Abstract
Although peroxisomes oxidize lipids, the metabolism of lipid bodies and peroxisomes is thought to be largely uncoupled from one another. In this study, using oleic acid–cultured Saccharomyces cerevisiae as a model system, we provide evidence that lipid bodies and peroxisomes have a close physiological relationship. Peroxisomes adhere stably to lipid bodies, and they can even extend processes into lipid body cores. Biochemical experiments and proteomic analysis of the purified lipid bodies suggest that these processes are limited to enzymes of fatty acid β oxidation. Peroxisomes that are unable to oxidize fatty acids promote novel structures within lipid bodies (“gnarls”), which may be organized arrays of accumulated free fatty acids. However, gnarls are suppressed, and fatty acids are not accumulated in the absence of peroxisomal membranes. Our results suggest that the extensive physical contact between peroxisomes and lipid bodies promotes the coupling of lipolysis within lipid bodies with peroxisomal fatty acid oxidation.
Introduction
Many pathways of intermediary metabolism, macromolecular biosynthesis, energy transduction, motility, protein trafficking, and intracellular signaling require collaboration among organelles. Examples include the conversion of fatty acids to gluconeogenic substrates (Masters, 1997; Penfield et al., 2005), sterol biosynthesis (Zinser et al., 1993; Krisans, 1996), membrane lipid synthesis (Voelker, 2005), the unfolded protein response (Zhang and Kaufman, 2004), vesicular transport (Beraud-Dufour and Balch, 2002; Gundelfinger et al., 2003), the retrograde pathway (Butow and Avadhani, 2004), and calcium signaling (Ma and Pan, 2003; Rizzuto et al., 2004).
Intermediates en route between organelles generally pass through the cytoplasm, but this does not always have to be the case. Organellar compartments are usually densely packed within the cytoplasm, and organelles often associate with one another, raising the possibility of communication that bypasses the cytoplasmic compartment.
Intracellular lipid bodies store neutral lipids, mainly triglycerides and sterol esters (Zweytick et al., 2000). The organelles are derived from the ER, although the details of this pathway are not well understood (Murphy and Vance, 1999; Robenek et al., 2004; for review see Murphy, 2001). They are bounded by a phospholipid monolayer (Tauchi-Sato et al., 2002) into which are embedded a specific subset of proteins. Many of these have been identified through recent proteomic efforts (Athenstaedt et al., 1999; Brasaemle et al., 2004; Liu et al., 2004). This compartment is comprised of enzymes that promote the synthesis or mobilization of lipids, structural proteins, and signaling molecules.
Lipid bodies can be in close proximity to mitochondria or ER (Blanchette-Mackie et al., 1995; Cohen et al., 2004). The ER can virtually surround lipid bodies, and mitochondria have been detected in the cortical layer surrounding lipid bodies. There are also reports of the close proximity of peroxisomes to lipid bodies in etiolated cotyledons, mammalian cells, and yeast (Hayashi et al., 2001; Schrader, 2001; Bascom et al., 2003). Because lipid bodies are an obvious source for fatty acids, the substrates for mitochondrial and peroxisomal oxidation, which are contacts between lipid bodies and peroxisomes or mitochondria, could indicate the direct transfer of fatty acids across organellar boundaries.
The yeast Saccharomyces cerevisiae is an apt model system to explore peroxisome–lipid body interactions. This organism can be cultured on oleic acid, which generates large lipid bodies and causes peroxisomes to become engorged with enzymes of fatty acid oxidation and the glyoxylate cycle (Veenhuis et al., 1987; McCammon et al., 1990).
In this study, we show that peroxisomes stably adhere to lipid bodies when grown in oleic acid and that they can extend processes into their core. Enzymes of peroxisomal β oxidation are selectively enriched in purified lipid bodies. Our data suggest that peroxisomal contact may stimulate neutral lipid breakdown in lipid bodies: fatty acids accumulate within lipid bodies if peroxisomes are present but are unable to metabolize them, generating novel structures we term “gnarls.”
Results
Peroxisomes associate extensively with lipid bodies
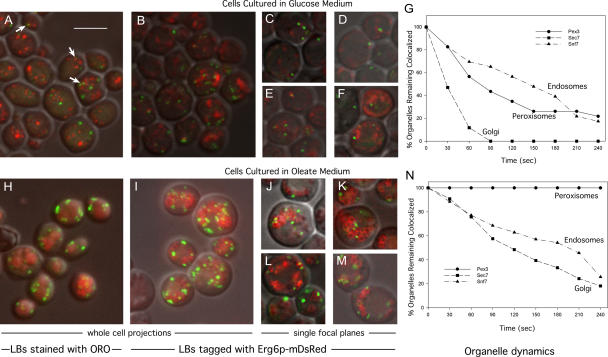
To determine the extent to which peroxisomes associate with lipid bodies in S. cerevisiae, Pot1p (peroxisomal 3-ketoacyl-CoA thiolase) was tagged with GFP, and cells were stained for lipid bodies with oil red O. Cells cultured in glucose had relatively small lipid bodies and few peroxisomes (Fig. 1 A), although peroxisomes associating with lipid bodies could easily be seen (Fig. 1 A, arrows). Cells cultured in oleic acid contained large clusters of lipid bodies and more peroxisomes, many of which seemed to adhere to the lipid bodies (Fig. 1 H).
Figure 1.
Peroxisomes decorate lipid bodies. MMYO11α expressing Pot1p (thiolase)-GFP (A–F and H–M) or other GFP-tagged proteins (G and N) was grown in SD medium (2% glucose; A–G) or oleate medium (H–N). Cells were either fixed and stained with oil red O (ORO; A and H) or expressed Erg6p-mDsRed (B–G and I–N). A, B, H, and I represent projections through entire cells, whereas C–F and J–M are images of single planes. Lipid bodies (LBs) are larger and more abundant in oleate medium (J–M). Examples of peroxisomes associating with lipid bodies are indicated by arrows in A. For G and N, Erg6p-mDsRed was introduced into strains expressing GFP-tagged Pex3p, Sec7p, or Snf7p as indicated. Individual GFP-tagged organelles (20–30 per strain) that were associated with lipid bodies in glucose (G) or oleate (N) cultures were observed continually over 4 min and scored for release from lipid bodies. For clarity, the graphs show loss of organelles at 30-s intervals. Bar, 5 μm (applies to all images).
To determine whether the same pattern was seen in living cells, cells containing Pot1p-GFP were also tagged with Erg6p-mDsRed (Fig. 1, B–F and I–M); Erg6p is an abundant protein of lipid bodies (Athenstaedt et al., 1999). Because Erg6p is on the periphery of the organelle, the large lipid bodies from oleate cultures often appear hollow. Again, peroxisomes were seen to decorate lipid bodies, especially in cells grown on oleic acid. To quantify this effect and compare it with the extent of association of other organelles with lipid bodies, we obtained matched strains (Huh et al., 2003) in which markers of peroxisomes, late Golgi, or endosomes (Pex3p, Sec7p, or Snf7p, respectively) were tagged with GFP. We then labeled their lipid bodies with Erg6p-mDsRed as before. Random images from single focal planes (such as those in Fig. 1, C–F and J–M) were used to score the organelles that appeared to be associated with lipid bodies. We found in glucose cultures that 64.0 ± 2.9% of peroxisomes associated with lipid bodies compared with 49.5 ± 1.2 and 43.3 ± 3.7% of late Golgi and endosomal particles, respectively. Although a fraction of these interactions undoubtedly reflect adventitious proximity, these data may also reflect a role of lipid bodies in providing membrane lipids to all of these organelles. In oleate cultures, >90% of all three organelles seemed to associate, although much of this reflects the large volume within the cytoplasm that is taken up by the lipid bodies and the limited spatial resolution of this technique.
To compare the dynamics of these organellar associations, we tracked the dissociation of individual organelles from lipid bodies using spinning disk confocal microscopy, and the results are shown for glucose and oleate cultures in Fig. 1 (G and N, respectively). In glucose culture, both peroxisomes and endosomes dissociated from lipid bodies more slowly than did Golgi, although only 20% of peroxisomes remained bound after 4 min. In contrast, all observed peroxisomes in cells from oleate culture remained bound during the experiment, whereas 74% of endosomes and 82% of Golgi dissociated (see Videos 1–3 of representative cells from the three tagged strains, available at http://www.jcb.org/cgi/content/full/jcb.200511125/DC1).
These data indicate that peroxisomes are tightly bound to lipid bodies in oleate cultures, where fatty acids could potentially be transferred. In contrast, in cells cultured in glucose where fatty acid oxidation is repressed, the relationship is more transient.
