus (Sancho et al., 2004). However, obtaining homogeneous populations of differentiating cells from tissues for biochemical studies is difficult. Therefore, detailed analyses of this process has turned to defined cell lines, such as Caco-2, which have retained the ability to develop structural and functional cell polarity in tissue culture (Grasset et al., 1985; Wice et al., 1985; Le Bivic et al., 1990; Matter et al., 1990). Although some transcriptional regulation may have been considered, neither the degree of temporal regulation nor the extent of transcriptional programming has been studied, and instead past research focused almost exclusively on posttranslational mechanisms.
Here, we present for the first time a detailed genomic portrait of changes in transcript patterns during development of epithelial cell polarity in vitro. We found that the transition from proliferating, nonpolarized cells to postmitotic polarizing cells involves a switch in gene expression programs. Our analysis provides striking evidence for similar gene expression patterns in proliferating, nonpolarized Caco-2 cells and colon tumor cells on the one hand, and postmitotic polarized Caco-2 cells and normal healthy colonic tissue on the other. Equally intriguing were the parallels between the temporal program of gene expression during Caco-2 cell polarization in vitro and the temporal and spatial gene expression program of intestinal cell differentiation along the crypt-villus axis in vivo. From this perspective, the transcriptional program of Caco-2 cell polarization suggests a complex and multilayered interplay among developmental signaling activities including Wnt, BMPs, hedgehog, FGF, and Notch, in which an intrinsic transcriptional program in the developing enterocyte participates in a molecular conversation with the stromal cells in its microenvironment to provide a robust and precise specification of the cellular architecture of the villus. Note that the time course itself was consistently reproducible, indicating both a synchrony within the monolayer and an innate timing to the differentiation process.
The well-described phenomenon of decreased cell division as epithelial cells form extensive cell–cell contacts, termed “contact inhibition” (Abercrombie, 1970), is reflected in this model at the genomic level at day 4 by a marked decrease in expression of a large set of genes with roles in cell division. That this effect is synchronous for a large number of genes suggests a global mechanism for repression of these genes upon establishment of cell–cell contact. What is the specific trigger for this switch in expression of these genes? It has been suggested that some transcriptional regulators are controlled by formation of stable cell-adhesions to other cells or to the ECM (Beckerle, 1997; Balda and Matter, 2000; Kanungo et al., 2000; Mertens et al., 2001; Wang and Gilmore, 2001; Miravet et al., 2002; Woods et al., 2002; Nelson and Nusse, 2004), but the molecular mechanisms remain to be defined. For example, it is not clear whether this is similar to mechanisms involved in cessation of cell division in progenitor cells. Thus, our data bring into focus the need and the opportunities for new analysis of this process in this model system.
The gene expression profile of the Wnt signaling pathway was a prominent feature of the early transcriptional program in Caco-2 cells. Local Wnt signaling in the intestinal crypt is required to maintain the proliferating stem cell population (Clevers, 2006). However, as postmitotic differentiating cells migrate away from the crypt region and into villus, Wnt activity gradually decreases. Multiple mechanisms participate in regulating Wnt-signaling activity, including secreted inhibitors of ligand binding, the β-catenin destruction complex, activators and repressors of β-catenin/TCF-induced transcription, and activity of the noncanonical Wnt pathway and other growth factor signaling pathways (Clevers, 2006). The classical Wnt pathway acts by controlling the cytoplasmic/nuclear level of β-catenin through the APC/Axin/GSK3β destruction complex (Polakis, 2000). In vivo, decreasing Wnt levels along the crypt-villus axis results in progressive activation of the destruction complex and targeting of β-catenin for degradation as the enterocytes migrate up the villus (Clevers, 2006). Mutations in APC are common in early stages of colorectal cancers and are thought to result in β-catenin stabilization and constitutive Wnt signaling activity, which then promotes cell proliferation and restricts differentiation (Fodde et al., 2001). Caco-2 cells, derived from a human colorectal cancer, have a truncated APC that can bind β-catenin but cannot target β-catenin for degradation (Ilyas et al., 1997). A strong prediction, therefore, was that β-catenin–dependent transcription would be independent of Wnt signals in Caco-2 cells and correspondingly high in polarized cells. On the contrary, our global transcriptional analysis revealed that Wnt target gene expression was high in mitotic nonpolarized Caco-2 cells but decreased coordinately over time as cells became postmitotic and polarized. An opposite trend was found for a set of genes normally repressed by β-catenin/TCF activity. Thus, a transcriptional program normally controlled by the canonical Wnt signaling pathway was regulated in Caco-2 cells in a pattern that mimics the gradient of high-to-low Wnt signaling activity between mitotic cells in the crypt and postmitotic differentiating cells migrating up the villus in vivo. These results differ from an earlier study (Bertucci et al., 2004) in which Caco-2 cells were grown to confluency on plastic. However, cell growth on plastic results in poor cell polarization and nutrient and growth factor starvation because access to basal-lateral receptors is excluded by the tight junction; in contrast, we grew Caco-2 cells on Transwell filters upon which they develop full polarity and nutrients can access the basal-lateral surface through the filter support.