Peroxisomes form extensive contacts with lipid bodies and can penetrate them
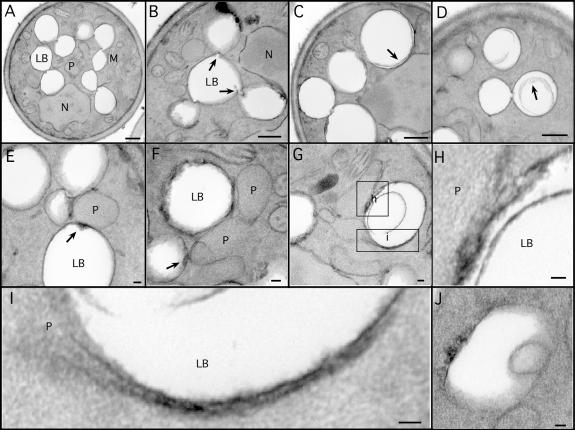
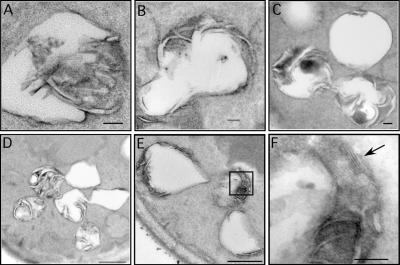
To visualize the physical association of peroxisomes and lipid bodies at higher resolution, cells were imaged by transmission electron microscopy. The large fraction of cytoplasmic volume taken up by lipid bodies is obvious in these cells (Fig. 2 A). Lipid bodies had extensive contacts with each other. In Fig. 2 A, all nine lipid bodies in the section are connected in a linear array. Less often, branches were observed; the lipid body in the center of Fig. 2 B contacts three other lipid bodies. The individual units were connected through novel structures that resemble nipples or valves (Fig. 2 B, arrows).
Figure 2.
Ultrastructure of lipid bodies and contacts with peroxisomes. Cells were grown for 18–21 h in oleate medium and processed for EM. (A–D) Aspects of lipid body structure are illustrated, particularly their connections to each other (A and B) and inclusions (C and D). Arrows in B illustrate valvelike structures; arrows in C and D illustrate inclusions. (E–H) Lipid bodies in contact with peroxisomes are shown, illustrating electron-dense material at the site of contact (arrows). Note a peroxisome in contact with three lipid bodies (E). A lipid body making contact with two peroxisomes, one of which has extended a tail around it, and the other has breached the periphery and is extending a process (pexopodium) into the core (G). The boxed areas are shown in higher resolution in H and I. Another putative pexopodium is shown in J. LB, lipid body; M, mitochondria; N, nucleus, P, peroxisome. Bars (A–D), 500 nm; (E–J), 50 nm.
We frequently observed whisps or ribbons of electron-dense material within lipid bodies either close to the periphery or more internally (Fig. 2, C, D, and H). The composition of this material is unknown, although they may represent structures induced by free fatty acids (see Fatty acids accumulate…in pex3Δ).
As expected from the fluorescently tagged cells, contacts between peroxisomes and lipid bodies were easily detected (Fig. 2, E–G). The contact sites usually were accompanied by an increase in electron density on the lipid body surface (Fig. 2, E and F; arrows), suggesting an interface more complex than the three phospholipid leaflets of the apposing organelles.
Occasionally, lipid bodies were detected with extensions of peroxisomes that we term pexopodia, which penetrate into the core of the organelles. Fig. 2 G shows a lipid body that is both enwrapped by a peroxisomal tail (bottom rectangle, enlarged in Fig. 2 I) and is also penetrated by a separate peroxisome (top square, enlarged in Fig. 2 H). The tail contains a midline structure that is similar to peroxisomal extensions that extend from the ER in mouse dendritic cells (Geuze et al., 2003). The pexopodium extends beyond the periphery of the organelle, appearing to make contact with the lipid core. In other lipid bodies, we see inclusions that may be the result of pexopodial invasion (an example is shown in Fig. 2 J).
Pexopodia occurred in 3.2% of cell sections in which both lipid bodies and peroxisomes were visible. We saw a general correlation between the number of pexopodia and the nutritional state of the cells. For example, when cells growing in oleate were starved of carbon source for 8 h, the occurrence of pexopodia rose to 9.1%. An intermediate response was seen when cells growing on oleate were switched to low (0.1%) raffinose: 7.2% of cells had pexopodia. These results suggest that pexopodial formation was stimulated when cells were forced to use fatty acids from lipid bodies. As reported elsewhere for other systems (Blanchette-Mackie et al., 1995; Cohen et al., 2004), we also observed frequent contacts of lipid bodies with ER and mitochondria (unpublished data).
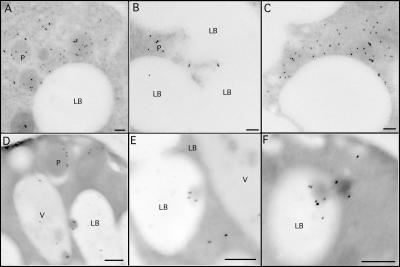
To examine with more certainty the physical interactions of peroxisomes and lipid bodies, we used antibodies to Pot1p and Pox1p (acyl-CoA oxidase), two abundant core enzymes of peroxisomal β oxidation, in an immunogold analysis (Fig. 3). Both antibodies stained peroxisomes, although antithiolase typically stained throughout the peroxisomal matrix (Fig. 3, A–C), whereas antioxidase more often stained closer to the membrane (Fig. 3 D). Thiolase often appeared extensively around the periphery of lipid bodies, representing peroxisomes bound to these organelles. In contrast, acyl-CoA oxidase staining also was seen in the lipid body inclusions (Fig. 3, D–F).
Figure 3.
Lipid body inclusions contain Pox1p. MMYO11α was grown overnight in oleate medium and processed for immunogold EM. (A–C) Anti-Pot1p antibody illustrates peroxisomes making extensive contacts with the lipid body. (D–F) Anti-Pox1p antibodies stain peroxisomes (D) but also stain inclusions (pexopodia) in lipid bodies. We have not detected the immunoreactivity of pexopodia with anti-Pot1p. LB, lipid body; P, peroxisome; V, vacuole. Bars, 100 μm.
Our images indicate that peroxisomes frequently associate with lipid bodies in S. cerevisiae, suggesting a close metabolic relationship. The contacts between lipid bodies and peroxisomes was more than mere apposition; peroxisomes extended processes into the cores of lipid bodies. The immunogold evidence raises the possibility that the contents of pexopodia and lipid body inclusions may be somewhat different to that of intact peroxisomes and are perhaps specialized structures for the rapid and efficient mobilization/oxidation of stored lipid.
Pox1p and other enzymes of β oxidation associate tightly with lipid bodies
Biochemical evidence provided further support for lipid body–peroxisome associations. For these experiments, we tagged lipid bodies by introducing the myc epitope into the chromosomal copy of ERG6.
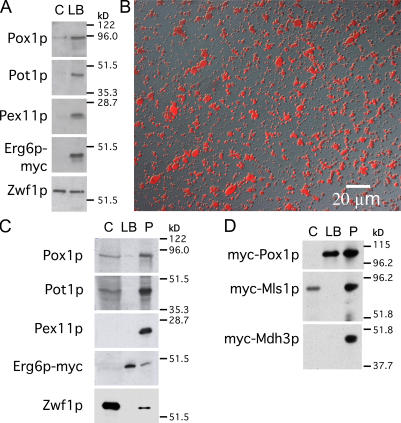
The binding of peroxisomes to lipid bodies was confirmed by gently floating lipid bodies from a postnuclear supernatant (PNS). The Erg6p-myc marker was clearly enriched in this fraction compared with the lipid body–depleted cytoplasm underneath (Fig. 4 A). Similarly, three peroxisomal markers, Pox1p, Pot1p, and Pex11p (an abundant peroxisomal membrane protein), were also enriched in this fraction. In contrast, the cytoplasmic marker Zwf1p was not enriched. (The lipid body fraction was not purified further to remove contaminating cytoplasm for fear of disrupting this association).
Figure 4.
Purified lipid bodies contain Pox1p. (A) A PNS from cells expressing Erg6p-myc was grown in oleic acid and gently centrifuged to float lipid bodies. The lipid body fraction and underlying supernatant were analyzed for organelle markers. Peroxisomal proteins but not cytosolic Zwf1p were enriched in the lipid body fraction. (B) Lipid bodies were purified and stained with oil red O. (C) Purified lipid bodies and an organellar pellet were analyzed for peroxisome markers. An equal percentage of cytosol, washed lipid bodies, and pellet (C, LB, and P, respectively) were analyzed by immunoblotting. Pox1p but not Pot1p or Pex11p is visible in the lipid body fraction. (D) As in C, except cells expressed myc-tagged Pox1p, Mls1p, or Mdh3p as indicated, and they were cultured and processed in parallel.
Next, lipid bodies were highly purified by centrifuging the organelles from a PNS through a buffer cushion and washing them several times, which is similar to the procedure previously described for mammalian lipid bodies (Liu et al., 2004). The quality of the preparation was assessed by staining with oil red O. Virtually all particles stained with this dye, suggesting a high level of lipid body purity and integrity (Fig. 4 B). These organelles, along with the cytosol and organellar pellet (containing peroxisomes), were analyzed by immunoblotting (Fig. 4 C). Zwf1p was almost exclusively localized to the cytosol fraction, whereas Pex11p was found exclusively in the organellar pellet. Pox1p and Pot1p were also localized to the pellet, although in this experiment, there was leakage of Pot1p into the supernatant. However, a small fraction of Pox1p (10% or more of total Pox1p in some experiments) clearly migrated with the purified lipid bodies in all experiments, which is consistent with our immunogold results. We next compared the localization of Pox1p with that of two peroxisomal proteins that function outside the β-oxidation pathway: Mls1p and Mdh3p (both myc tagged). Neither enzyme associated with lipid bodies (Fig. 4 D). We considered the possibility that Pox1p traffics to lipid bodies independent of its targeting to peroxisomes. The localization of Pox1p to peroxisomes depends on the shuttling receptor Pex5p. We found that Pox1p associated with neither peroxisomes nor lipid bodies but remained in the cytosol in a pex5Δ strain (unpublished data), indicating that Pox1p trafficking to lipid bodies is Pex5p dependent. However, because Pox1p binds to a location on Pex5p far removed from the normal PTS1 (peroxisomal targeting signal 1)-binding site (Klein et al., 2002), we considered the possibility that Pex5p might shuttle Pox1p to lipid bodies in some unusual way that bypassed peroxisomes. To address this possibility, we forced Pox1p to traffic via the PTS1 pathway by attaching a PTS1 to Pox1p and eliminating the normal site of interaction on Pex5p for Pox1p binding (see Materials and methods; Klein et al., 2002). With these modifications, Pox1p still localized to peroxisomes and lipid bodies as it did in control cells (unpublished data). We conclude that the binding of Pox1p to lipid bodies occurs subsequent to its targeting to peroxisomes. However, the binding of Pot1p, Pex11p, Mls1p, or Mdh3p to these organelles is somehow excluded or is at least much weaker so that they are removed upon fractionation.