As neither Wnts nor Wnt antagonists were provided to Caco-2 cells because of the absence of surrounding stroma and as APC in these cells is not functional for β-catenin degradation, these results prompted us to investigate how Wnt signaling activity might be regulated in vitro. Examination of other key regulatory steps of the Wnt pathway identified several additional possible points of control that we suspect may be important for regulation of colonic epithelium in vivo. In Caco-2 cells, we observed a complex temporal regulatory program of genes important at several stages of Wnt signaling including regulators of β-catenin stability (Axin, Axin2, APC, GSK3β, Casein kinase, Deversin), Wnt antagonists (DKK1, SFRP1), modifiers of β-catenin transcriptional activity (TLEs, ICAT, HBP1, RuvBL1), and the noncanonical Wnt pathway (CamKII, TAK1/MAP3K7, NLK). Although all of these regulators may be important in precise control of self-renewal and differentiation of the colonic epithelium in vivo, most could be excluded in the regulation of the Wnt pathway–controlled transcriptional program in Caco-2 cells. We narrowed the list by determining the subcellular localization of β-catenin in polarized Caco-2 cells. Surprisingly, β-catenin levels in the nucleus increased after polarization, as did association of β-catenin with TCF4. Because nuclear localization of β-catenin is downstream of both ligand binding and activity of the destruction complex, our finding of β-catenin in the nucleus of polarized cells supports the conclusion that Caco-2 cells have a mechanism for regulating the transcriptional output of the Wnt pathway during polarization even in the absence of temporal regulation of Wnt and fully functional APC. TLEs also do not appear to be essential for this regulation, as they would be expected to disrupt the β-catenin–TCF interaction. The expression profiles of HBP1 and RuvBL1 are still consistent with their reported roles in inactivating the Wnt pathway during Caco-2 polarization, but the change in HBP1 expression level was relatively small, and RuvBL1 is a known component of a nonspecific chromatin remodeling complex as well, further complicating the interpretation of its temporal expression during Caco-2 polarization. We suggest that as cell polarization proceeds, β-catenin–TCF complexes are unable to activate transcription of Wnt target genes. Possible mechanisms include inhibition of the ability of the TCF4–β-catenin complex to associate with its DNA targets, perhaps by HBP1, repression of transcriptional activation by this complex or repression of the target genes by a mechanism independent of TCF4–β-catenin. Further studies beyond the scope of the current analysis will be required to identify the underlying mechanism in detail.