To identify other peroxisomal proteins that were tightly associated with lipid bodies and to detect other tightly associated organelles, highly purified lipid bodies from oleate cultures were subjected to proteolysis and mass spectrometric analysis of peptides. The identified proteins corresponding to the peptides are listed in Table I. All but three of them fall into the following three categories: (1) previously identified proteins of lipid bodies (Athenstaedt et al., 1999); (2) proteins of mitochondria, peroxisomes, and/or ER (organelles that we and others have seen associating with lipid bodies; Blanchette-Mackie et al., 1995; Cohen et al., 2004; unpublished data); and (3) cytosolic proteins (assignments of localization based on www.yeastgenome.org). We did not consider the three remaining identifications, Pma2p (the plasma membrane hydrogen-exporting ATPase), Hnf1/2p, and Htb1/2p (histones), of likely physiological significance. Among cytoskeletal elements, only actin was identified. No proteins of other cellular compartments (including vacuoles or other endocytic or exocytic organelles) were found, suggesting that adventitious binding of cellular elements during organelle isolation was minimal.
Table I.
Proteins identified with purified lipid bodies from oleate-cultured cells
| Category | Proteins | ||
|---|---|---|---|
| Known lipid body proteins | Ayr1p, Eht1p, Erg1p, Erg6p, Erg7p, Erg27p, Faa1p, Faa4p, Fat1p, Hfd1p, Pdr16p, Pet10p, Tg11p Tg13p, Slc1p | ||
| Peroxisomal, mitochondrial, or ER proteins |
|||
| Peroxisome proteins | Matrix | Membrane | |
| Cta1p, Faa2p, Fox2p, Idp3p, Pcs60p, Pox1p, Sps19p, Tes1p |
None | ||
| Mitochondrial proteins | Matrix | Inner/outer membrane or nucleoid | |
| Bat1p and Cit1p Ald4p, Hsp60p, Ilv5p, and Kgd1p are matrix proteins localized to nucleoids |
Aac2, Ady2p, Ald4p, Atp1p, Atp2p, Atp4p, Atp6p, Atp7p, Cor1p, Dld1p, Fmp52, Gut2p, Hsp60p, Ilv5p, Kgd1p, Mcr1p, Mir1p, Nde1p, Nde2p, Ndi1p, Om45p, Pet9p, Phb1p, Por1p, Qcr2p, Qcr7p, Sdh1p, Tom40p |
||
| ER proteins | Resident | Trafficking | |
| Dpm1p, Erg2p, Gpi8p, Gtt1p, Kar2p, Pdi1p, Pmt1p, Sec63p |
Arf1p, Erv14p, Sar1p, Sec21p, Ypt1p | ||
| Partially in peroxisomes, mitochondria, and/or ER |
Acs1p (c, m), Cat2p (m, p), Hxt(6a/o7)pa (pm, m), Msc1p (e, m), Nce2p (c, m, e), Rtn1p (e, m), Rho1p (pm, m, p), Yhb1p (c, m) |
||
| Cytosolic proteins | ||||
| Cytosolic proteins | Translation factors | Chaperones | Others | |
| Eft1/2p, Rpl4A, Rpl7A, Rpl5p, Rps7A, Rps5p, Tef1/2p, Tif1/2p |
Hsc82p, Hsp104p, Ssa3p, Ssb1p |
Act1p, Adh2p Fba1p, Gvp36p, Hom3p, Pgk1p | ||
| Proteins also in nucleus | Bmh2p, Crm1p, Pdc1p, Rib1p, Ynl101w, Ypr127w | |||
| Others | Hhf1/2p (n), Htb(1a/o2)p (n), Pma2p (pm) | |||
Organelle assignments are based on data accumulated at www.yeastgenome.org. Underlined identifications correspond to the 20 best (lowest) logE scores (Fenyo and Beavis, 2003). All identifications in the table have a score below −3, corresponding to at least 99.9% certainty. c, cytosol; e, ER; m, mitochondria; p, peroxisome; pm, plasma membrane; n, nucleus.
For protein1 and/or protein2, identified peptides occur in both proteins, and a unique assignment cannot be made. For protein1/protein2, the two proteins have identical sequences.
Of the 24 cytosolic proteins identified, eight were ribosomal components or translational factors, and four others were molecular chaperones. Among the remaining 12 are several other abundant metabolic enzymes. The significance of these associated proteins, if any, will require further work.
We assume that the ER and mitochondrial proteins identified in the lipid body isolation were derived from tightly attached organelles, as is the case for mammalian lipid bodies (Brasaemle et al., 2004). A region of the ER is known to tightly associate with mitochondria (Vance, 1990). Nearly all of the identified mitochondrial proteins were derived from membranes or nucleoids (25 of 27), suggesting leakage of soluble proteins during fractionation.
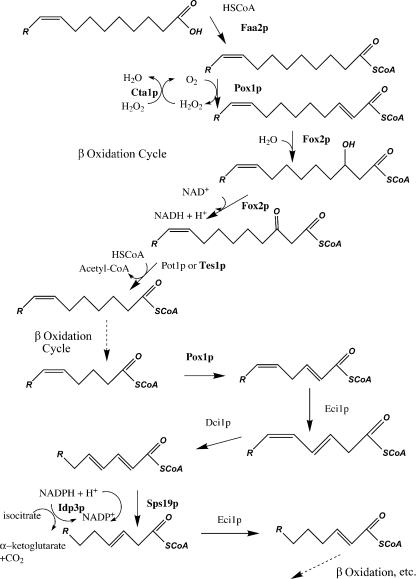
In contrast, all of the eight peroxisomal proteins in the fraction were matrix components. Furthermore, all eight proteins function in the β-oxidation pathway of oleic acid (Fig. 5). Besides the core enzymes of β oxidation (including Pox1p), two proteins involved in the rearrangement of the double bond in oleic acid (Sps19p and Idp3p) and one protein that detoxifies H2O2 (Cta1p) were identified. In fact, the entire β-oxidation pathway was represented in the lipid body proteome except Dci1p and Eci1p. Interestingly, although Pot1p was not detected (consistent with our biochemical and immunogold data), the other peroxisomal ketoacyl-CoA thiolase, Tes1p, was identified even though it is much less abundant in isolated peroxisomes than Pot1p (unpublished data). Other abundant matrix proteins, such as the glyoxylate cycle enzymes Mls1p and Mdh3p, were not identified, which is consistent with our fractionation results, nor were any peroxisomal membrane proteins, including Pex11p, the most abundant of them. These data suggest that pexopodia and lipid body inclusions are enriched in the enzymes of fatty acid oxidation (with Tes1p catalyzing the thiolase reaction) and not those proteins with other peroxisomal functions.
Figure 5.
Peroxisomal β oxidation of oleic acid in S. cerevisiae. The first round is shown in detail. The enzymes in bold are the only peroxisomal proteins identified in the lipid body proteome. These may represent the major proteins of pexopodia.
As a control, we also performed identical analyses on lipid bodies purified from a glucose culture. Only one peroxisomal protein was detected in lipid bodies from this source, Pex30p, and it ranked 74th by logE score (only one Pex30p peptide was confirmed; Fenyo and Beavis, 2003), suggesting a very low abundance in lipid bodies. This result is consistent with the more transient binding of peroxisomes to lipid bodies in glucose cultures, which is shown in Fig. 1 G.
To confirm that the peroxisomal identifications were caused primarily by the binding of organelles rather than random cytosolic adsorption, we also repeated the analysis in the pex5Δ strain, which lacks the PTS1 transporter. Three peroxisomal proteins were identified (Pox1p, Fox2p, and Faa2p; all contain PTS1), but all were less abundant than in the wild-type strain based on ranking by logE scores. Thus, Pox1p dropped from second to the 42nd position (immunoblots of total cell lysates showed similar levels of Pox1p in the two strains; unpublished data), Fox2p dropped from fourth to 12th, and Faa2p dropped from 19th to 46th. The other five peroxisomal proteins identified in wild-type lipid bodies were not detectable at all in lipid bodies from the pex5Δ sample even though more proteins were identified in lipid bodies in this strain (176 compared with 105 for wild type). We conclude that the localization of peroxisomal proteins with lipid bodies shown in Table I principally represents the binding of peroxisomal particles rather than adventitious binding.