Together, our results establish the existence of a complex, autonomous transcriptional program under fine temporal control during polarization of intestinal epithelial cells in vitro with striking similarities to the spatial and temporal patterns of gene expression during enterocyte differentiation in vivo, independent of the extracellular signals present in the intestinal niche. For example, a large set of Wnt target genes was tightly regulated during Caco-2 cell polarization, in a pattern that closely paralleled their expression along the intestinal crypt-villus axis in vivo. Because Caco-2 cells do not have a functional APC destruction complex, the decreased activation of Wnt target genes cannot be explained by classical β-catenin degradation, but suggest a novel mechanism of Wnt target gene regulation upon establishment of an epithelial layer in vitro. These surprising results suggest that the program of enterocyte gene expression along the crypt-villus axis in vivo is at least in part intrinsic to the enterocytes themselves—perhaps the complex spatially regulated interplay of developmental signals in this microenvironment functions mainly to entrain this cell-autonomous program to a precise spatial pattern and thereby patterning the physical arrangement of the differentiating cells along the crypt and villus. Our studies highlight the usefulness of Caco-2 cell polarization as an in vitro model for investigating these signaling pathways and suggest potential roles of specific components that will guide more detailed biochemical studies in the future.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Brown and Nelson laboratories, and Ronald S. Rock for helpful discussions. We also want to thank members of the Stanford Functional Genomics Facility (SFGF) and Stanford Microarray Database (SMD) for their help and advice. We gratefully acknowledge financial assistance from the Swedish Research Council, “Vetenskapsrådet” (A.S). J.M.H. is supported by a Howard Hughes Medical Institute Predoctoral fellowship. This work was supported by National Cancer Institute Grant CA77097 (P.O.B.) and by the Howard Hughes Medical Institute. P.O.B. is an Investigator of the Howard Hughes Medical Institute. Work from the Nelson laboratory is supported by the National Institutes of Health Grant GM35527.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-04-0309) on August 15, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Abercrombie M. Contact inhibition in tissue culture. In Vitro. 1970;6:128–142. doi: 10.1007/BF02616114. [DOI] [PubMed] [Google Scholar]
- Aberle H., Bauer A., Stappert J., Kispert A., Kemler R. beta-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasan A., Ashkenazi A. Apo2L/TRAIL: apoptosis signaling, biology, and potential for cancer therapy. Cytokine Growth Factor Rev. 2003;14:337–348. doi: 10.1016/s1359-6101(03)00029-7. [DOI] [PubMed] [Google Scholar]
- Anderson C. B., Neufeld K. L., White R. L. Subcellular distribution of Wnt pathway proteins in normal and neoplastic colon. Proc. Natl. Acad. Sci. USA. 2002;99:8683–8688. doi: 10.1073/pnas.122235399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda M. S., Matter K. The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. EMBO J. 2000;19:2024–2033. doi: 10.1093/emboj/19.9.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N., Huls G., Korinek V., Clevers H. Restricted high level expression of Tcf-4 protein in intestinal and mammary gland epithelium. Am. J. Pathol. 1999;154:29–35. doi: 10.1016/S0002-9440(10)65247-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle E., et al. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- Beckerle M. C. Zyxin: zinc fingers at sites of cell adhesion. Bioessays. 1997;19:949–957. doi: 10.1002/bies.950191104. [DOI] [PubMed] [Google Scholar]
- Behrens J., Jerchow B. A., Wurtele M., Grimm J., Asbrand C., Wirtz R., Kuhl M., Wedlich D., Birchmeier W. Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- Behrens J., von Kries J. P., Kuhl M., Bruhn L., Wedlich D., Grosschedl R., Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Bertucci F., et al. Gene expression profiling of colon cancer by DNA microarrays and correlation with histoclinical parameters. Oncogene. 2004;23:1377–1391. doi: 10.1038/sj.onc.1207262. [DOI] [PubMed] [Google Scholar]
- Bitgood M. J., McMahon A. P. Hedgehog and Bmp genes are coexpressed at many diverse sites of cell-cell interaction in the mouse embryo. Dev. Biol. 1995;172:126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- Blache P., van de Wetering M., Duluc I., Domon C., Berta P., Freund J. N., Clevers H., Jay P. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J. Cell Biol. 2004;166:37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z., Llinas M., Pulliam B. L., Wong E. D., Zhu J., DeRisi J. L. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantjes H., Roose J., van De Wetering M., Clevers H. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 2001;29:1410–1419. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan K. M., Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Cavallo R. A., Cox R. T., Moline M. M., Roose J., Polevoy G. A., Clevers H., Peifer M., Bejsovec A. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–608. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- Chang W. C., Everley L. C., Pfeiffer G. R., 2nd, Cooper H. S., Barusevicius A., Clapper M. L. Sulindac sulfone is most effective in modulating beta-catenin-mediated transcription in cells with mutant APC. Ann. NY Acad. Sci. 2005;1059:41–55. doi: 10.1196/annals.1339.020. [DOI] [PubMed] [Google Scholar]
- Chen H., Thiagalingam A., Chopra H., Borges M. W., Feder J. N., Nelkin B. D., Baylin S. B., Ball D. W. Conservation of the Drosophila lateral inhibition pathway in human lung cancer: a hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. Proc. Natl. Acad. Sci. USA. 1997;94:5355–5360. doi: 10.1073/pnas.94.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Crosnier C., Stamataki D., Lewis J. Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat. Rev. Genet. 2006;7:349–359. doi: 10.1038/nrg1840. [DOI] [PubMed] [Google Scholar]
- Dailey L., Ambrosetti D., Mansukhani A., Basilico C. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 2005;16:233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- el-Deiry W. S., et al. Topological control of p21WAF1/CIP1 expression in normal and neoplastic tissues. Cancer Res. 1995;55:2910–2919. [PubMed] [Google Scholar]
- Fagotto F., Jho E., Zeng L., Kurth T., Joos T., Kaufmann C., Costantini F. Domains of axin involved in protein-protein interactions, Wnt pathway inhibition, and intracellular localization. J. Cell Biol. 1999;145:741–756. doi: 10.1083/jcb.145.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon E. R., Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Fodde R. The APC gene in colorectal cancer. Eur. J. Cancer. 2002;38:867–871. doi: 10.1016/s0959-8049(02)00040-0. [DOI] [PubMed] [Google Scholar]
- Fodde R., Smits R., Clevers H. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer. 2001;1:55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- Fre S., Huyghe M., Mourikis P., Robine S., Louvard D., Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- Fukuda K., Yasugi S. Versatile roles for sonic hedgehog in gut development. J. Gastroenterol. 2002;37:239–246. doi: 10.1007/s005350200030. [DOI] [PubMed] [Google Scholar]
- Gao Z. H., Seeling J. M., Hill V., Yochum A., Virshup D. M. Casein kinase I phosphorylates and destabilizes the beta-catenin degradation complex. Proc. Natl. Acad. Sci. USA. 2002;99:1182–1187. doi: 10.1073/pnas.032468199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasset E., Bernabeu J., Pinto M. Epithelial properties of human colonic carcinoma cell line Caco-2: effect of secretagogues. Am. J. Physiol. 1985;248:C410–C418. doi: 10.1152/ajpcell.1985.248.5.C410. [DOI] [PubMed] [Google Scholar]