The presence of mitochondrial, ER, and peroxisomal proteins (but virtually no proteins of other organelles) confirms the morphological observations that we and others have reported (Blanchette-Mackie et al., 1995; Cohen et al., 2004; unpublished data) and suggests that these organelles can be tightly coupled for metabolic or ontogenic reasons. The simplest interpretation is that these organelles dock on lipid bodies to either deliver lipids to this compartment or withdraw lipid metabolites from it.
Gnarls are novel structures within lipid bodies
Although lipid bodies usually have a simple morphology (Fig. 2 A), we sometimes observed electron-dense material within the core (Fig. 2, C and D), and a few tubular elements at the periphery were rarely seen. Electron-dense inclusions were much more abundant in the pex5Δ strain using our laboratory strain MMY011α (McCammon et al., 1990) as background. Lipid bodies in 10–20% of cell sections from this strain contained a series of elongated, curled, and often tangled electron-dense tubules (Fig. 6). The diameter of the tubules varied but was usually between 10 and 30 nm. The tubules radiated from the periphery, extending outward from the lipid body (Fig. 6, A and B) or inward (Fig. 6 D, central lipid body). Lipid bodies that were connected to each other were often filled by similar structures (Fig. 6 D). We termed these lipid body inclusions gnarls. The lumen of the tubules often emptied into the core of the lipid body. There was usually an electron-dense ground substance associated with the tubules (Fig. 6, C and E). In certain sections, the dense material resolved into an ordered pattern of electron-dense rows 7 nm apart (Fig. 6 F, arrow).
Figure 6.
Gnarls. (A–F) Gnarled lipid bodies from Δpex5 are illustrated. A higher resolution image from the boxed area in E is shown in F. Arrow points to the pattern of electron-dense planes, 7 μm apart, which may be sheets of fatty acid–enriched membranes. Bars (A–C and F), 100 nm; (D and E), 500 nm.
We attempted to isolate gnarled lipid bodies from pex5Δ using density gradient centrifugation to identify specific proteins associated with the structure, but the profile of lipid bodies (identified with Erg6p-myc) in gradients was indistinguishable from wild-type organelles (unpublished data). There were no abundant proteins found on SDS gels from lipid body fractions that were specific to the pex5Δ strain, and the aforementioned proteomics data failed to show the presence of any peroxisomal membrane proteins, ruling out the possibility that the gnarls represented peroxisomal membranes. We also tried to visualize gnarls using different stains but found that potassium permanganate, which stains polar lipids (Song et al., 1991), was essential to visualize the gnarls. We concluded that the gnarls were probably not proteinaceous but might instead represent arrays of lipid-derived material such as free fatty acids.
We reasoned that gnarls may accumulate in the pex5Δ strain because an important Pex5p substrate failed to be imported into peroxisomes and was therefore inactive. Both Pox1p and Fox2p (encoding the two initial enzymes of the core β-oxidation pathway) are imported via Pex5p. We found that gnarls accumulated in both pox1Δ and fox2Δ strains as well as in a strain with a point mutation (E488Q) in Pox1p that should render it catalytically dead (Battaile et al., 2002; Nakajima et al., 2002). Gnarls were present but less developed in pot1Δ (probably because Tes1p can substitute as a thiolase). Thus, several single peroxisomal enzyme deficiencies were sufficient to promote gnarl formation.
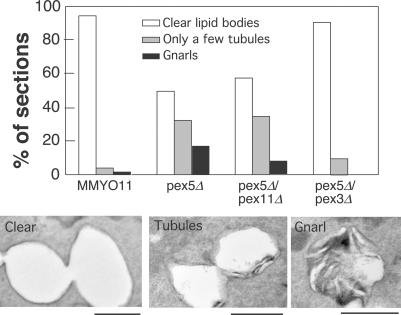
Because peroxisomes and lipid bodies can associate extensively, we determined whether the presence of peroxisomal membranes was necessary to promote gnarls. Peroxisomes do not form in a pex3Δ strain because PEX3 is essential for formation of the peroxisomal membrane (Baerends et al., 1996). Indeed, gnarls were rarely observed if PEX3 was disrupted (unpublished data). We next decided to use pex5Δ as the genetic background because gnarls are abundant in it and disrupted PEX3 in it. As a control, we also disrupted PEX11, which controls peroxisomal fission (Erdmann and Blobel, 1995; Marshall et al., 1995, 1996), in the pex5Δ background. We scored the resulting strains growing in oleic acid medium for gnarls, clear lipid bodies, or lipid bodies that contained up to a few tubules at the periphery, which is probably a precursor to gnarls (Fig. 7). Gnarls and tubules were present in wild-type cells but were quite rare (1.9% of cells had them, and another 3.8% had lipid bodies with a few tubules or less). In contrast, 50% of pex5Δ cells had either gnarls or tubules (17.3% gnarls and 32.7% tubules). Introducing pex11Δ in the pex5Δ background diminished the frequency of gnarls to 7.4%, although the number of tubules was slightly higher (35.2%). A much more dramatic effect was seen upon introducing pex3Δ in the pex5Δ background. Gnarls were completely suppressed in this strain, and the frequency of tubules was greatly diminished (to 9.6%). We conclude that gnarls form as a result of peroxisomes that are unable to metabolize fatty acids. However, gnarls require peroxisomal membranes (present in pex5Δ), suggesting that contact with peroxisomes stimulates gnarl formation. The relatively minor effect of pex11Δ may be explained by the presence of fewer peroxisomes to stimulate gnarl formation (Erdmann and Blobel, 1995; Marshall et al., 1995). A simple explanation for the nature of gnarls is that they are free fatty acids, the synthesis of which is stimulated by contact with peroxisomal membranes. If they are not metabolized by peroxisomes, they accumulate and form structures that are visualized with permanganate. Experiments are underway to identify the lipase that may be stimulated by contact with peroxisomes; there are several in lipid bodies, including the newly identified Tgl4p and Tgl5p (Athenstaedt and Daum, 2005; Kurat et al., 2006).
Figure 7.
Pex3p is essential to promote gnarls. 50 cells of the indicated strains (blinded) were scored for lipid bodies that were clear, contained at most a few tubules, or contained gnarls. Examples of each are shown below. Bars, 500 nm.
Fatty acids accumulate in lipid bodies in pex5Δ but not in pex3Δ
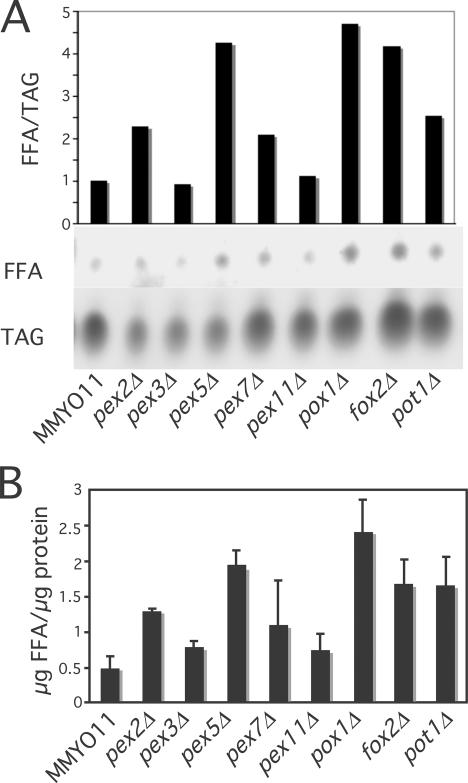
To determine whether an increase in free fatty acids within lipid bodies correlates with the development of gnarls, we determined their levels in various yeast strains, normalizing the data to the amount of lipid body triacylglycerols (Fig. 8 A) or to lipid body protein (Fig. 8 B). Similar to our detection of gnarls, levels of free fatty acids were highest in pex5Δ and in disruptions of the core β-oxidation genes. They were the lowest in wild type and pex3Δ (absence of peroxisomes) and also low in pex11Δ, where the peroxisomal fatty acid oxidation pathway is intact. The relatively lower level of fatty acids in pex7Δ (PEX7 encodes the PTS2 receptor and is required for Pot1p import) may be caused by the sharing of thiolase activity by Pot1p and Tes1p. Thus, these biochemical results are consistent with our hypothesis that gnarls result from the accumulation of fatty acids within lipid bodies, as illustrated in Fig. 9 (top and left). We predict that fatty acids generated by the interaction of lipid bodies and peroxisomes can directly enter peroxisomes, perhaps by diffusing into the pexopodia, where they can be quickly oxidized without passing through the cytosolic compartment. In the absence of peroxisomal catabolism, they form gnarls.
Figure 8.
Peroxisomal mutants cause fatty acid accumulation in lipid bodies. Lipid bodies were purified from the indicated strains and subjected to thin layer chromatography. (A) Spots were visualized and quantified after charring, and the ratio of free fatty acids to triacylglycerols are plotted. (B) Free fatty acid amounts were quantified by densitometric scanning relative to free oleic acid standards and were normalized to total lipid body protein. Results shown were based on three independent experiments. Means and SEM (error bars) are plotted.
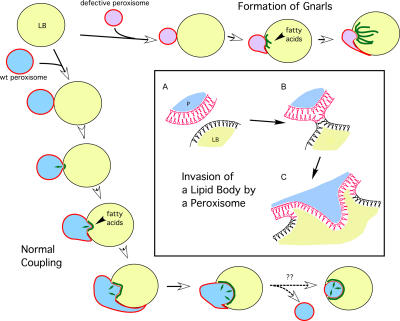
Figure 9.