- Gregorieff A., Pinto D., Begthel H., Destree O., Kielman M., Clevers H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Groden J., Thliveris A., Samowitz W., Carlson M., Gelbert L., Albertsen H., Joslyn G., Stevens J., Spirio L., Robertson M. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- Halbleib J. M., Sääf A. M., Brown P. O., Nelson W. J. Transcriptional modulation of genes encoding structural characteristics of differentiating enterocytes during development of a polarized epithelium in vitro. Mol. Biol. Cell. 2007;18:4261–4278. doi: 10.1091/mbc.E07-04-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick J. C., Van Den Brink G. R., Bleuming S. A., Ballester I., Van Den Brande J. M., Keller J. J., Offerhaus G. J., Van Deventer S. J., Peppelenbosch M. P. Bone morphogenetic protein 2 is expressed by, and acts upon, mature epithelial cells in the colon. Gastroenterology. 2004;126:111–121. doi: 10.1053/j.gastro.2003.10.067. [DOI] [PubMed] [Google Scholar]
- Hart M. J., de los Santos R., Albert I. N., Rubinfeld B., Polakis P. Downregulation of beta-catenin by human Axin and its association with the APC tumor suppressor, beta-catenin and GSK3 beta. Curr. Biol. 1998;8:573–581. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- Heath J. P. Epithelial cell migration in the intestine. Cell Biol. Int. 1996;20:139–146. doi: 10.1006/cbir.1996.0018. [DOI] [PubMed] [Google Scholar]
- Hinck L., Nathke I. S., Papkoff J., Nelson W. J. Dynamics of cadherin/catenin complex formation: novel protein interactions and pathways of complex assembly. J. Cell Biol. 1994a;125:1327–1340. doi: 10.1083/jcb.125.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinck L., Nelson W. J., Papkoff J. Wnt-1 modulates cell-cell adhesion in mammalian cells by stabilizing beta-catenin binding to the cell adhesion protein cadherin. J. Cell Biol. 1994b;124:729–741. doi: 10.1083/jcb.124.5.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S., Kishida M., Matsuura Y., Usui H., Kikuchi A. GSK-3beta-dependent phosphorylation of adenomatous polyposis coli gene product can be modulated by beta-catenin and protein phosphatase 2A complexed with Axin. Oncogene. 2000;19:537–545. doi: 10.1038/sj.onc.1203359. [DOI] [PubMed] [Google Scholar]
- Ikeda S., Kishida S., Yamamoto H., Murai H., Koyama S., Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J. 1998;17:1371–1384. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyas M., Tomlinson I. P., Rowan A., Pignatelli M., Bodmer W. F. Beta-catenin mutations in cell lines established from human colorectal cancers. Proc. Natl. Acad. Sci. USA. 1997;94:10330–10334. doi: 10.1073/pnas.94.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani T., Kishida S., Hyodo-Miura J., Ueno N., Yasuda J., Waterman M., Shibuya H., Moon R. T., Ninomiya-Tsuji J., Matsumoto K. The TAK1-NLK mitogen-activated protein kinase cascade functions in the Wnt-5a/Ca(2+) pathway to antagonize Wnt/beta-catenin signaling. Mol. Cell. Biol. 2003;23:131–139. doi: 10.1128/MCB.23.1.131-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani T., et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature. 1999;399:798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- Kanungo J., Pratt S. J., Marie H., Longmore G. D. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol. Biol. Cell. 2000;11:3299–3313. doi: 10.1091/mbc.11.10.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh Y., Katoh M. Hedgehog signaling pathway and gastrointestinal stem cell signaling network (review) Int. J. Mol. Med. 2006a;18:1019–1023. [PubMed] [Google Scholar]
- Katoh Y., Katoh M. WNT antagonist, SFRP1, is Hedgehog signaling target. Int. J. Mol. Med. 2006b;17:171–175. [PubMed] [Google Scholar]
- Kitagawa M., Hatakeyama S., Shirane M., Matsumoto M., Ishida N., Hattori K., Nakamichi I., Kikuchi A., Nakayama K., Nakayama K. An F-box protein, FWD1, mediates ubiquitin-dependent proteolysis of beta-catenin. EMBO J. 1999;18:2401–2410. doi: 10.1093/emboj/18.9.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolligs F. T., et al. ITF-2, a downstream target of the Wnt/TCF pathway, is activated in human cancers with beta-catenin defects and promotes neoplastic transformation. Cancer Cell. 2002;1:145–155. doi: 10.1016/s1535-6108(02)00035-1. [DOI] [PubMed] [Google Scholar]
- Kuhl M., Sheldahl L. C., Malbon C. C., Moon R. T. Ca(2+)/calmodulin-dependent protein kinase II is stimulated by Wnt and Frizzled homologs and promotes ventral cell fates in Xenopus. J. Biol. Chem. 2000;275:12701–12711. doi: 10.1074/jbc.275.17.12701. [DOI] [PubMed] [Google Scholar]
- Le Bivic A., Quaroni A., Nichols B., Rodriguez-Boulan E. Biogenetic pathways of plasma membrane proteins in Caco-2, a human intestinal epithelial cell line. J. Cell Biol. 1990;111:1351–1361. doi: 10.1083/jcb.111.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gall A. H., Yeaman C., Muesch A., Rodriguez-Boulan E. Epithelial cell polarity: new perspectives. Semin. Nephrol. 1995;15:272–284. [PubMed] [Google Scholar]
- Lens S. M., Vader G., Medema R. H. The case for Survivin as mitotic regulator. Curr. Opin. Cell Biol. 2006;18:616–622. doi: 10.1016/j.ceb.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Madison B. B., Braunstein K., Kuizon E., Portman K., Qiao X. T., Gumucio D. L. Epithelial hedgehog signals pattern the intestinal crypt-villus axis. Development. 2005;132:279–289. doi: 10.1242/dev.01576. [DOI] [PubMed] [Google Scholar]
- Marikawa Y., Elinson R. P. beta-TrCP is a negative regulator of Wnt/beta-catenin signaling pathway and dorsal axis formation in Xenopus embryos. Mech. Dev. 1998;77:75–80. doi: 10.1016/s0925-4773(98)00134-8. [DOI] [PubMed] [Google Scholar]
- Mason J. M., Morrison D. J., Basson M. A., Licht J. D. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 2006;16:45–54. doi: 10.1016/j.tcb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Massague J., Gomis R. R. The logic of TGFbeta signaling. FEBS Lett. 2006;580:2811–2820. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Matter K., Brauchbar M., Bucher K., Hauri H. P. Sorting of endogenous plasma membrane proteins occurs from two sites in cultured human intestinal epithelial cells (Caco-2) Cell. 1990;60:429–437. doi: 10.1016/0092-8674(90)90594-5. [DOI] [PubMed] [Google Scholar]
- Melhem M. F., Meisler A. I., Finley G. G., Bryce W. H., Jones M. O., Tribby I. I., Pipas J. M., Koski R. A. Distribution of cells expressing myc proteins in human colorectal epithelium, polyps, and malignant tumors. Cancer Res. 1992;52:5853–5864. [PubMed] [Google Scholar]
- Meneghini M. D., Ishitani T., Carter J. C., Hisamoto N., Ninomiya-Tsuji J., Thorpe C. J., Hamill D. R., Matsumoto K., Bowerman B. MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature. 1999;399:793–797. doi: 10.1038/21666. [DOI] [PubMed] [Google Scholar]
- Mertens C., Hofmann I., Wang Z., Teichmann M., Sepehri Chong S., Schnolzer M., Franke W. W. Nuclear particles containing RNA polymerase III complexes associated with the junctional plaque protein plakophilin 2. Proc. Natl. Acad. Sci. USA. 2001;98:7795–7800. doi: 10.1073/pnas.141219498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikels A. J., Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miravet S., Piedra J., Miro F., Itarte E., Garcia de Herreros A., Dunach M. The transcriptional factor Tcf-4 contains different binding sites for beta-catenin and plakoglobin. J. Biol. Chem. 2002;277:1884–1891. doi: 10.1074/jbc.M110248200. [DOI] [PubMed] [Google Scholar]
- Molenaar M., van de Wetering M., Oosterwegel M., Peterson-Maduro J., Godsave S., Korinek V., Roose J., Destree O., Clevers H. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Nelson C. M., Bissell M. J. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 2006;22:287–309. doi: 10.1146/annurev.cellbio.22.010305.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson W. J., Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi H., Pardali K., Vanlandewijck M., Heldin C. H., Moustakas A. Notch signaling is necessary for epithelial growth arrest by TGF-β. J. Cell Biol. 2007;176:695–707. doi: 10.1083/jcb.200612129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S., Wakabayashi N., Toyoda K., Kashima K., Mitsufuji S. Expression of Musashi-1 in human normal colon crypt cells: a possible stem cell marker of human colon epithelium. Dig. Dis. Sci. 2003;48:1523–1529. doi: 10.1023/a:1024763723240. [DOI] [PubMed] [Google Scholar]