Scheme depicting the development and dynamics of peroxisome–lipid body interactions. (left and bottom) Peroxisomes bind to lipid bodies (LBs) and stimulate the breakdown of triglycerides, releasing fatty acids (green lines) that get imported and oxidized by peroxisomes. Peroxisome invasion into the lipid body is also shown. (top) Peroxisomes that are unable to perform β oxidation cause the accumulation of fatty acids, resulting in gnarls. (box) Peroxisome invasion into lipid bodies. (A) A peroxisome (P) with its phospholipid bilayer approaches the lipid body monolayer. (B) A hemifusion event occurs, resulting in invasion of the lipid core by the peroxisomal membrane inner leaflet (C). Small arrows indicate the flux of fatty acids into peroxisomes for oxidation.
Discussion
In this study, we report novel interactions between peroxisomes and lipid bodies. These two organelles physically associate to a degree not previously seen outside of germinating oil seeds, where extensive contacts between glyoxysomes and lipid bodies presumably allow the efficient conversion of fatty acids to TCA cycle substrates (Hayashi et al., 2001). We provide evidence that the association of peroxisomes and lipid bodies results in the synthesis of free fatty acids from neutral lipids, providing substrate for peroxisomal oxidation. Transfer of fatty acids from the lipid body to the peroxisome may occur across the extensive lipid body–peroxisome interface, including the pexopodia.
Our work shows that peroxisomes are not solitary organelles; many associate with lipid bodies even when cells are cultured on glucose, and peroxisomes are not required for growth. It is known that peroxisomes can acquire their membrane lipids from lipid bodies (Chapman and Trelease, 1991). As our images indicate, contact may involve a large surface area. What attracts these organelles to each other? It is reasonable to think that surface proteins interact to provide the apparently high affinity. In glucose culture, peroxisomes dock on lipid bodies for a short time and then disengage; in oleate culture, interactions are more permanent. The molecular basis for this difference probably resides in the nature of the interacting surface proteins or intervening factors.
We also report the formation of pexopodia that extend into lipid bodies. Fig. 9 (box) illustrates our hypothesis that this occurs as the result of a hemifusion of the single leaflet of the lipid body membrane and the outer leaflet of the peroxisomal membrane. A stalk forms according to current fusion theory (Markin et al., 1984; Markin and Albanesi, 2002), and its enlargement leads to direct contact of the inner peroxisomal leaflet with the core of the lipid body. This is energetically favorable because the hydrophobic tails of the leaflet would interact with the neutral lipids of the core. Moreover, fatty acids from the lipid body should be able to easily diffuse across the monolayer to be available for peroxisomal oxidation.
What is the nature of these peroxisomal insertions? Evidence that they are physiologically important is provided by the list of peroxisomal proteins that are associated with purified lipid bodies (Table I). The group comprises nearly the entire β-oxidation pathway and nothing else even though there are several other peroxisomal proteins that are at least as abundant as some of the proteins identified (unpublished data). Therefore, it is tempting to speculate that these inclusions consist of sacs of β-oxidation enzymes that somehow are segregated from the other peroxisomal enzymes. Whether the pexopodia are physiologically important or are simply fragments of peroxisomes that tightly adhere to isolated lipid bodies, it is clear that the β-oxidation enzymes have a high affinity for lipid bodies compared with other peroxisomal proteins. The lack of peroxisomal membrane proteins in the preparation may reflect active exclusion from the lipid body–peroxisome interface.
Complementary proteomic results were recently obtained from purified peroxisomes, where the lipid body proteins Faa1p, Erg1p, and Erg6p were identified (Marelli et al., 2004). It is unclear whether this reflects residual lipid body monolayer that is tightly associated with purified peroxisomes. These investigators also detected a significant amount of Rho1p, as did we with lipid bodies. We do not yet know if Rho1p, which was shown in other work to be important for peroxisomal biogenesis (Marelli et al., 2004), is appearing in our analysis as a result of peroxisomal (or other endomembrane) attachment or if Rho1p has a separate function for lipid body dynamics.
Our entry into this project was the visualization of gnarls in pex5Δ. We originally hypothesized that gnarls were the result of aberrant peroxisomal membrane assembly in association with lipid bodies, but we looked in vain for peroxisomal proteins associating with these structures. We then considered that they were lipid structures. This is consistent with the increase in free fatty acids that we see in these aberrant lipid bodies. The gnarl tubules could represent inverted fatty acid structures where the polar head groups have self-assembled (perhaps through cation bridges provided by potassium permanganate) within the lipid core. We also see gnarled lipid bodies associating with vacuoles that may represent lipid hydrolysis during autophagy (unpublished data). Although these structures have not been previously reported in yeast cells, they are somewhat similar to the whorls of lipid lamina that penetrate cholesteryl ester droplets in mouse macrophages (McGookey and Anderson, 1983). The gnarls in our study were not seen without the use of permanganate stain, and their delicate nature may depend on early prefixation steps described by the method (Wright, 2000) and the extended infiltration time we used. There may also be differences in their frequency in various background strains. In fact, we observed them more frequently in our MMYO11α background than in BY4741 or BY4742, two other frequently used background strains. However, gnarled lipid bodies occur as usual in an atg1Δ strain (in which autophagy does not occur) that is also disrupted in PEX5. Thus, autophagy cannot account for gnarls in the peroxisomal mutants.
Gnarls are scarce in wild-type cells. In this sense, they are unphysiological. They represent a failure of peroxisomes to metabolize fatty acids, the synthesis of which was stimulated by their physical interaction with lipid bodies.
We also observed several interesting elements of lipid body organization. These organelles are often clustered with connections between them, across which core and peripheral material may flow. This may explain the frequent appearance of gnarls in adjacent organelles. The valvelike structures we see between them may represent lipid body communication either as a passive filtration system or a more active and regulated barrier. Perhaps they are related to the stringy material observed when lipid bodies are isolated from mammalian cells (Liu et al., 2004). Regardless, it is evident that lipid bodies are not independent of each other but form a larger structure—an adiposomal reticulum.
Although lipid bodies are often depicted as isolated intracellular fat globules, it is clear that they are at the center of metabolism in many ways. Our work focused on their interactions with peroxisomes, but the proteomics data suggest that they also have essential interactions with mitochondria and ER. Lipid intermediates certainly can flow through the cytoplasm (in vesicles or bound to transfer proteins), but physical interactions may be essential for the efficient transfer of some metabolites, as suggested for other organellar interactions (Voelker, 2005). The lipid body surface may be considered as a source for diffusible substrates and also as a dock receiving the heavy traffic of cargo going in both directions and on which organelles can affect the availability of cargo from the lipid body core. Organelles such as peroxisomes are likely to collaborate with lipid bodies in a controlled manner (for example, in regulating fatty acid production), and the rules governing this regulation will be interesting to work out.
Materials and methods
Reagents and DNA manipulation
Restriction enzymes, DNA ligases, and other reagents for DNA manipulation were purchased from New England Biolabs, Inc., Roche Diagnostics, and Bio-Rad Laboratories. All other chemical reagents were obtained from Sigma-Aldrich or Fisher Scientific. Antisera against Pox1p was generated in rabbits from SDS gel–purified protein from isolated peroxisomes; antisera against Pot1p was a gift of R. Rachubinski (University of Alberta, Edmonton, Canada). The expression and cloning plasmids consisted of pRS313-316 (Sikorski and Hieter, 1989) or pBluescript KS(−) (equivalent to pBluescript II KS(−) in the current catalog except the bases ATT immediately upstream from the T7 promoter has been replaced by GCGCGC in the II series; Stratagene). The PGK promoter and/or terminator used in many plasmid constructions was cloned from pRS315-PGK (Wang et al., 2001). Standard recombinant DNA techniques were used (Maniatis et al., 1982). Changes to chromosomal genes were performed using PCR products containing ∼50 bases of chromosomal sequence at each end for homologous recombination. DNA manipulations resulting in changes to the coding sequence as well as changes to the genome were verified by sequencing performed by the McDermott Center for Human Growth and Development on campus (University of Texas Southwestern Medical School, Dallas, TX). Yeast strains were transformed by the lithium acetate method (Ito et al., 1983).
Strains and culturing
The bacterial strain DH10β (Invitrogen) was used for all bacterial transformations. For yeast experiments, ATCC 201388 (American Type Culture Collection) was used as the background strain for the organelle dynamics experiment shown in Fig. 1 (G and N), and BY4741 (Open Biosystems) was used for the thin-section EMs shown in Fig. 2. For all other yeast experiments, MMYO11α, which was developed for growth in oleic acid (McCammon et al., 1990), or derived mutants or transformants (see below) were used. The ATG1 disruption strain in MMYO11α was a gift of D. Klionsky (University of Michigan, Ann Arbor, MI). Cells were cultured either in 2% glucose or in oleic acid as previously described (Hettema et al., 2000).
Gene knockout strains.
The disruption of PEX11 was described previously (Marshall et al., 1995). For the others, open reading frames were replaced by the HIS3 or LEU2 genes, which were cloned from pRS313 or pRS315, respectively.
ERG6-myc knock-in strain.
The ERG6-myc knock-in strain was created similarly to the method previously described (Wang et al., 2004). In brief, a cassette was assembled in pBluescript pKS(−) consisting of a myc(×2) tag with a stop codon (GAACAAAAATTGATTTCTGAAGAAGATTTGGAGCAAAAGTTAATTTCAGAAGAGGACTTATAA) between the SacII and NotI restriction sites, the PGK 3′ terminator between the BamHI and HindIII sites, and TRP1 between the SalI and ApaI sites. The myc-term TRP1 cassette was amplified with primers containing sequence on either side of the end of the ERG6 open reading frame for homologous recombination into the chromosome.