- Noseda M., Chang L., McLean G., Grim J. E., Clurman B. E., Smith L. L., Karsan A. Notch activation induces endothelial cell cycle arrest and participates in contact inhibition: role of p21Cip1 repression. Mol. Cell. Biol. 2004;24:8813–8822. doi: 10.1128/MCB.24.20.8813-8822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou C. M., et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Pinto D., Clevers H. Wnt control of stem cells and differentiation in the intestinal epithelium. Exp. Cell Res. 2005;306:357–363. doi: 10.1016/j.yexcr.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Pinto D., Gregorieff A., Begthel H., Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- Powell S. M., Zilz N., Beazer-Barclay Y., Bryan T. M., Hamilton S. R., Thibodeau S. N., Vogelstein B., Kinzler K. W. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- Qi R., An H., Yu Y., Zhang M., Liu S., Xu H., Guo Z., Cheng T., Cao X. Notch1 signaling inhibits growth of human hepatocellular carcinoma through induction of cell cycle arrest and apoptosis. Cancer Res. 2003;63:8323–8329. [PubMed] [Google Scholar]
- Rangarajan A., et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robitaille J., et al. Mutant frizzled-4 disrupts retinal angiogenesis in familial exudative vitreoretinopathy. Nat. Genet. 2002;32:326–330. doi: 10.1038/ng957. [DOI] [PubMed] [Google Scholar]
- Rocheleau C. E., Yasuda J., Shin T. H., Lin R., Sawa H., Okano H., Priess J. R., Davis R. J., Mello C. C. WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell. 1999;97:717–726. doi: 10.1016/s0092-8674(00)80784-9. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., Kreitzer G., Musch A. Organization of vesicular trafficking in epithelia. Nat. Rev. 2005;6:233–247. doi: 10.1038/nrm1593. [DOI] [PubMed] [Google Scholar]
- Roose J., Huls G., van Beest M., Moerer P., van der Horn K., Goldschmeding R., Logtenberg T., Clevers H. Synergy between tumor suppressor APC and the beta-catenin-Tcf4 target Tcf1. Science. 1999;285:1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- Roose J., Molenaar M., Peterson J., Hurenkamp J., Brantjes H., Moerer P., van de Wetering M., Destree O., Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- Royet J., Bouwmeester T., Cohen S. M. Notchless encodes a novel WD40-repeat-containing protein that modulates Notch signaling activity. EMBO J. 1998;17:7351–7360. doi: 10.1093/emboj/17.24.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagara N., Kirikoshi H., Terasaki H., Yasuhiko Y., Toda G., Shiokawa K., Katoh M. FZD4S, a splicing variant of frizzled-4, encodes a soluble-type positive regulator of the WNT signaling pathway. Biochem. Biophys. Res. Commun. 2001;282:750–756. doi: 10.1006/bbrc.2001.4634. [DOI] [PubMed] [Google Scholar]
- Sampson E. M., Haque Z. K., Ku M. C., Tevosian S. G., Albanese C., Pestell R. G., Paulson K. E., Yee A. S. Negative regulation of the Wnt-beta-catenin pathway by the transcriptional repressor HBP1. EMBO J. 2001;20:4500–4511. doi: 10.1093/emboj/20.16.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho E., Batlle E., Clevers H. Signaling pathways in intestinal development and cancer. Annu. Rev. Cell Dev. Biol. 2004;20:695–723. doi: 10.1146/annurev.cellbio.20.010403.092805. [DOI] [PubMed] [Google Scholar]
- Sekiya T., Nakamura T., Kazuki Y., Oshimura M., Kohu K., Tago K., Ohwada S., Akiyama T. Overexpression of Icat induces G(2) arrest and cell death in tumor cell mutants for adenomatous polyposis coli, beta-catenin, or Axin. Cancer Res. 2002;62:3322–3326. [PubMed] [Google Scholar]
- Sheldahl L. C., Park M., Malbon C. C., Moon R. T. Protein kinase C is differentially stimulated by Wnt and Frizzled homologs in a G-protein-dependent manner. Curr. Biol. 1999;9:695–698. doi: 10.1016/s0960-9822(99)80310-8. [DOI] [PubMed] [Google Scholar]
- Smit L., Baas A., Kuipers J., Korswagen H., van de Wetering M., Clevers H. Wnt activates the Tak1/Nemo-like kinase pathway. J. Biol. Chem. 2004;279:17232–17240. doi: 10.1074/jbc.M307801200. [DOI] [PubMed] [Google Scholar]
- Sriuranpong V., Borges M. W., Strock C. L., Nakakura E. K., Watkins D. N., Blaumueller C. M., Nelkin B. D., Ball D. W. Notch signaling induces rapid degradation of achaete-scute homolog 1. Mol. Cell. Biol. 2002;22:3129–3139. doi: 10.1128/MCB.22.9.3129-3139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tago K., Nakamura T., Nishita M., Hyodo J., Nagai S., Murata Y., Adachi S., Ohwada S., Morishita Y., Shibuya H., Akiyama T. Inhibition of Wnt signaling by ICAT, a novel beta-catenin-interacting protein. Genes Dev. 2000;14:1741–1749. [PMC free article] [PubMed] [Google Scholar]
- Taipale J., Beachy P. A. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- Topol L., Jiang X., Choi H., Garrett-Beal L., Carolan P. J., Yang Y. Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent beta-catenin degradation. J. Cell Biol. 2003;162:899–908. doi: 10.1083/jcb.200303158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V. G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M., et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- van den Brink G. R., et al. Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nat. Genet. 2004;36:277–282. doi: 10.1038/ng1304. [DOI] [PubMed] [Google Scholar]
- van der Heyden M. A., Rook M. B., Hermans M. M., Rijksen G., Boonstra J., Defize L. H., Destree O. H. Identification of connexin43 as a functional target for Wnt signalling. J. Cell Sci. 1998;111(Pt 12):1741–1749. doi: 10.1242/jcs.111.12.1741. [DOI] [PubMed] [Google Scholar]
- van Es J. H., et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- Van Gelder R. N., von Zastrow M. E., Yool A., Dement W. C., Barchas J. D., Eberwine J. H. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl. Acad. Sci. USA. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman M. T., Axelrod J. D., Moon R. T. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev. Cell. 2003;5:367–377. doi: 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- Wang Y., Gilmore T. D. LIM domain protein Trip6 has a conserved nuclear export signal, nuclear targeting sequences, and multiple transactivation domains. Biochim. Biophys. Acta. 2001;1538:260–272. doi: 10.1016/s0167-4889(01)00077-5. [DOI] [PubMed] [Google Scholar]
- Whitfield M. L., et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wice B. M., Trugnan G., Pinto M., Rousset M., Chevalier G., Dussaulx E., Lacroix B., Zweibaum A. The intracellular accumulation of UDP-N-acetylhexosamines is concomitant with the inability of human colon cancer cells to differentiate. J. Biol. Chem. 1985;260:139–146. [PubMed] [Google Scholar]
- Wielenga V. J., Smits R., Korinek V., Smit L., Kielman M., Fodde R., Clevers H., Pals S. T. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am. J. Pathol. 1999;154:515–523. doi: 10.1016/S0002-9440(10)65297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W. M., Wright N. A. Epidermal growth factor, epidermal growth factor receptors, intestinal growth, and adaptation. J. Parenter. Enteral Nutr. 1999;23:S83–S88. doi: 10.1177/014860719902300521. [DOI] [PubMed] [Google Scholar]
- Woods A. J., Roberts M. S., Choudhary J., Barry S. T., Mazaki Y., Sabe H., Morley S. J., Critchley D. R., Norman J. C. Paxillin associates with poly(A)-binding protein 1 at the dense endoplasmic reticulum and the leading edge of migrating cells. J. Biol. Chem. 2002;277:6428–6437. doi: 10.1074/jbc.M109446200. [DOI] [PubMed] [Google Scholar]
- Yeaman C., Grindstaff K. K., Nelson W. J. New perspectives on mechanisms involved in generating epithelial cell polarity. Physiol. Rev. 1999;79:73–98. doi: 10.1152/physrev.1999.79.1.73. [DOI] [PubMed] [Google Scholar]
- Yeaman C., Grindstaff K. K., Nelson W. J. Mechanism of recruiting Sec6/8 (exocyst) complex to the apical junctional complex during polarization of epithelial cells. J. Cell Sci. 2004;117:559–570. doi: 10.1242/jcs.00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisberg M., Hanai J., Sugimoto H., Mammoto T., Charytan D., Strutz F., Kalluri R. BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat. Med. 2003;9:964–968. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.