PEX5-HA Y253N and N393D knock-in strains.
Fragments of the 3′ open reading frame of PEX5 corresponding to amino acids 254 or 394 to the end (before the stop codon) were first cloned separately into the SacII site of Bluescript using an oligonucleotide that destroyed SacII at the 5′ end. The HAx2-term HIS2 cassette (Wang et al., 2004) was then cloned directly behind into the SacII–Xho1 sites. The cloned PEX5 fragments HA tag–HIS2 sequences were then introduced into the chromosomal PEX5 site using mutagenic oligonucleotides to create the Y253N and N393D mutations. The corresponding mutant proteins both contained PRFYPYDVPDYAGYPYDVPDYAG (HAx2) at their carboxy termini.
Plasmids for yeast transformation
Pot1p-GFP plasmid.
The POT1 open reading frame (from genomic DNA) and the GFP reading frame and stop codon (from the P27-GFP stop plasmid; Dyer et al., 1996) were subjected to overlap extension PCR using oligonucleotides that included a spacer encoding three glycines between Pot1p and GFP and BglII ends. The vector was generated by transferring the PGK promoter-terminator from pRS315-PGK to pRS316, creating pRS316-PGK. Finally, the Pot1p-GFP fragment was cloned into the BglII site of pRS316-PGK such that expression of the PGK promoter was upstream of the coding region.
Erg6p monomeric DsRed plasmid.
The PGK promoter and terminator were cloned from pRS315-PGK such that the 5′ and 3′ ends of the promoter contained new SacII and XbaI sites, respectively, and 5′ and 3′ ends of the terminator contained HindIII and XhoI sites, respectively. These fragments were then inserted into pRS315 at these sites, generating pRS315-PGKII.
The ERG6 open reading frame (from genomic DNA) and the sequence for monomeric DsRed (mDsRed, from pDsRed-monomer-N1; a gift from B. Glick, University of Chicago, Chicago, IL) were used in an overlap extension PCR using primers that inserted the sequence for a two-glycine spacer in between the protein elements and conferred BamHI and HindIII ends to the ERG6-mDsRed fusion sequence. This fragment was then inserted between the promoter and terminator of pRS315-PGKII. The resulting vector is termed pERGmDsRed.
Plasmids containing wild-type and mutated Pox1p.
The POX1 reading frame and 652 bp of upstream sequence was subcloned into pRS316 at the SacII site. The PGK terminator was subcloned downstream at the Xho1 and Kpn1 sites, generating Pox1-316, which was used for mutagenesis (see the following paragraph) and to transform yeast for the production of Pox1p at normal levels as a control.
The POX1 E488Q mutation and the GGAKL addition were performed using a Pfu polymerase-based method. The E488Q mutation should yield a catalytically dead acyl-CoA oxidase by comparison with other acyl-CoA oxidases that have been crystallized and characterized more fully (Battaile et al., 2002; Nakajima et al., 2002), and the addition of GGAKL to the carboxy terminus of Pox1p adds a peroxisomal targeting signal-1 upstream of the stop codon. Primers and their complements containing the sequences for both the E488Q mutation and the GGAKL addition were used in a PCR reaction on Pox1-316 plasmid using Pfu polymerase. The extension time was determined by the size of the Pox1-316 plasmid (>2 min/kb). The PCR reactions were then treated by direct addition of the restriction enzyme DpnI at 37°C for 3 h to cleave the template DNA (the Pox1-316 plasmid). DpnI only cleaves its recognition site, GATC, when it is methylated, and this site is methylated in the template DNA because it was produced in DH10β, a strain that is dam+. After treatment with DpnI, the PCR reaction was used to transform the bacteria. Plasmid DNA was harvested, the combination of mutation and addition was screened by PCR, and the correct plasmid was used for yeast transformations.
For the construction of myc-tagged Pox1p, a plasmid copy of the POX1 promoter (652 bp of upstream sequence) and open reading frame was constructed with an XbaI site between them and was subcloned into the Pox1-316 plasmid at the SacII and XbaI restriction sites (which removed the POX1 sequences originally present in Pox1-316) to create the plasmid Pox1-2. The POX1 open reading frame was then amplified and subcloned into Pox1-2 at the XbaI and Xho1 restriction sites to create Pox1-3. Double-stranded DNA encoding the myc epitope were then subcloned into Pox1-3 using XbaI to create plasmid myc–Pox1-316, encoding Pox1p with two copies of myc (MEQKLISEEDLEQKLISEEDLSR) at its amino terminus.
Plasmids encoding Mdh3p and Mls1p with the myc epitope at their amino termini.
The MDH3 open reading frame and 608 bp of downstream sequence (to serve as a terminator) was subcloned into pRS316 at the EcoR1 and XbaI restriction sites to create plasmid Mdh3-1. The MDH3 promoter region (582 bp upstream of the ORF) was then subcloned into Mdh3-1 using the Kpn1 and Xho1 restriction sites to create plasmid Mdh3-2. The myc tag was prepared similarly as in the preceding paragraph and subcloned into Mdh3-2 using Xho1 and EcoR1 to create plasmid myc–Mdh3-316 such that the myc ×2 sequence MEQKLISEEDLEQKLISEEDLEF was inserted in frame before the MDH3 open reading frame.
A similar strategy was used to generate myc-Mls1p. The MLS1 ORF and 318 bp of downstream sequence were subcloned into the pRS316 plasmid at the EcoR1 and SacII restriction sites to create plasmid Mls1-1. The MLS1 promoter region (793 bp upstream of the open reading frame) was amplified and subcloned into Mls1-1 using the Kpn1 and Xho1 restriction sites to create plasmid Mls1-2. The myc tag was added exactly as described for the myc–Mdh3-316 plasmid.
Fluorescence microscopy
For the acquisition of static images, the MMY011α strain was transformed either with pPOT1GFP alone if they were to be stained with oil red O or with pPOT1GFP along with pERG6DsRed or pERG6mDsRed. For cells expressing both GFP- and DsRed/mDsRed fusion proteins, cells in log phase were removed from growth medium (either SD or oleate medium), concentrated by centrifugation (3,000 g for 5 min at room temperature), and suspended on microscope slides in growth medium containing 1% agar (Kron, 2002). Images were acquired in Slidebook (version 4.1.0.3; Intelligent Imaging Innovations) using a microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.) with a 100× 1.3 NA oil immersion objective lens (plan-Neofluar) equipped with a digital camera (Sensicam; Cooke). GFP images were acquired using the fluorescein isothiocyanate filter set, and DsRed/mDsRed images were acquired with the CY3 filter set (Chroma Technology Corp.). For Fig. 1 (A, B, G, and H), z-axis stacks were obtained at 0.1-μm spacing, and the Slidebook commands of deconvolution (nearest neighbor option) and projection were used to build images through the cell. Further processing of images used Slidebook and Photoshop software.
For cultures transformed only with the GFP-containing plasmid, lipid bodies were visualized with oil red O. To prepare the stain, a few milliliters of a stock of 1 g oil red O (Sigma-Aldrich) in 100 ml isopropanol was diluted with water (3:2 vol/vol) and filtered through 0.45- and 0.22-μm syringe filters connected in series. Yeast were pelleted by centrifugation (3,000 g for 5 min at room temperature), washed with water, and resuspended in the freshly filtered oil red O solution (1 A600 unit of yeast/200 μl of filtered oil red O solution) in a microfuge tube. The yeast were vortexed briefly and incubated in the dark at room temperature for 10 min. The stained yeast were washed twice with 1 ml of water, resuspended in 25 μL of water, and mounted on slides for microscopy as in the previous paragraph. For z stacks of oil red O–stained cells, 0.5-μm spacing between images was used.
To observe the dissociation of organelles from lipid bodies, strains (background ATCC 201388) that expressed the GFP-tagged proteins Pex3p, Sec7p, or Snf7p from the genome were purchased from Invitrogen and transformed with pERG6mDsRed to tag lipid bodies. They were grown in glucose or oleate culture and were subjected to spinning disk confocal microscopy using a spinning disk confocal system (Ultraview ERS; PerkinElmer) set up on a microscope (Axiovert 200M; Carl Zeiss MicroImaging, Inc.) with a camera (Orca; Hamamatsu). At least 30 of each type of organelle that bound to lipid bodies in each strain at the start of observation were followed for 4 min by saving images over 26 planes (0.2 μm between planes), cycling every 5 s. The time at which they dissociated from lipid bodies was recorded. Organelles that dissociated were not scored again if they reassociated with a lipid body at a later time. To generate the QuickTime videos, data were collected over three continuous planes (0.2 μm between them) every 4–5 s and projected into one image per time point.
EM
A detailed protocol for processing yeast cells for transmission EM (Wright, 2000) was followed closely except that the infiltration procedure was extended. After washing the samples in 100% ethanol as described previously (Wright, 2000), samples were subjected to 1% Spurr (in ethanol) overnight, 2% Spurr for 8 h, 10% overnight, 33% for 8 h, 66% overnight, and 100% for 36 h with three to four changes. After embedding, samples of 70–90-nm thickness were placed on 200 mesh copper Formvar grids and poststained. The thin sections were observed in a transmission electron microscope (1200 EX; JEOL) at 80 kV using a CCD camera (C4742-95; Hamamatsu) and AMT410 software (Advanced Microscopy Techniques) for image capture.
For immunogold analysis, cells cultured on oleic acid were high pressure frozen, freeze substituted, and embedded in LR White (London Resin Company; Humbel et al., 2001). Thin sections (70–90 nm) were cut and placed on Formvar nickel grids. The grids were placed in an automated EM immunogold labeler (Leica) such that samples were exposed to primary antibody for 4.5 h and protein A–gold (GE Healthcare) for 90 min, both at room temperature. Grids were contrast stained with 2% aqueous uranyl acetate.
Organelle fractionation
Yeast cells were converted to spheroplasts, osmotically lysed, and a PNS was prepared as previously described (Dyer et al., 1996). To determine peroxisomal association with crude lipid bodies, the PNS was centrifuged at 2,500 g ave for 45 min at 4°C in a SW60 rotor (Beckman Coulter). Approximately 100–250 μl containing the lipid bodies was removed from the top of the gradient. An identical volume of infranatant below the lipid bodies was also removed. Cytosol fractions were TCA precipitated, and all samples were subjected to SDS gel electrophoresis and immunoblotting.
For experiments involving purified lipid bodies, the organelles were purified from PNS as described previously (Liu et al., 2004). In brief, PNS prepared as above (in the previous paragraph) was overlaid with 3–5 ml Hepes buffer and centrifuged at 198K g ave for 50 min in a SW41 rotor (Beckman Coulter). Lipid bodies were removed from the top of the tubes and washed several times in Hepes buffer. They were visualized by embedding in collagen and staining with oil red O (Liu et al., 2004). To compare organelle markers among fractions, the pellet and the cytosol from the SW41 spin were harvested. Equal amounts (typically 2%) of purified lipid bodies, cytosol (concentrated by TCA precipitation and dissolved in SDS sample buffer), and pellet fractions were analyzed by PAGE and immunoblotting.
Protein identification by nano-HPLC/mass spectrometry
Dried protein extracts from yeast lipid droplets were dissolved in 50 μl of 50 mM NH4CO3 buffer. Porcine modified trypsin (Promega) was added to the solution at the ratio of 1:50 (wt/wt) followed by overnight incubation at 37°C. The solution was then dried in SpeedVac (ThermoFinnigan), and tryptic peptides were desalted by μ-C18 Ziptip (Millipore) according the manufacturer's instructions before HPLC/mass spectrometry analysis for protein identification.
HPLC/tandem mass spectrometry was performed on a mass spectrometer (LTQ; ThermoFinnigan) coupled with a nano-flow HPLC system (Agilent 1100; Agilent Technologies). Tryptic peptides were dissolved in HPLC buffer A (2% acetonitrile, 97.9% water, and 0.1% acetic acid [vol/vol/vol]), and 2 μl of the peptide solution was loaded onto an in-house packed HPLC capillary column (10 cm × 75 μm) equilibrated with 6% buffer B (90% acetonitrile, 9.9% water, and 0.1% acetic acid [vol/vol/vol]). Peptides were eluted from the column using the following gradient conditions: 38–42% B in 2 min, 42–56% B in 0.1 min, 56–64% B in 7 min, and 64–90% B in 0.1 min. Initial flow rate was 0.6 μl/min; at 8 min, the flow rate was dropped from 0.6 to 0.1 μl/min.
Data from tandem mass spectra were searched against a nonredundant yeast protein sequence database from the National Center for Biotechnology Information by an in-house X! Tandem database search engine (The Global Proteome Machine Organization, http://www.thegpm.org; Craig and Beavis, 2004). Peptide mass spectra identified with a log (expectation value) less than or equal to −2.0 were manually evaluated (Chen et al., 2005).
Lipid analysis
Frozen lipid bodies purified from 500 or 1,000 ml of cultures were thawed, adjusted with H2O to 500 μl, and extracted by a modified Bligh and Dyer method (Bligh and Dyer, 1959) using hot isopropanol (70°C) instead of methanol (Chapman and Moore, 1993). Lipids were extracted into chloroform and washed with 1 M KCl. The purified lipids were then dried under N2 and dissolved in a small volume, typically 50 μl of chloroform. Samples were separated by thin layer chromatography on 20-cm K6 silica gel plates (Whatman) in 80:20:1 hexane/diethyl ether/acetic acid (vol/vol/vol). Lipids were visualized after staining with I2 and quantified after scanning the plate with ImageJ software (National Institutes of Health [NIH]) using a standard curve and quantitative reference standards (Nu-check Prep, Inc.). In the experiment shown in Fig. 8 A, the plate was charred to visualize lipids. Samples from at least three independent cultures of each strain were analyzed.
Other methods
Protein concentration of lipid body samples was determined by amido black staining (Schaffner and Weissmann, 1973).
Online supplemental material
Strains expressing Erg6p-mDsRed and GFP-tagged Pex3p, Sec7p, or Snf7p were monitored over a 4-min period by spinning disk confocal microscopy at room temperature. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200511125/DC1.
Supplementary Material
Acknowledgments
We thank René Bartz, John Zehmer, Pingsheng Liu, and Yunshu Ying for useful conversations and technical assistance with the lipid body isolations; Kate Phelps for help with the spinning disk confocal microscope; and Kimberly Szymanski for excellent technical help.
This work was supported by NIH grants HL 20948 and GM 52016 to R.G.W. Anderson, a Perot Foundation grant to R.G.W. Anderson, a Cecil H. Green Distinguished Chair in Cellular and Molecular Biology grant to R.G.W. Anderson, Welch Foundation grant I-1085 to J.M. Goodman, American Heart Association Texas Affiliate grant 0555043Y to J.M. Goodman, and National Science Foundation grant MCB-0455329 to J.M. Goodman.
Abbreviations used in this paper: PNS, postnuclear supernatant.
References
- Athenstaedt, K., and G. Daum. 2005. Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J. Biol. Chem. 280:37301–37309. [DOI] [PubMed] [Google Scholar]
- Athenstaedt, K., D. Zweytick, A. Jandrositz, S.D. Kohlwein, and G. Daum. 1999. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J. Bacteriol. 181:6441–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerends, R.J., S.W. Rasmussen, R.E. Hilbrands, M. van der Heide, K.N. Faber, P.T. Reuvekamp, J.A. Kiel, J.M. Cregg, I.J. van der Klei, and M. Veenhuis. 1996. The Hansenula polymorpha PER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J. Biol. Chem. 271:8887–8894. [DOI] [PubMed] [Google Scholar]
- Bascom, R.A., H. Chan, and R.A. Rachubinski. 2003. Peroxisome biogenesis occurs in an unsynchronized manner in close association with the endoplasmic reticulum in temperature-sensitive Yarrowia lipolytica Pex3p mutants. Mol. Biol. Cell. 14:939–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaile, K.P., J. Molin-Case, R. Paschke, M. Wang, D. Bennett, J. Vockley, and J.J. Kim. 2002. Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA: comparison with other acyl-CoA dehydrogenases. J. Biol. Chem. 277:12200–12207. [DOI] [PubMed] [Google Scholar]
- Beraud-Dufour, S., and W. Balch. 2002. A journey through the exocytic pathway. J. Cell Sci. 115:1779–1780. [DOI] [PubMed] [Google Scholar]
- Blanchette-Mackie, E.J., N.K. Dwyer, T. Barber, R.A. Coxey, T. Takeda, C.M. Rondinone, J.L. Theodorakis, A.S. Greenberg, and C. Londos. 1995. Perilipin is located on the surface layer of intracellular lipid droplets in adipocytes. J. Lipid Res. 36:1211–1226. [PubMed] [Google Scholar]
- Bligh, E.G., and W.J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917. [DOI] [PubMed] [Google Scholar]
- Brasaemle, D.L., G. Dolios, L. Shapiro, and R. Wang. 2004. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J. Biol. Chem. 279:46835–46842. [DOI] [PubMed] [Google Scholar]
- Butow, R.A., and N.G. Avadhani. 2004. Mitochondrial signaling: the retrograde response. Mol. Cell. 14:1–15. [DOI] [PubMed] [Google Scholar]
- Chapman, K.D., and R.N. Trelease. 1991. Acquisition of membrane lipids by differentiating glyoxysomes: role of lipid bodies. J. Cell Biol. 115:995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, K.D., and T.S. Moore Jr. 1993. N-acylphosphatidylethanolamine synthesis in plants: occurrence, molecular composition, and phospholipid origin. Arch. Biochem. Biophys. 301:21–33. [DOI] [PubMed] [Google Scholar]
- Chen, Y., S.C. Kim, and Y. Zhao. 2005. High-throughput identification of ingel digested proteins by rapid, isocratic HPLC/MS/MS. Anal. Chem. 77:8179–8184. [DOI] [PubMed] [Google Scholar]
- Cohen, A.W., B. Razani, W. Schubert, T.M. Williams, X.B. Wang, P. Iyengar, D.L. Brasaemle, P.E. Scherer, and M.P. Lisanti. 2004. Role of caveolin-1 in the modulation of lipolysis and lipid droplet formation. Diabetes. 53:1261–1270. [DOI] [PubMed] [Google Scholar]
- Craig, R., and R.C. Beavis. 2004. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 20:1466–1467. [DOI] [PubMed] [Google Scholar]
- Dyer, J.M., J.A. McNew, and J.M. Goodman. 1996. The sorting sequence of the peroxisomal integral membrane protein PMP47 is contained within a short hydrophilic loop. J. Cell Biol. 133:269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann, R., and G. Blobel. 1995. Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J. Cell Biol. 128:509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenyo, D., and R.C. Beavis. 2003. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Anal. Chem. 75:768–774. [DOI] [PubMed] [Google Scholar]
- Geuze, H.J., J.L. Murk, A.K. Stroobants, J.M. Griffith, M.J. Kleijmeer, A.J. Koster, A.J. Verkleij, B. Distel, and H.F. Tabak. 2003. Involvement of the endoplasmic reticulum in peroxisome formation. Mol. Biol. Cell. 14:2900–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger, E.D., M.M. Kessels, and B. Qualmann. 2003. Temporal and spatial coordination of exocytosis and endocytosis. Nat. Rev. Mol. Cell Biol. 4:127–139. [DOI] [PubMed] [Google Scholar]
- Hayashi, Y., M. Hayashi, H. Hayashi, I. Hara-Nishimura, and M. Nishimura. 2001. Direct interaction between glyoxysomes and lipid bodies in cotyledons of the Arabidopsis thaliana ped1 mutant. Protoplasma. 218:83–94. [DOI] [PubMed] [Google Scholar]
- Hettema, E.H., W. Girzalsky, M. van Den Berg, R. Erdmann, and B. Distel. 2000. Saccharomyces cerevisiae Pex3p and Pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 19:223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W.K., J.V. Falvo, L.C. Gerke, A.S. Carroll, R.W. Howson, J.S. Weissman, and E.K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691. [DOI] [PubMed] [Google Scholar]
- Humbel, B.M., M. Konomi, T. Takagi, N. Kamasawa, S.A. Ishijima, and M. Osumi. 2001. In situ localization of beta-glucans in the cell wall of Schizosaccharomyces pombe. Yeast. 18:433–44. [DOI] [PubMed] [Google Scholar]
- Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, A.T., M. van den Berg, G. Bottger, H.F. Tabak, and B. Distel. 2002. Saccharomyces cerevisiae acyl-CoA oxidase follows a novel, non-PTS1, import pathway into peroxisomes that is dependent on Pex5p. J. Biol. Chem. 277:25011–25019. [DOI] [PubMed] [Google Scholar]
- Krisans, S.K. 1996. Cell compartmentalization of cholesterol biosynthesis. Ann. NY Acad. Sci. 804:142–164. [DOI] [PubMed] [Google Scholar]
- Kron, S.J. 2002. Digital time-lapse microscopy of yeast cell growth. Methods Enzymol. 351:3–15. [DOI] [PubMed] [Google Scholar]
- Kurat, C.F., K. Natter, J. Petschnigg, H. Wolinski, K. Scheuringer, H. Scholz, R. Zimmermann, R. Leber, R. Zechner, and S.D. Kohlwein. 2006. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J. Biol. Chem. 281:491–500. [DOI] [PubMed] [Google Scholar]
- Liu, P., Y. Ying, Y. Zhao, D.I. Mundy, M. Zhu, and R.G. Anderson. 2004. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 279:3787–3792. [DOI] [PubMed] [Google Scholar]
- Ma, J., and Z. Pan. 2003. Retrograde activation of store-operated calcium channel. Cell Calcium. 33:375–384. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., E.F. Fritsch, and J. Sambrook. 1982. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 545 pp.
- Marelli, M., J.J. Smith, S. Jung, E. Yi, A.I. Nesvizhskii, R.H. Christmas, R.A. Saleem, Y.Y. Tam, A. Fagarasanu, D.R. Goodlett, et al. 2004. Quantitative mass spectrometry reveals a role for the GTPase Rho1p in actin organization on the peroxisome membrane. J. Cell Biol. 167:1099–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markin, V.S., and J.P. Albanesi. 2002. Membrane fusion: stalk model revisited. Biophys. J. 82:693–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markin, V.S., M.M. Kozlov, and V.L. Borovjagin. 1984. On the theory of membrane fusion. The stalk mechanism. Gen. Physiol. Biophys. 3:361–377. [PubMed] [Google Scholar]
- Marshall, P.A., Y.I. Krimkevich, R.H. Lark, J.M. Dyer, M. Veenhuis, and J.M. Goodman. 1995. Pmp27 promotes peroxisomal proliferation. J. Cell Biol. 129:345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, P.A., J.M. Dyer, M.E. Quick, and J.M. Goodman. 1996. Redox-sensitive homodimerization of Pex11p: a proposed mechanism to regulate peroxisomal division. J. Cell Biol. 135:123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters, C. 1997. Gluconeogenesis and the peroxisome. Mol. Cell. Biochem. 166:159–168. [DOI] [PubMed] [Google Scholar]
- McCammon, M.T., M. Veenhuis, S.B. Trapp, and J.M. Goodman. 1990. Association of glyoxylate and beta-oxidation enzymes with peroxisomes of Saccharomyces cerevisiae. J. Bacteriol. 172:5816–5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGookey, D.J., and R.G. Anderson. 1983. Morphological characterization of the cholesteryl ester cycle in cultured mouse macrophage foam cells. J. Cell Biol. 97:1156–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, D.J. 2001. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 40:325–438. [DOI] [PubMed] [Google Scholar]
- Murphy, D.J., and J. Vance. 1999. Mechanisms of lipid-body formation. Trends Biochem. Sci. 24:109–115. [DOI] [PubMed] [Google Scholar]
- Nakajima, Y., I. Miyahara, K. Hirotsu, Y. Nishina, K. Shiga, C. Setoyama, H. Tamaoki, and R. Miura. 2002. Three-dimensional structure of the flavoenzyme acyl-CoA oxidase-II from rat liver, the peroxisomal counterpart of mitochondrial acyl-CoA dehydrogenase. J. Biochem. (Tokyo). 131:365–374. [DOI] [PubMed] [Google Scholar]
- Penfield, S., S. Graham, and I.A. Graham. 2005. Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochem. Soc. Trans. 33:380–383. [DOI] [PubMed] [Google Scholar]
- Rizzuto, R., M.R. Duchen, and T. Pozzan. 2004. Flirting in little space: the ER/mitochondria Ca2+ liaison. Sci. STKE. 10.1126/stke.2152004re1. [DOI] [PubMed]
- Robenek, M.J., N.J. Severs, K. Schlattmann, G. Plenz, K.P. Zimmer, D. Troyer, and H. Robenek. 2004. Lipids partition caveolin-1 from ER membranes into lipid droplets: updating the model of lipid droplet biogenesis. FASEB J. 18:866–868. [DOI] [PubMed] [Google Scholar]
- Schaffner, W., and C. Weissmann. 1973. A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal. Biochem. 56:502–514. [DOI] [PubMed] [Google Scholar]
- Schrader, M. 2001. Tubulo-reticular clusters of peroxisomes in living COS-7 cells: dynamic behavior and association with lipid droplets. J. Histochem. Cytochem. 49:1421–1429. [DOI] [PubMed] [Google Scholar]
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J., C. Lee, C.H. Lin, and L.B. Chen. 1991. Electron microscopic studies of the endoplasmic reticulum in whole-mount cultured cells fixed with potassium permanganate. J. Struct. Biol. 107:106–115. [DOI] [PubMed] [Google Scholar]
- Tauchi-Sato, K., S. Ozeki, T. Houjou, R. Taguchi, and T. Fujimoto. 2002. The surface of lipid droplets is a phospholipid monolayer with a unique Fatty Acid composition. J. Biol. Chem. 277:44507–44512. [DOI] [PubMed] [Google Scholar]
- Vance, J.E. 1990. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265:7248–7256. [PubMed] [Google Scholar]
- Veenhuis, M., M. Mateblowski, W.H. Kunau, and W. Harder. 1987. Proliferation of microbodies in Saccharomyces cerevisiae. Yeast. 3:77–84. [DOI] [PubMed] [Google Scholar]
- Voelker, D.R. 2005. Bridging gaps in phospholipid transport. Trends Biochem. Sci. 30:396–404. [DOI] [PubMed] [Google Scholar]
- Wang, X., M.J. Unruh, and J.M. Goodman. 2001. Discrete targeting signals direct Pmp47 to oleate-induced peroxisomes in Saccharomyces cerevisiae. J. Biol. Chem. 276:10897–10905. [DOI] [PubMed] [Google Scholar]
- Wang, X., M.A. McMahon, S.N. Shelton, M. Nampaisansuk, J.L. Ballard, and J.M. Goodman. 2004. Multiple targeting modules on peroxisomal proteins are not redundant: discrete functions of targeting signals within Pmp47 and Pex8p. Mol. Biol. Cell. 15:1702–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, R. 2000. Transmission electron microscopy of yeast. Microsc. Res. Tech. 51:496–510. [DOI] [PubMed] [Google Scholar]
- Zhang, K., and R.J. Kaufman. 2004. Signaling the unfolded protein response from the endoplasmic reticulum. J. Biol. Chem. 279:25935–25938. [DOI] [PubMed] [Google Scholar]
- Zinser, E., F. Paltauf, and G. Daum. 1993. Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J. Bacteriol. 175:2853–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweytick, D., K. Athenstaedt, and G. Daum. 2000. Intracellular lipid particles of eukaryotic cells. Biochim. Biophys. Acta. 1469:101–120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.